Extracellular Vesicles from Plasma of Patients with Glioblastoma Promote Invasion of Glioblastoma Cells Even After Tumor Resection
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
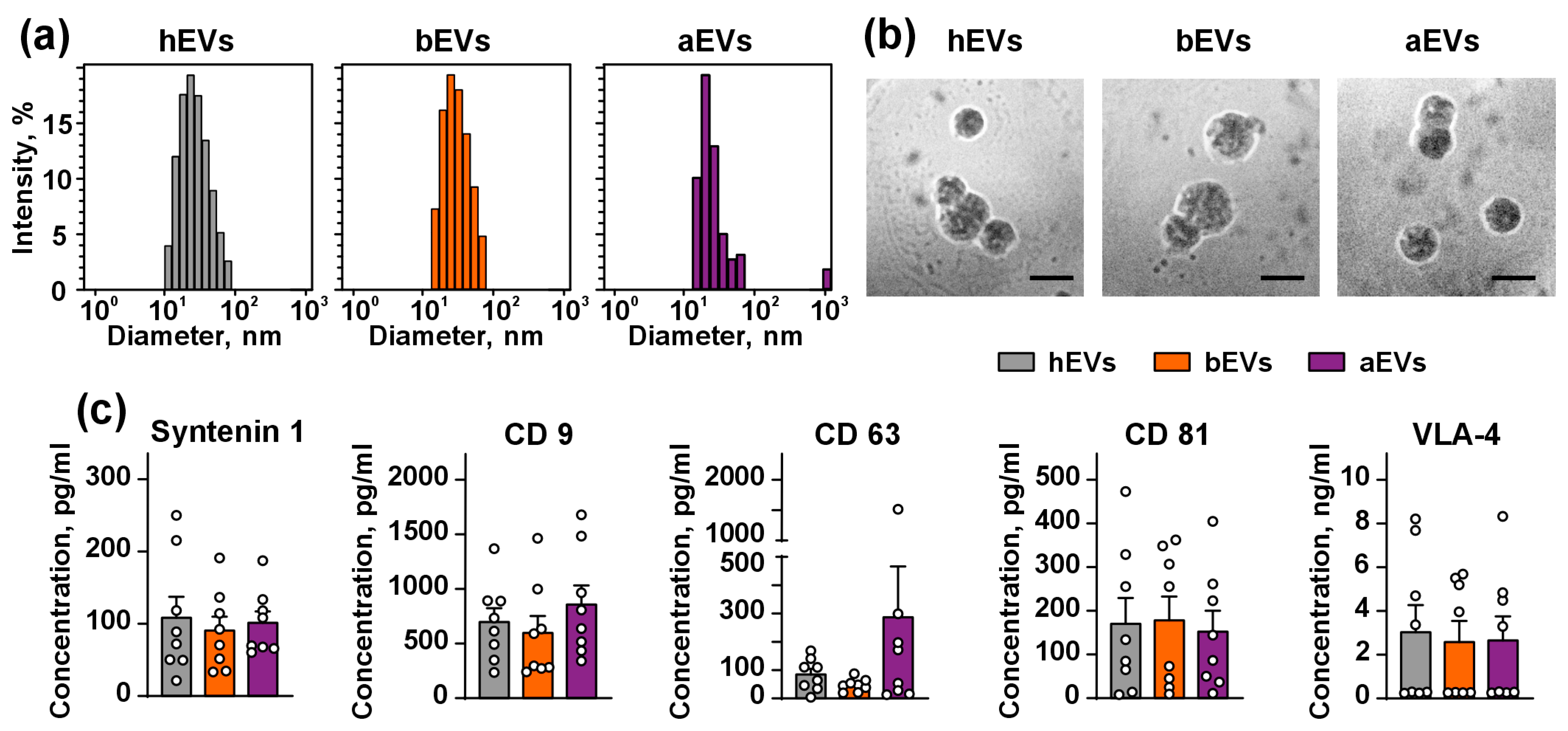
2.2. Isolation and Characterization of EVs
2.3. Cell Cultivation and Isolation of Primary Astrocytes
2.4. Transwell Invasion Assay
2.5. Western Blotting
2.6. Flow Cytometry
2.7. Confocal Microscopy
2.8. Analysis of Inflammation and Adhesion Regulators Secretion
2.9. Statistical Analysis
3. Results
3.1. aEVs and bEVs from the Plasma of GB Patients Enhance Invasion of GB Cells
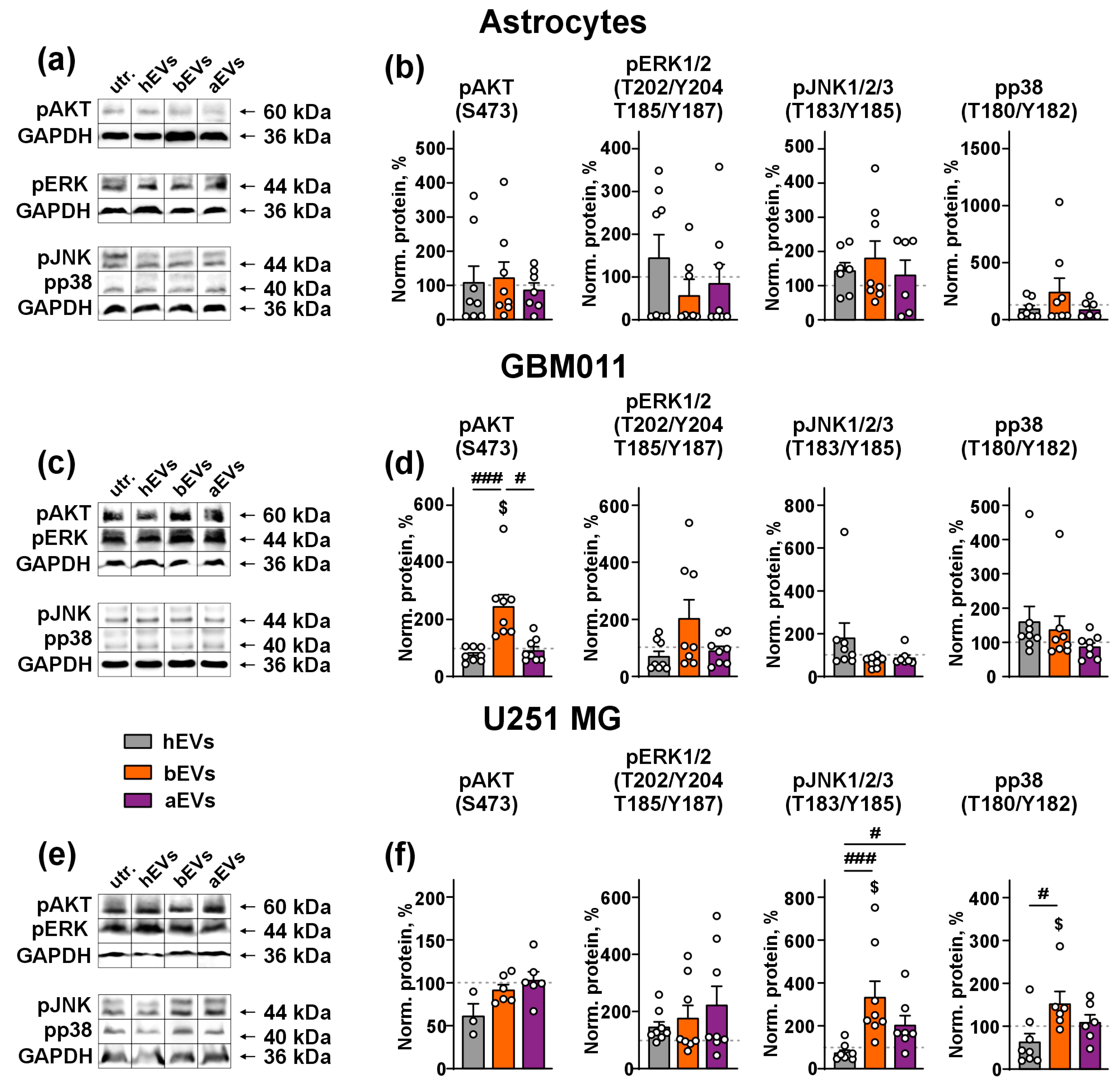
3.2. aEVs and bEVs Activate MAP Kinases in GB Cells
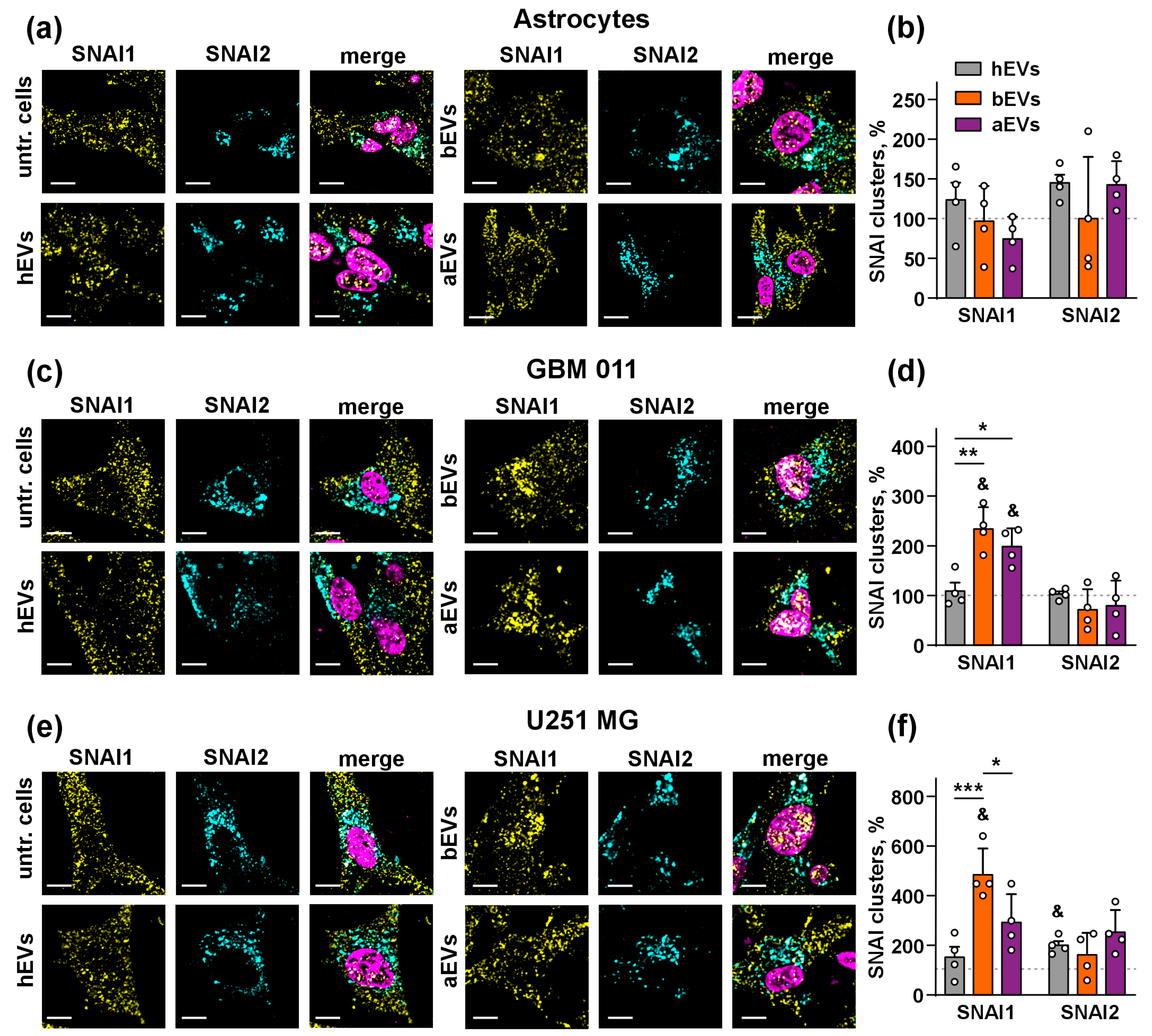
3.3. aEVs and bEVs Drive Localization of SNAI1 to the Nuclei in GB Cells
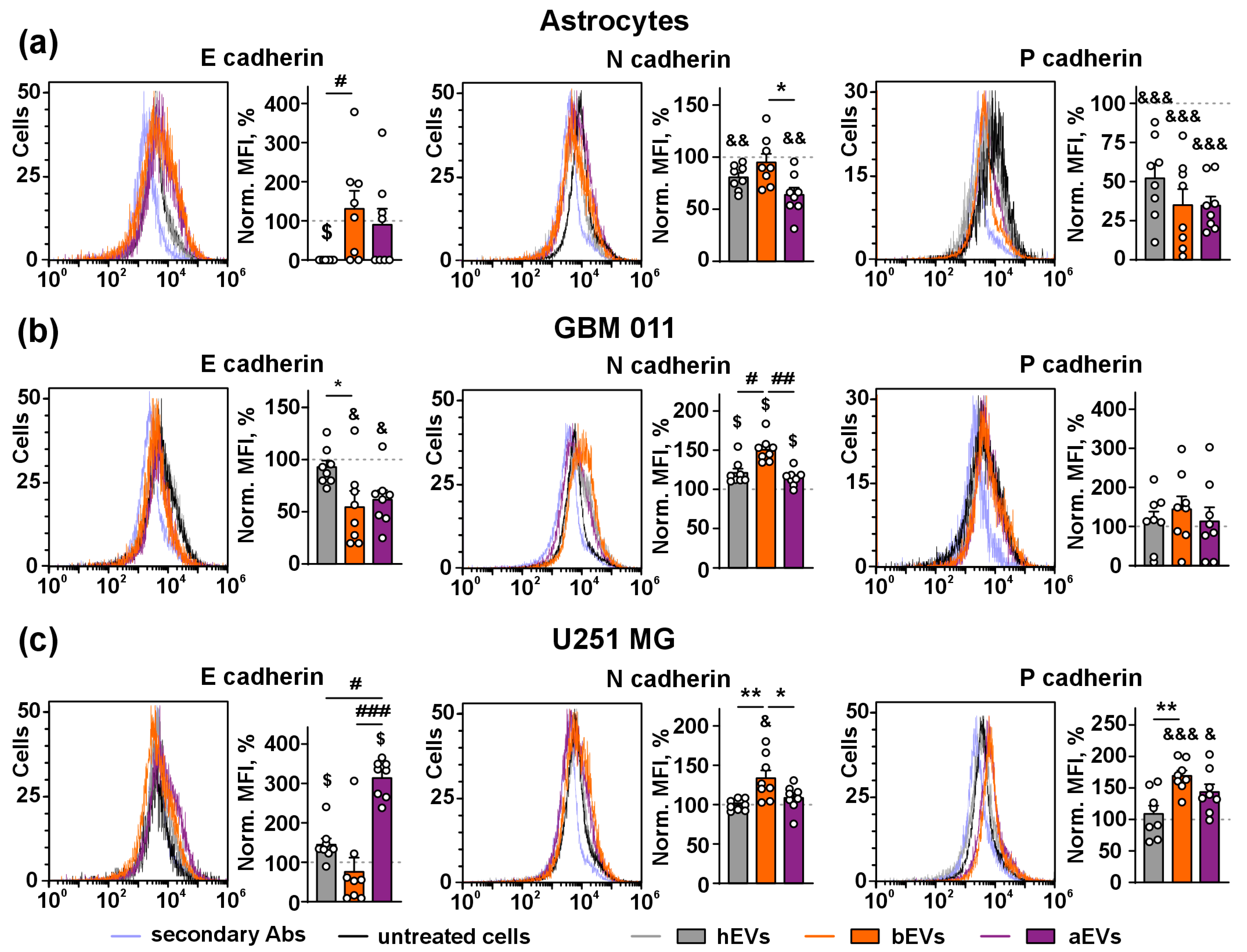
3.4. aEVs and bEVs Induce E/N Cadherin Switch in GB Cells
3.5. aEVs and bEVs Stimulate Secretion of Inflammation and Adhesion Regulators by GB Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular Signature and Crossroads with Tumor Microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Almiron Bonnin, D.A.; Havrda, M.C.; Israel, M.A. Glioma Cell Secretion: A Driver of Tumor Progression and a Potential Therapeutic Target. Cancer Res. 2018, 78, 6031–6039. [Google Scholar] [CrossRef] [PubMed]
- Davidson, C.L.; Vengoji, R.; Jain, M.; Batra, S.K.; Shonka, N. Biological, Diagnostic and Therapeutic Implications of Exosomes in Glioma. Cancer Lett. 2024, 582, 216592. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Sun, Y.; Liu, W.; Zhang, Y.; Sun, G.; Xiang, B.; Yang, J. Exosomes in Glioma: Unraveling Their Roles in Progression, Diagnosis, and Therapy. Cancers 2024, 16, 823. [Google Scholar] [CrossRef]
- Guo, X.; Sui, R.; Piao, H. Exosomes-Mediated Crosstalk between Glioma and Immune Cells in the Tumor Microenvironment. CNS Neurosci. Ther. 2023, 29, 2074–2085. [Google Scholar] [CrossRef]
- Osaid, Z.; Haider, M.; Hamoudi, R.; Harati, R. Exosomes Interactions with the Blood-Brain Barrier: Implications for Cerebral Disorders and Therapeutics. Int. J. Mol. Sci. 2023, 24, 15635. [Google Scholar] [CrossRef]
- Spelat, R.; Jihua, N.; Sánchez Triviño, C.A.; Pifferi, S.; Pozzi, D.; Manzati, M.; Mortal, S.; Schiavo, I.; Spada, F.; Zanchetta, M.E.; et al. The Dual Action of Glioma-Derived Exosomes on Neuronal Activity: Synchronization and Disruption of Synchrony. Cell Death Dis. 2022, 13, 1–15. [Google Scholar] [CrossRef]
- Banerjee, S.; Sharma, V.; Das Mukhopadhyay, C. Exploring Emerging Concepts of Exosomes for the Diagnosis, Prognosis, and Therapeutics of Brain Cancers. Extracell. Vesicle 2024, 3, 100038. [Google Scholar] [CrossRef]
- Pancholi, S.; Tripathi, A.; Bhan, A.; Acharya, M.M.; Pillai, P. Emerging Concepts on the Role of Extracellular Vesicles and Its Cargo Contents in Glioblastoma-Microglial Crosstalk. Mol. Neurobiol. 2022, 59, 2822–2837. [Google Scholar] [CrossRef]
