PPP3R1 Promoter Polymorphism (Allelic Variation) Affects Tacrolimus Treatment Efficacy by Modulating E2F6 Binding Affinity
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture
2.2. Jurkat T Cell Activation
2.3. Plasmid Construction
2.4. Transient Transfection and Dual-Luciferase Reporter Gene Assays
2.5. In Silico Bioinformatic Prediction
2.6. Quantitative Real-Time PCR Analysis (qRT-PCR)
2.7. Electrophoretic Mobility Shift Assay (EMSA)
2.8. Western Blotting
2.9. Establishment of Stable Cell Lines
2.10. Detection of IL-2 and GM-CSF Production
2.11. Statistical Analysis
3. Results
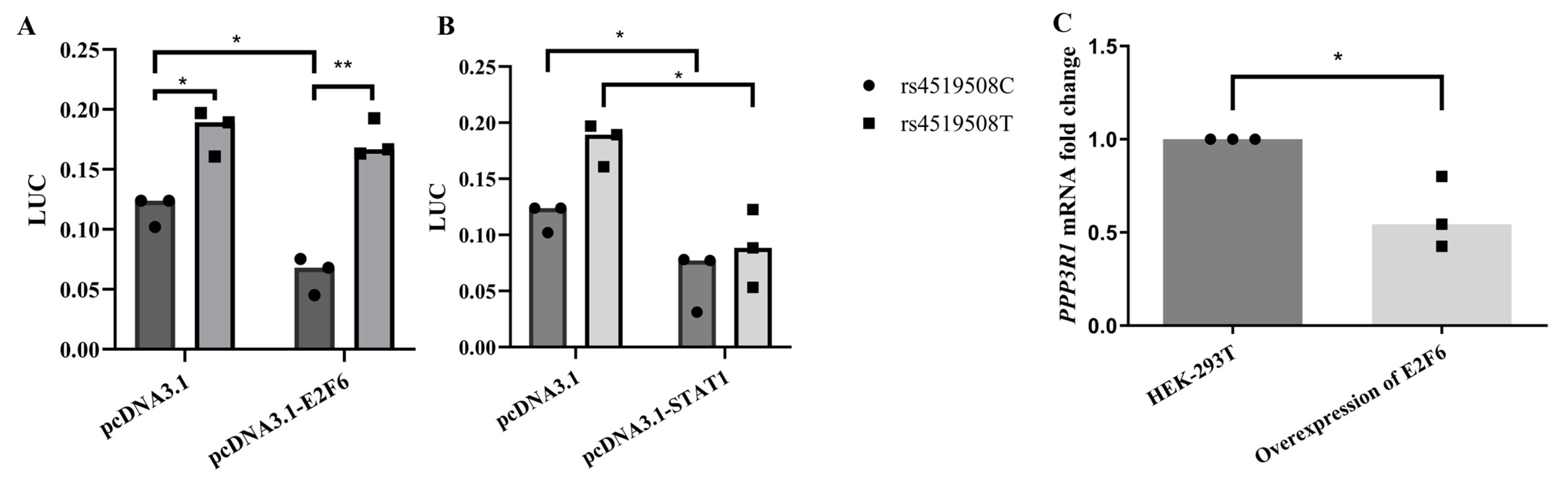
3.1. Rs4519508 (C > T) and rs875 (T > C) in Strong LD Increased Luciferase Reporter Activity Comparably
3.2. In Silico Functional Prediction Found SNP Allelic Variation Change the Binding Affinity of TF
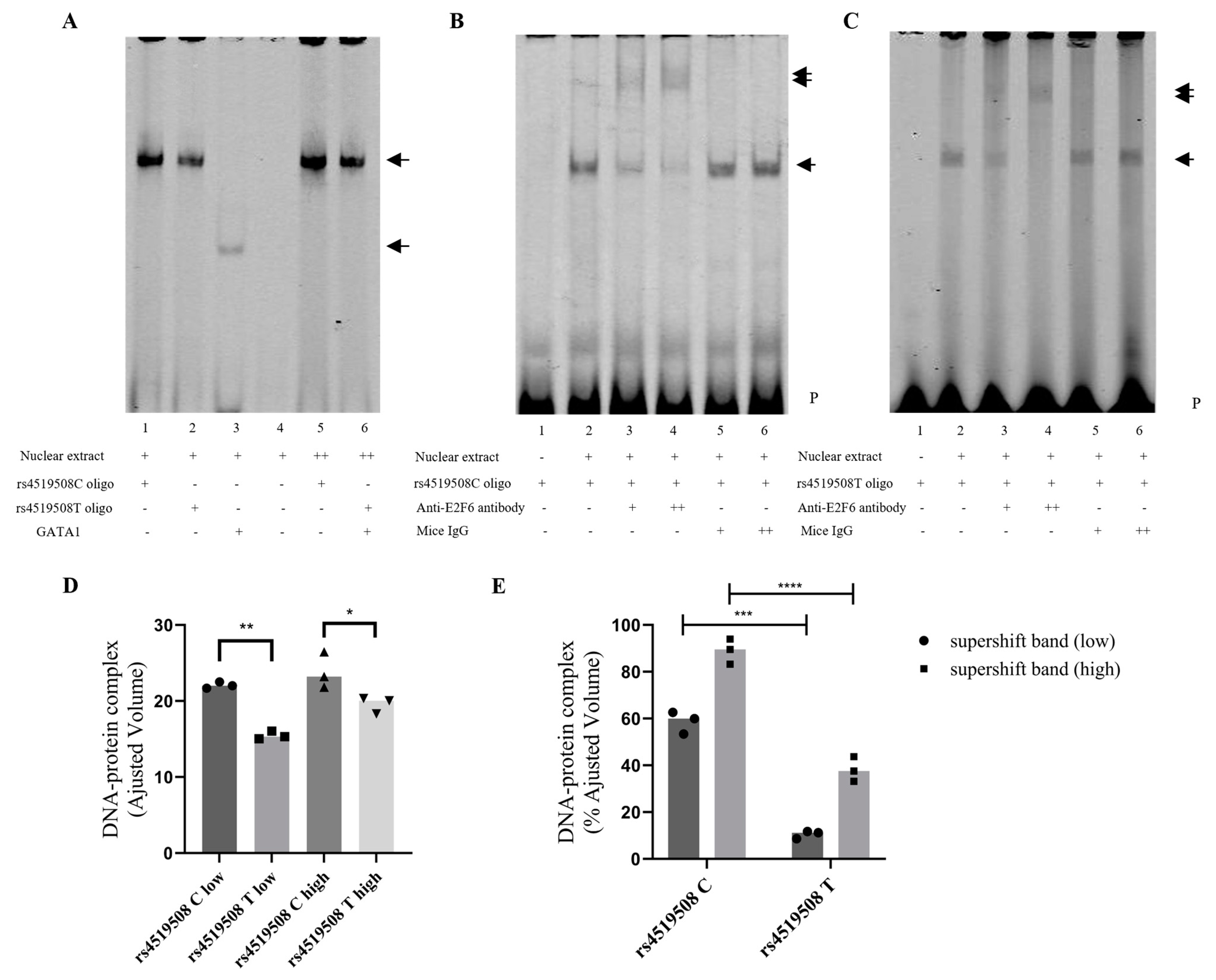
3.3. Analysis of Candidate TF Binding Elements Altered by rs4519508 C > T
3.4. E2F6 Binds to the PPP3R1 rs4519508 Site
3.5. E2F6 Knockdown Reduced the Immunosuppressive Effect of TAC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Supplementary Details for Plasmid Construction
References
- Barbarino, J.M.; Staatz, C.E.; Venkataramanan, R.; Klein, T.E.; Altman, R.B. PharmGKB summary: Cyclosporine and tacrolimus pathways. Pharmacogenet. Genom. 2013, 23, 563–585. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Liu, M.; Zhang, W.; Ming, Y. Pharmacokinetics, Pharmacodynamics and Pharmacogenetics of Tacrolimus in Kidney Transplantation. Curr. Drug Metab. 2018, 19, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Huai, C.; Xu, Q.; Xu, L.; Zhang, M.; Zhong, M.; Qiu, X. FKBP-CaN-NFAT pathway polymorphisms selected by in silico biological function prediction are associated with tacrolimus efficacy in renal transplant patients. Eur. J. Pharm. Sci. 2020, 160, 105694. [Google Scholar] [CrossRef]
- Noceti, O.M.; Woillard, J.B.; Boumediene, A.; Esperon, P.; Taupin, J.L.; Gerona, S.; Valverde, M.; Tourino, C.; Marquet, P. Tacrolimus pharmacodynamics and pharmacogenetics along the calcineurin pathway in human lymphocytes. Clin. Chem. 2014, 60, 1336–1345. [Google Scholar] [CrossRef] [PubMed]
- Pouche, L.; Stojanova, J.; Marquet, P.; Picard, N. New challenges and promises in solid organ transplantation pharmacogenetics: The genetic variability of proteins involved in the pharmacodynamics of immunosuppressive drugs. Pharmacogenomics 2016, 17, 277–296. [Google Scholar] [CrossRef]
- Brunet, M.; van Gelder, T.; Åsberg, A.; Haufroid, V.; Hesselink, D.A.; Langman, L.; Lemaitre, F.; Marquet, P.; Seger, C.; Shipkova, M.; et al. Therapeutic Drug Monitoring of Tacrolimus-Personalized Therapy: Second Consensus Report. Ther. Drug Monit. 2019, 41, 261–307. [Google Scholar] [CrossRef]
- Van Gelder, T.; Van Schaik, R.H.; Hesselink, D.A. Pharmacogenetics and immunosuppressive drugs in solid organ transplantation. Nat. Rev. Nephrol. 2014, 10, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Fioretto, P.; Najafian, B.; Sutherland, D.E.; Mauer, M. Tacrolimus and cyclosporine nephrotoxicity in native kidneys of pancreas transplant recipients. Clin. J. Am. Soc. Nephrol. 2011, 6, 101–106. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, Q.; Qiu, X.; Jiao, Z.; Zhang, M.; Zhong, M. FOXP3 rs3761548 polymorphism is associated with tacrolimus-induced acute nephrotoxicity in renal transplant patients. Eur. J. Clin. Pharmacol. 2017, 73, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Seyhun, Y.; Ciftci, H.S.; Kekik, C.; Karadeniz, M.S.; Tefik, T.; Nane, I.; Turkmen, A.; Oguz, F.S.; Aydin, F. Genetic association of interleukin-2, interleukin-4, interleukin-6, transforming growth factor-beta, tumour necrosis factor-alpha and blood concentrations of calcineurin inhibitors in Turkish renal transplant patients. Int. J. Immunogenet. 2015, 42, 147–160. [Google Scholar] [CrossRef]
- Elens, L.; Sombogaard, F.; Hesselink, D.; van Schaik, R.; van Gelder, T. Single-nucleotide polymorphisms in P450 oxidoreductase and peroxisome proliferator-activated receptor-α are associated with the development of new-onset diabetes after transplantation in kidney transplant recipients treated with tacrolimus. Pharmacogenet. Genom. 2013, 23, 649–657. [Google Scholar] [CrossRef]
- Kang, C.B.; Hong, Y.; Dhe-Paganon, S.; Yoon, H.S. FKBP family proteins: Immunophilins with versatile biological functions. Neurosignals 2008, 16, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Farmer Jr, J.D.; Lane, W.S.; Friedman, J.; Weissman, I.; Schreiber, S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 1991, 66, 807–815. [Google Scholar] [CrossRef]
- Shaw, K.; Ho, A.M.; Raghavan, A.; Kim, J.; Jain, J.; Park, J.; Sharma, S.; Rao, A.; Hogan, P.G. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11205–11209. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, M.; Wang, F.; Lu, J.; Chen, R.; Xie, Q.; Sun, J.; Xue, J.; Hao, C.; Lin, S. The calcineurin regulatory subunit polymorphism and the treatment efficacy of tacrolimus for idiopathic membranous nephropathy. Int. Immunopharmacol. 2018, 65, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Rusnak, F.; Mertz, P. Calcineurin: Form and function. Physiol. Rev. 2000, 80, 1483–1521. [Google Scholar] [CrossRef] [PubMed]