- Zeng, A.; Wei, Z.; Rabinovsky, R.; Jun, H.J.; El Fatimy, R.; Deforzh, E.; Arora, R.; Yao, Y.; Yao, S.; Yan, W.; et al. Glioblastoma-Derived Extracellular Vesicles Facilitate Transformation of Astrocytes via Reprogramming Oncogenic Metabolism. iScience 2020, 23, 101420. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.; Yao, B.; Sun, P.; Hao, Y.; Piao, H.; Zhao, X. Role of Exosomes in the Progression, Diagnosis, and Treatment of Gliomas. Med. Sci. Monit. 2020, 26, e924023. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Musatova, O.E.; Rubtsov, Y.P. Effects of Glioblastoma-Derived Extracellular Vesicles on the Functions of Immune Cells. Front. Cell Dev. Biol. 2023, 11, 1060000. [Google Scholar] [CrossRef] [PubMed]
- Low, J.J.W.; Sulaiman, S.A.; Johdi, N.A.; Abu, N. Immunomodulatory Effects of Extracellular Vesicles in Glioblastoma. Front. Cell Dev. Biol. 2022, 10, 996805. [Google Scholar] [CrossRef]
- Oushy, S.; Hellwinkel, J.E.; Wang, M.; Nguyen, G.J.; Gunaydin, D.; Harland, T.A.; Anchordoquy, T.J.; Graner, M.W. Glioblastoma Multiforme-Derived Extracellular Vesicles Drive Normal Astrocytes towards a Tumour-Enhancing Phenotype. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160477. [Google Scholar] [CrossRef]
- Indira Chandran, V.; Gopala, S.; Venkat, E.H.; Kjolby, M.; Nejsum, P. Extracellular Vesicles in Glioblastoma: A Challenge and an Opportunity. npj Precis. Oncol. 2024, 8, 1–8. [Google Scholar] [CrossRef]
- Foster, J.B.; Koptyra, M.P.; Bagley, S.J. Recent Developments in Blood Biomarkers in Neuro-Oncology. Curr. Neurol. Neurosci. Rep. 2023, 23, 857–867. [Google Scholar] [CrossRef]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.A.; Webb, D.J.; Dear, J.W. Identification and Proteomic Profiling of Exosomes in Human Cerebrospinal Fluid. J. Transl. Med. 2012, 10, 5. [Google Scholar] [CrossRef]
- Chen, W.W.; Balaj, L.; Liau, L.M.; Samuels, M.L.; Kotsopoulos, S.K.; Maguire, C.A.; Loguidice, L.; Soto, H.; Garrett, M.; Zhu, L.D.; et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol. Ther. Nucleic Acids 2013, 2, e109. [Google Scholar] [CrossRef]
- Figueroa, J.M.; Skog, J.; Akers, J.; Li, H.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of Wild-Type EGFR Amplification and EGFRvIII Mutation in CSF-Derived Extracellular Vesicles of Glioblastoma Patients. Neuro Oncol. 2017, 19, 1494–1502. [Google Scholar] [CrossRef]
- Treps, L.; Edmond, S.; Harford-Wright, E.; Galan-Moya, E.M.; Schmitt, A.; Azzi, S.; Citerne, A.; Bidère, N.; Ricard, D.; Gavard, J. Extracellular Vesicle-Transported Semaphorin3A Promotes Vascular Permeability in Glioblastoma. Oncogene 2016, 35, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.J.; Lustig, R.A.; Yang, X.-Y.; Jenkins, W.T.; Wolf, R.L.; Martinez-Lage, M.; Desai, A.; Williams, D.; Evans, S.M. Microvesicles as a Biomarker for Tumor Progression versus Treatment Effect in Radiation/Temozolomide-Treated Glioblastoma Patients. Transl. Oncol. 2014, 7, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-Based Analysis of Exosomal mRNA Mediating Drug Resistance in Glioblastoma. Nat. Commun. 2015, 6, 6999. [Google Scholar] [CrossRef]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Skog, J.; Nakano, I.; Pingle, S.; Kalinina, J.; Hua, W.; Kesari, S.; Mao, Y.; et al. MiR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): A Platform for Glioblastoma Biomarker Development. PLoS ONE 2013, 8, e78115. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes Reflect the Hypoxic Status of Glioma Cells and Mediate Hypoxia-Dependent Activation of Vascular Cells during Tumor Development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef]
- Tzaridis, T.; Weller, J.; Bachurski, D.; Shakeri, F.; Schaub, C.; Hau, P.; Buness, A.; Schlegel, U.; Steinbach, J.-P.; Seidel, C.; et al. A Novel Serum Extracellular Vesicle Protein Signature to Monitor Glioblastoma Tumor Progression. Int. J. Cancer 2023, 152, 308–319. [Google Scholar] [CrossRef]
- Lunavat, T.R.; Nieland, L.; Vrijmoet, A.B.; Zargani-Piccardi, A.; Samaha, Y.; Breyne, K.; Breakefield, X.O. Roles of Extracellular Vesicles in Glioblastoma: Foes, Friends and Informers. Front. Oncol. 2023, 13, 1291177. [Google Scholar] [CrossRef]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune Evasion Mediated by PD-L1 on Glioblastoma-Derived Extracellular Vesicles. Sci. Adv. 2018, 4, eaar2766. [Google Scholar] [CrossRef]
- Serpe, C.; Monaco, L.; Relucenti, M.; Iovino, L.; Familiari, P.; Scavizzi, F.; Raspa, M.; Familiari, G.; Civiero, L.; D’Agnano, I.; et al. Microglia-Derived Small Extracellular Vesicles Reduce Glioma Growth by Modifying Tumor Cell Metabolism and Enhancing Glutamate Clearance through miR-124. Cells 2021, 10, 2066. [Google Scholar] [CrossRef]
- Whitehead, C.A.; Luwor, R.B.; Morokoff, A.P.; Kaye, A.H.; Stylli, S.S. Cancer Exosomes in Cerebrospinal Fluid. Transl. Cancer Res. 2017, 6, S1352–S1370. [Google Scholar] [CrossRef]
- Wang, M.; Cai, Y.; Peng, Y.; Xu, B.; Hui, W.; Jiang, Y. Exosomal LGALS9 in the Cerebrospinal Fluid of Glioblastoma Patients Suppressed Dendritic Cell Antigen Presentation and Cytotoxic T-Cell Immunity. Cell Death Dis. 2020, 11, 896. [Google Scholar] [CrossRef] [PubMed]
- Jafari, D.; Tiyuri, A.; Rezaei, E.; Moradi, Y.; Jafari, R.; Jokar Shoorijeh, F.; Barati, M. Diagnostic Accuracy of Cerebrospinal Fluid and Serum-Isolated Extracellular Vesicles for Glioblastoma: A Systematic Review and Meta-Analysis. Expert Rev. Mol. Diagn. 2020, 20, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Del Bene, M.; Osti, D.; Faletti, S.; Beznoussenko, G.V.; DiMeco, F.; Pelicci, G. Extracellular Vesicles: The Key for Precision Medicine in Glioblastoma. Neuro Oncol. 2022, 24, 184–196. [Google Scholar] [CrossRef]
- Simionescu, N.; Zonda, R.; Petrovici, A.R.; Georgescu, A. The Multifaceted Role of Extracellular Vesicles in Glioblastoma: microRNA Nanocarriers for Disease Progression and Gene Therapy. Pharmaceutics 2021, 13, 988. [Google Scholar] [CrossRef] [PubMed]