- Kung, L.; Batiuk, T.D.; Palomo-Pinon, S.; Noujaim, J.; Helms, L.M.; Halloran, P.F. Tissue distribution of calcineurin and its sensitivity to inhibition by cyclosporine. Am. J. Transplant. 2001, 1, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Klee, C.B.; Ren, H.; Wang, X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 1998, 273, 13367–13370. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, Q.; Qiu, X.; Xu, L.; Jiao, Z.; Zhang, M.; Zhong, M. FKBP1A rs6041749 polymorphism is associated with allograft function in renal transplant patients. Eur. J. Clin. Pharmacol. 2019, 75, 33–40. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, H.; Si, S.; Han, Z.; Tao, J.; Chen, H.; Ge, Y.; Guo, M.; Wang, K.; Tan, R.; et al. Polymorphisms of nucleotide factor of activated T cells cytoplasmic 2 and 4 and the risk of acute rejection following kidney transplantation. World J. Urol. 2018, 36, 111–116. [Google Scholar] [CrossRef]
- Li, D.; Zhu, G.; Lou, S.; Ma, L.; Zhang, C.; Pan, Y.; Wang, L. The functional variant of NTN1 contributes to the risk of nonsyndromic cleft lip with or without cleft palate. Europ. J. Hum. Genet. 2020, 28, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Musunuru, K. From Genotype to Phenotype: A Primer on the Functional Follow-up of Genome-Wide Association Studies in Cardiovascular Disease. Circ. Genom. Precis. Med. 2018, 11, e001946. [Google Scholar] [CrossRef] [PubMed]
- Mesbah-Uddin, M.; Elango, R.; Banaganapalli, B.; Shaik, N.; Al-Abbasi, F. In-silico analysis of inflammatory bowel disease (IBD) GWAS loci to novel connections. PLoS ONE 2015, 10, e0119420. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Li, W.; Liyanarachchi, S.; Srinivas, M.; Wang, Y.; Akagi, K.; Wang, Y.; Wu, D.; Wang, Q.; Jin, V.; et al. Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 6128–6133. [Google Scholar] [CrossRef]
- Liu, H.; Leslie, E.; Carlson, J.; Beaty, T.; Marazita, M.; Lidral, A.; Cornell, R. Identification of common non-coding variants at 1p22 that are functional for non-syndromic orofacial clefting. Nat. Commun. 2017, 8, 14759. [Google Scholar] [CrossRef] [PubMed]