- Muller, L.; Muller-Haegele, S.; Mitsuhashi, M.; Gooding, W.; Okada, H.; Whiteside, T.L. Exosomes Isolated from Plasma of Glioma Patients Enrolled in a Vaccination Trial Reflect Antitumor Immune Activity and Might Predict Survival. Oncoimmunology 2015, 4, e1008347. [Google Scholar] [CrossRef]
- Papadimitrakis, D.; Perdikakis, M.; Gargalionis, A.N.; Papavassiliou, A.G. Biomarkers in Cerebrospinal Fluid for the Diagnosis and Monitoring of Gliomas. Biomolecules 2024, 14, 801. [Google Scholar] [CrossRef]
- Dobra, G.; Bukva, M.; Szabo, Z.; Bruszel, B.; Harmati, M.; Gyukity-Sebestyen, E.; Jenei, A.; Szucs, M.; Horvath, P.; Biro, T.; et al. Small Extracellular Vesicles Isolated from Serum May Serve as Signal-Enhancers for the Monitoring of CNS Tumors. Int. J. Mol. Sci. 2020, 21, 5359. [Google Scholar] [CrossRef]
- Cunha Silva, L.; Branco, F.; Cunha, J.; Vitorino, C.; Gomes, C.; Carrascal, M.A.; Falcão, A.; Miguel Neves, B.; Teresa Cruz, M. The Potential of Exosomes as a New Therapeutic Strategy for Glioblastoma. Eur. J. Pharm. Biopharm. 2024, 203, 114460. [Google Scholar] [CrossRef]
- Zhan, Q.; Yi, K.; Qi, H.; Li, S.; Li, X.; Wang, Q.; Wang, Y.; Liu, C.; Qiu, M.; Yuan, X.; et al. Engineering Blood Exosomes for Tumor-Targeting Efficient Gene/Chemo Combination Therapy. Theranostics 2020, 10, 7889–7905. [Google Scholar] [CrossRef]
- Verweij, F.J.; Balaj, L.; Boulanger, C.M.; Carter, D.R.F.; Compeer, E.B.; D’Angelo, G.; El Andaloussi, S.; Goetz, J.G.; Gross, J.C.; Hyenne, V.; et al. The Power of Imaging to Understand Extracellular Vesicle Biology in Vivo. Nat. Methods 2021, 18, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lyu, P.; Ze, Y.; Li, P.; Zeng, X.; Shi, Y.; Qiu, B.; Gong, P.; Yao, Y. Exosomes Derived from Plasma: Promising Immunomodulatory Agents for Promoting Angiogenesis to Treat Radiation-Induced Vascular Dysfunction. PeerJ 2021, 9, e11147. [Google Scholar] [CrossRef] [PubMed]
- Arraud, N.; Linares, R.; Tan, S.; Gounou, C.; Pasquet, J.-M.; Mornet, S.; Brisson, A.R. Extracellular Vesicles from Blood Plasma: Determination of Their Morphology, Size, Phenotype and Concentration. J. Thromb. Haemost. 2014, 12, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-B.; Xia, M.; Gao, Z.; Zhou, H.; Liu, M.; Huang, S.; Zhen, R.; Wu, J.Y.; Roth, W.W.; Bond, V.C.; et al. Characterization of Exosomes in Plasma of Patients with Breast, Ovarian, Prostate, Hepatic, Gastric, Colon, and Pancreatic Cancers. J. Cancer Ther. 2019, 10, 382–399. [Google Scholar] [CrossRef]
- Muller, L.; Hong, C.-S.; Stolz, D.B.; Watkins, S.C.; Whiteside, T.L. Isolation of Biologically-Active Exosomes from Human Plasma. J. Immunol. Methods 2014, 411, 55–65. [Google Scholar] [CrossRef]
- Bychkov, M.L.; Kirichenko, A.V.; Mikhaylova, I.N.; Paramonov, A.S.; Yastremsky, E.V.; Kirpichnikov, M.P.; Shulepko, M.A.; Lyukmanova, E.N. Extracellular Vesicles Derived from Acidified Metastatic Melanoma Cells Stimulate Growth, Migration, and Stemness of Normal Keratinocytes. Biomedicines 2022, 10, 660. [Google Scholar] [CrossRef]