- Dubois, P.; Trynka, G.; Franke, L.; Hunt, K.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.; Adány, R.; Aromaa, A.; et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Pagès, H. BSgenome: Software Infrastructure for Efficient Representation of Full Genomes and Their SNPs, R Package Version 1.74.0; Hervé Pagès: Waihi beach, New Zealand, 2017. [Google Scholar] [CrossRef]
- Tan, G.; Lenhard, B. TFBSTools: An R/bioconductor package for transcription factor binding site analysis. Bioinformatics 2016, 32, 1555–1556. [Google Scholar] [CrossRef]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [PubMed]
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient Manipulation of Biological Strings, R Package Version 2.74.0; Hervé Pagès: Waihi beach, New Zealand, 2017. [Google Scholar] [CrossRef]
- Zheng, R.; Wan, C.; Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Chen, C.H.; Brown, M.; Zhang, X.; Meyer, C.A.; et al. Cistrome Data Browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019, 47, D729–D735. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Qin, Q.; Wu, Q.; Sun, H.; Zheng, R.; Zang, C.; Zhu, M.; Wu, J.; Shi, X.; Taing, L.; et al. Cistrome Data Browser: A data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017, 45, D658–D662. [Google Scholar] [CrossRef]
- Xia, L.Q.; Chen, J.L.; Zhang, H.L.; Cai, J.; Zhou, S.; Lu, Y.S. Identification of virion-associated transcriptional transactivator (VATT) of SGIV ICP46 promoter and their binding site on promoter. Virol. J. 2019, 16, 110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, M.F.; Chen, F.Y.; Cui, Q.; Yang, B.Z.; Liu, Y.; Peng, H.; Wang, K.J. The transcriptional regulation of an antimicrobial peptide hepcidin1 in Oryzias melastigma upon EE2 exposure involved in a new pathway with a novel transcriptional regulatory element HepERE. Fish Shellfish Immunol. 2018, 82, 421–431. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 127, 269–287. [Google Scholar] [CrossRef]
- Suzuki, A.; Guerrini, M.M.; Yamamoto, K. Functional genomics of autoimmune diseases. Ann. Rheum. Dis. 2021, 80, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liao, J.; Tang, M.; Wang, Y.; Wei, X.; Mao, L.; Zeng, C. A functional single nucleotide polymorphism in the tyrosinase gene promoter affects skin color and transcription activity in the black-boned chicken. Poult. Sci. 2017, 96, 4061–4067. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; McDonough, C.W.; Gong, Y.; Bailey, K.R.; Boerwinkle, E.; Chapman, A.B.; Gums, J.G.; Turner, S.T.; Cooper-DeHoff, R.M.; Johnson, J.A. Genome Wide Analysis Approach Suggests Chromosome 2 Locus to be Associated with Thiazide and Thiazide Like-Diuretics Blood Pressure Response. Sci. Rep. 2019, 9, 17323. [Google Scholar] [CrossRef] [PubMed]