- Holcar, M.; Ferdin, J.; Sitar, S.; Tušek-Žnidarič, M.; Dolžan, V.; Plemenitaš, A.; Žagar, E.; Lenassi, M. Enrichment of Plasma Extracellular Vesicles for Reliable Quantification of Their Size and Concentration for Biomarker Discovery. Sci. Rep. 2020, 10, 21346. [Google Scholar] [CrossRef]
- Chen, X.; Luo, Y.; Zhu, Q.; Zhang, J.; Huang, H.; Kan, Y.; Li, D.; Xu, M.; Liu, S.; Li, J.; et al. Small Extracellular Vesicles from Young Plasma Reverse Age-Related Functional Declines by Improving Mitochondrial Energy Metabolism. Nat. Aging 2024, 4, 814–838. [Google Scholar] [CrossRef]
- Ren, J.; Li, L.; Yu, B.; Xu, E.; Sun, N.; Li, X.; Xing, Z.; Han, X.; Cui, Y.; Wang, X.; et al. Extracellular Vesicles Mediated Proinflammatory Macrophage Phenotype Induced by Radiotherapy in Cervical Cancer. BMC Cancer 2022, 22, 88. [Google Scholar] [CrossRef]
- Albino, D.; Falcione, M.; Uboldi, V.; Temilola, D.O.; Sandrini, G.; Merulla, J.; Civenni, G.; Kokanovic, A.; Stürchler, A.; Shinde, D.; et al. Circulating Extracellular Vesicles Release Oncogenic miR-424 in Experimental Models and Patients with Aggressive Prostate Cancer. Commun. Biol. 2021, 4, 1–13. [Google Scholar] [CrossRef]
- Cao, H.; Abd Aziz, N.H.; Xavier, J.R.; Shafiee, M.N.; Kalok, A.; Jee, B.; Salker, M.S.; Singh, Y. Dysregulated Exosomes Result in Suppression of the Immune Response of Pregnant COVID-19 Convalescent Women. Front. Mol. Biosci. 2022, 9, 869192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Chen, J.; Sheng, S.; Kleyman, T.R. Extracellular Intersubunit Interactions Modulate Epithelial Na+ Channel Gating. J. Biol. Chem. 2023, 299, 102914. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, Z.; Li, D.; Wang, X.; Dai, D.; Fu, H. Effect Study of Exosomes Derived from Platelet-Rich Plasma in the Treatment of Knee Cartilage Defects in Rats. J. Orthop. Surg. Res. 2023, 18, 160. [Google Scholar] [CrossRef] [PubMed]
- Pavlyukov, M.S.; Yu, H.; Bastola, S.; Minata, M.; Shender, V.O.; Lee, Y.; Zhang, S.; Wang, J.; Komarova, S.; Wang, J.; et al. Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma via Intercellular Transfer of Splicing Factors. Cancer Cell 2018, 34, 119–135.e10. [Google Scholar] [CrossRef] [PubMed]
- Schildge, S.; Bohrer, C.; Beck, K.; Schachtrup, C. Isolation and Culture of Mouse Cortical Astrocytes. J. Vis. Exp. 2013, 71, e50079. [Google Scholar] [CrossRef]
- Oeck, S.; Malewicz, N.M.; Hurst, S.; Rudner, J.; Jendrossek, V. The Focinator—A New Open-Source Tool for High-Throughput Foci Evaluation of DNA Damage. Radiat. Oncol. 2015, 10, 163. [Google Scholar] [CrossRef]
- Lyukmanova, E.; Kirichenko, A.; Kulbatskii, D.; Isaev, A.; Kukushkin, I.; Che, Y.; Kirpichnikov, M.; Bychkov, M. Water-Soluble Lynx1 Upregulates Dendritic Spine Density and Stimulates Astrocytic Network and Signaling. Mol. Neurobiol. 2024. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Z.; Lu, N. A New Role for the PI3K/Akt Signaling Pathway in the Epithelial-Mesenchymal Transition. Cell Adh Migr. 2015, 9, 317–324. [Google Scholar] [CrossRef]
- Meel, M.H.; Schaper, S.A.; Kaspers, G.J.L.; Hulleman, E. Signaling Pathways and Mesenchymal Transition in Pediatric High-Grade Glioma. Cell Mol. Life Sci. 2018, 75, 871–887. [Google Scholar] [CrossRef]
- Kudaravalli, S.; den Hollander, P.; Mani, S.A. Role of P38 MAP Kinase in Cancer Stem Cells and Metastasis. Oncogene 2022, 41, 3177–3185. [Google Scholar] [CrossRef]
- Kühnöl, C.D.; Würfel, C.; Staege, M.S.; Kramm, C. Snail Homolog 1 Is Involved in Epithelial-Mesenchymal Transition-like Processes in Human Glioblastoma Cells. Oncol. Lett. 2017, 13, 3882–3888. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.W.; Menon, L.G.; Black, P.M.; Carroll, R.S.; Johnson, M.D. SNAI2/Slug Promotes Growth and Invasion in Human Gliomas. BMC Cancer 2010, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Noronha, C.; Ribeiro, A.S.; Taipa, R.; Castro, D.S.; Reis, J.; Faria, C.; Paredes, J. Cadherin Expression and EMT: A Focus on Gliomas. Biomedicines 2021, 9, 1328. [Google Scholar] [CrossRef]
- Bălașa, A.; Șerban, G.; Chinezu, R.; Hurghiș, C.; Tămaș, F.; Manu, D. The Involvement of Exosomes in Glioblastoma Development, Diagnosis, Prognosis, and Treatment. Brain Sci. 2020, 10, 553. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Jackman, J.G.; Woodring, S.; McSherry, F.; Herndon, J.E.; Desjardins, A.; Friedman, H.S.; Peters, K.B. Second Primary Cancers in Long-Term Survivors of Glioblastoma. Neurooncol Pract. 2019, 6, 386–391. [Google Scholar] [CrossRef]
- Zhou, S.; Niu, R.; Sun, H.; Kim, S.-H.; Jin, X.; Yin, J. The MAP3K1/c-JUN Signaling Axis Regulates Glioblastoma Stem Cell Invasion and Tumor Progression. Biochem. Biophys. Res. Commun. 2022, 612, 188–195. [Google Scholar] [CrossRef]
- Grave, N.; Scheffel, T.B.; Cruz, F.F.; Rockenbach, L.; Goettert, M.I.; Laufer, S.; Morrone, F.B. The Functional Role of P38 MAPK Pathway in Malignant Brain Tumors. Front. Pharmacol. 2022, 13, 975197. [Google Scholar] [CrossRef]
- Muqbil, I.; Wu, J.; Aboukameel, A.; Mohammad, R.M.; Azmi, A.S. Snail Nuclear Transport: The Gateways Regulating Epithelial-to-Mesenchymal Transition? Semin. Cancer Biol. 2014, 27, 39–45. [Google Scholar] [CrossRef]
- Villarejo, A.; Cortés-Cabrera, A.; Molina-Ortíz, P.; Portillo, F.; Cano, A. Differential Role of Snail1 and Snail2 Zinc Fingers in E-Cadherin Repression and Epithelial to Mesenchymal Transition. J. Biol. Chem. 2014, 289, 930–941. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Iser, I.C.; Lenz, G.; Wink, M.R. EMT-like Process in Glioblastomas and Reactive Astrocytes. Neurochem. Int. 2019, 122, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Tuffin, L.J.; Rodriguez, F.; Giannini, C.; Scheithauer, B.; Necela, B.M.; Sarkaria, J.N.; Anastasiadis, P.Z. Misregulated E-Cadherin Expression Associated with an Aggressive Brain Tumor Phenotype. PLoS ONE 2010, 5, e13665. [Google Scholar] [CrossRef] [PubMed]