- Nishio, H.; Matsui, K.; Tsuji, H.; Tamura, A.; Suzuki, K. Immunolocalization of calcineurin and FKBP12, the FK506-binding protein, in Hassall’s corpuscles of human thymus and epidermis. Histochem. Cell Biol. 2000, 114, 9–14. [Google Scholar] [CrossRef]
- Trimarchi, J.M.; Fairchild, B.; Verona, R.; Moberg, K.; Andon, N.; Lees, J.A. E2F-6, a member of the E2F family that can behave as a transcriptionalrepressor. Proc. Natl. Acad. Sci. USA 1998, 95, 2850–2855. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wells, A.D. Comparative analysis of E2F family member oncogenic activity. PLoS ONE 2007, 2, e912. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, Y.; Nakamura, M.; Matsuda, M. Transcriptional repressor E2F-6 regulates apoptosis of hematopoietic stem cells. Exp. Hematol. 2000, 28, 1504–1505. [Google Scholar] [CrossRef]
- Kikuchi, J.; Shimizu, R.; Wada, T.; Ando, H.; Nakamura, M.; Ozawa, K.; Furukawa, Y. E2F-6 Suppresses Growth-Associated Apoptosis of Human Hematopoietic Progenitor Cells by Counteracting Proapoptotic Activity of E2F-1. Stem Cells 2007, 25, 2439–2447. [Google Scholar] [CrossRef]
- Cartwright, P.; Müller, H.; Wagener, C.; Holm, K.; Helin, K. E2F-6: A novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 1998, 17, 611–623. [Google Scholar] [CrossRef]
- Shi, X.; Ma, D.; Li, M.; Zeng, L.; Chen, J.; Yang, Y. Nuclear receptor TLX regulates islet beta cell proliferation via E2F6. Biochem. Biophys. Res. Commun. 2019, 513, 560–566. [Google Scholar] [CrossRef]
- Velasco, G.; Hubé, F.; Rollin, J.; Neuillet, D.; Philippe, C.; Bouzinba-Segard, H.; Galvani, A.; Viegas-Péquignot, E.; Francastel, C. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc. Natl. Acad. Sci. USA 2010, 107, 9281–9286. [Google Scholar] [CrossRef]
- Chen, H.; Ali, M.; Ruben, A.; Stelmakh, D.; Pak, M. E2F6-Mediated Downregulation of MIR22HG Facilitates the Progression of Laryngocarcinoma by Targeting the miR-5000-3p/FBXW7 Axis. Mol. Cell. Biol. 2020, 40, e00496-19. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, L.; Lv, S.; Xu, H.; Fan, Z.; He, Y.; Wen, H. E2F6-mediated lncRNA CASC2 down-regulation predicts poor prognosis and promotes progression in gastric carcinoma. Life Sci. 2019, 232, 116649. [Google Scholar] [CrossRef] [PubMed]


| Allele. | Start | End | Combination Score | Strand | TF | Brief Comment |
|---|---|---|---|---|---|---|