- Martins, E.P.; Gonçalves, C.S.; Pojo, M.; Carvalho, R.; Ribeiro, A.S.; Miranda-Gonçalves, V.; Taipa, R.; Pardal, F.; Pinto, A.A.; Custódia, C.; et al. Cadherin-3 Is a Novel Oncogenic Biomarker with Prognostic Value in Glioblastoma. Mol. Oncol. 2022, 16, 2611–2631. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.N.; Hong, J.Y.; Shim, D.H.; Sol, I.S.; Kim, Y.S.; Lee, J.H.; Kim, K.W.; Lee, J.M.; Sohn, M.H. Activated Leukocyte Cell Adhesion Molecule Stimulates the T-Cell Response in Allergic Asthma. Am. J. Respir. Crit. Care Med. 2018, 197, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Kijima, N.; Hosen, N.; Kagawa, N.; Hashimoto, N.; Nakano, A.; Fujimoto, Y.; Kinoshita, M.; Sugiyama, H.; Yoshimine, T. CD166/Activated Leukocyte Cell Adhesion Molecule Is Expressed on Glioblastoma Progenitor Cells and Involved in the Regulation of Tumor Cell Invasion. Neuro Oncol. 2012, 14, 1254–1264. [Google Scholar] [CrossRef]
- Inoue, A.; Ohnishi, T.; Nishikawa, M.; Ohtsuka, Y.; Kusakabe, K.; Yano, H.; Tanaka, J.; Kunieda, T. A Narrative Review on CD44’s Role in Glioblastoma Invasion, Proliferation, and Tumor Recurrence. Cancers 2023, 15, 4898. [Google Scholar] [CrossRef]
- Kolliopoulos, C.; Ali, M.M.; Castillejo-Lopez, C.; Heldin, C.-H.; Heldin, P. CD44 Depletion in Glioblastoma Cells Suppresses Growth and Stemness and Induces Senescence. Cancers 2022, 14, 3747. [Google Scholar] [CrossRef]
- Huang, L.; Yang, Y.; Yang, F.; Liu, S.; Zhu, Z.; Lei, Z.; Guo, J. Functions of EpCAM in Physiological Processes and Diseases (Review). Int. J. Mol. Med. 2018, 42, 1771–1785. [Google Scholar] [CrossRef]
- Mohtar, M.A.; Syafruddin, S.E.; Nasir, S.N.; Low, T.Y. Revisiting the Roles of Pro-Metastatic EpCAM in Cancer. Biomolecules 2020, 10, 255. [Google Scholar] [CrossRef]
- Piao, Y.; Henry, V.; Tiao, N.; Park, S.Y.; Martinez-Ledesma, J.; Dong, J.W.; Balasubramaniyan, V.; de Groot, J.F. Targeting Intercellular Adhesion Molecule-1 Prolongs Survival in Mice Bearing Bevacizumab-Resistant Glioblastoma. Oncotarget 2017, 8, 96970–96983. [Google Scholar] [CrossRef]
- Park, J.K.; Park, S.H.; So, K.; Bae, I.H.; Yoo, Y.D.; Um, H.-D. ICAM-3 Enhances the Migratory and Invasive Potential of Human Non-Small Cell Lung Cancer Cells by Inducing MMP-2 and MMP-9 via Akt and CREB. Int. J. Oncol. 2010, 36, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Xie, J.; Zhao, S.; Du, R.; Luo, X.; He, H.; Jiang, S.; Hao, N.; Chen, C.; Guo, C.; et al. ICAM3 Mediates Inflammatory Signaling to Promote Cancer Cell Stemness. Cancer Lett. 2018, 422, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Seker-Polat, F.; Pinarbasi Degirmenci, N.; Solaroglu, I.; Bagci-Onder, T. Tumor Cell Infiltration into the Brain in Glioblastoma: From Mechanisms to Clinical Perspectives. Cancers 2022, 14, 443. [Google Scholar] [CrossRef] [PubMed]
- Prag, S.; Lepekhin, E.A.; Kolkova, K.; Hartmann-Petersen, R.; Kawa, A.; Walmod, P.S.; Belman, V.; Gallagher, H.C.; Berezin, V.; Bock, E.; et al. NCAM Regulates Cell Motility. J. Cell Sci. 2002, 115, 283–292. [Google Scholar] [CrossRef]