| C | 5 | 15 | 0.8236 | + | E2F4 | E2F transcription factor 4, target of the transforming proteins of small DNA tumor viruses, important in the control of cell cycle and action of tumor suppressor proteins, altered by the mutation of C to T. The average binding score of the two predicted binding sites within rs4519508 C was equivalent to that within rs4519508 T, which suggested that the rs4519508 mutation has no major impact on the binding of E2F4. |
| C | 6 | 16 | 0.9264 | + | E2F4 | |
| T | 6 | 16 | 0.8724 | + | E2F4 | |
| C | 6 | 16 | 0.8769 | + | E2F6 | E2F transcription factor 6, abolished when in allele T. |
| C | 8 | 17 | 0.8256 | + | ERG | ETS transcription factor ERG, abolished when in allele T, while ERG shows no expression in both HEK 293 T cells and blood cells, which reduce its value to validate. |
| C | 8 | 17 | 0.8191 | + | ETS1 | ETS1: ETS proto-oncogene 1, transcription factor, two motifs in allele C and abolished when in allele T. ETS1 was predicted to bind to the genotype of the C allele alone, but ETS1 shows no expression in both HEK 293 T cells and blood cells, which reduce its value to validate. |
| C | 1 | 10 | 0.8022 | + | ETS1 | |
| C | 5 | 15 | 0.8559 | + | STAT1 | STAT1: signal transducer and activator of transcription 1, important for cell viability, altered by the mutation of C to T. |
| T | 5 | 15 | 0.8006 | + | STAT1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Qin, S.; Zhong, M.; Xu, Q.; Huai, C.; Qiu, X. PPP3R1 Promoter Polymorphism (Allelic Variation) Affects Tacrolimus Treatment Efficacy by Modulating E2F6 Binding Affinity. Biomedicines 2024, 12, 2896. https://doi.org/10.3390/biomedicines12122896
Zheng X, Qin S, Zhong M, Xu Q, Huai C, Qiu X. PPP3R1 Promoter Polymorphism (Allelic Variation) Affects Tacrolimus Treatment Efficacy by Modulating E2F6 Binding Affinity. Biomedicines. 2024; 12(12):2896. https://doi.org/10.3390/biomedicines12122896
Chicago/Turabian StyleZheng, Xinyi, Shengying Qin, Mingkang Zhong, Qinxia Xu, Cong Huai, and Xiaoyan Qiu. 2024. "PPP3R1 Promoter Polymorphism (Allelic Variation) Affects Tacrolimus Treatment Efficacy by Modulating E2F6 Binding Affinity" Biomedicines 12, no. 12: 2896. https://doi.org/10.3390/biomedicines12122896
APA StyleZheng, X., Qin, S., Zhong, M., Xu, Q., Huai, C., & Qiu, X. (2024). PPP3R1 Promoter Polymorphism (Allelic Variation) Affects Tacrolimus Treatment Efficacy by Modulating E2F6 Binding Affinity. Biomedicines, 12(12), 2896. https://doi.org/10.3390/biomedicines12122896