- Zhang, D.; Bi, J.; Liang, Q.; Wang, S.; Zhang, L.; Han, F.; Li, S.; Qiu, B.; Fan, X.; Chen, W.; et al. VCAM1 Promotes Tumor Cell Invasion and Metastasis by Inducing EMT and Transendothelial Migration in Colorectal Cancer. Front. Oncol. 2020, 10, 1066. [Google Scholar] [CrossRef]
- Wu, T.-C. The Role of Vascular Cell Adhesion Molecule-1 in Tumor Immune Evasion. Cancer Res. 2007, 67, 6003–6006. [Google Scholar] [CrossRef]
- Barthel, S.R.; Gavino, J.D.; Descheny, L.; Dimitroff, C.J. Targeting Selectins and Selectin Ligands in Inflammation and Cancer. Expert Opin. Ther. Targets 2007, 11, 1473. [Google Scholar] [CrossRef]
- Tvaroška, I.; Selvaraj, C.; Koča, J. Selectins-The Two Dr. Jekyll and Mr. Hyde Faces of Adhesion Molecules-A Review. Molecules 2020, 25, 2835. [Google Scholar] [CrossRef]
- Yeini, E.; Ofek, P.; Pozzi, S.; Albeck, N.; Ben-Shushan, D.; Tiram, G.; Golan, S.; Kleiner, R.; Sheinin, R.; Israeli Dangoor, S.; et al. P-Selectin Axis Plays a Key Role in Microglia Immunophenotype and Glioblastoma Progression. Nat. Commun. 2021, 12, 1912. [Google Scholar] [CrossRef]
- Akers, J.C.; Ramakrishnan, V.; Kim, R.; Phillips, S.; Kaimal, V.; Mao, Y.; Hua, W.; Yang, I.; Fu, C.-C.; Nolan, J.; et al. miRNA Contents of Cerebrospinal Fluid Extracellular Vesicles in Glioblastoma Patients. J. Neurooncol. 2015, 123, 205–216. [Google Scholar] [CrossRef]
- Exosomes: The Good, The Bad, and The Ugly. Available online: https://www.asra.com/news-publications/asra-newsletter/newsletter-item/asra-news/2023/05/12/exosomes-the-good-the-bad-and-the-ugly (accessed on 6 December 2024).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyukmanova, E.N.; Kirichenko, A.V.; Medyanik, I.A.; Yashin, K.S.; Kirpichnikov, M.P.; Bychkov, M.L. Extracellular Vesicles from Plasma of Patients with Glioblastoma Promote Invasion of Glioblastoma Cells Even After Tumor Resection. Biomedicines 2024, 12, 2834. https://doi.org/10.3390/biomedicines12122834
Lyukmanova EN, Kirichenko AV, Medyanik IA, Yashin KS, Kirpichnikov MP, Bychkov ML. Extracellular Vesicles from Plasma of Patients with Glioblastoma Promote Invasion of Glioblastoma Cells Even After Tumor Resection. Biomedicines. 2024; 12(12):2834. https://doi.org/10.3390/biomedicines12122834
Chicago/Turabian StyleLyukmanova, Ekaterina N., Artem V. Kirichenko, Igor A. Medyanik, Konstantin S. Yashin, Mikhail P. Kirpichnikov, and Maxim L. Bychkov. 2024. "Extracellular Vesicles from Plasma of Patients with Glioblastoma Promote Invasion of Glioblastoma Cells Even After Tumor Resection" Biomedicines 12, no. 12: 2834. https://doi.org/10.3390/biomedicines12122834
APA StyleLyukmanova, E. N., Kirichenko, A. V., Medyanik, I. A., Yashin, K. S., Kirpichnikov, M. P., & Bychkov, M. L. (2024). Extracellular Vesicles from Plasma of Patients with Glioblastoma Promote Invasion of Glioblastoma Cells Even After Tumor Resection. Biomedicines, 12(12), 2834. https://doi.org/10.3390/biomedicines12122834






