Mutations in the PIK3C2B, ERBB3, KIT, and MLH1 Genes and Their Relationship with Resistance to Temozolomide in Patients with High-Grade Gliomas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Clinical Information
2.3. Sample Collection for Molecular Biology Analysis
2.4. Sequencing 324 Cancer-Associated Genes
2.5. Mutated Genes Identification Related to Temozolomide Resistance Mechanisms
2.6. Bioinformatic Analysis of Mutations Effect on the Proteins
2.7. Prediction of Pathogenic SNPs
2.8. Prediction of Deleterious SNPs for the Protein
2.9. Predicting Protein Stability for Functionally Deleterious SNPs
2.10. Prediction of Protein Structural Alteration and Loss of Activity
2.11. Structural Comparison Between Normal and Mutated Residues
2.12. Group of Mutations
2.13. Observation of Amino Acid Change
2.14. Protein–Protein Interaction
3. Results
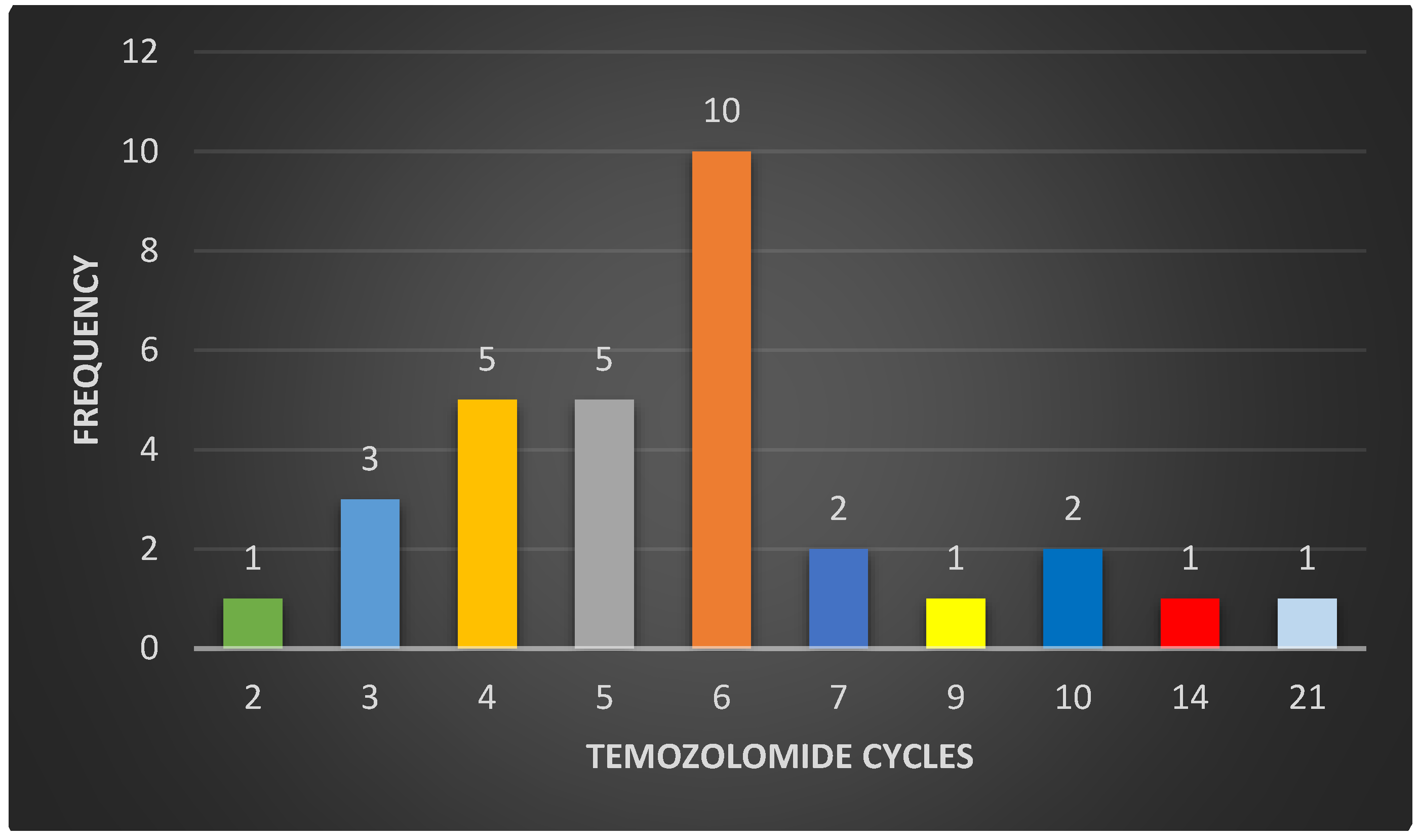
3.1. Study Population
3.2. Clinical Information
3.3. Sample Collection for Molecular Biology Analysis
3.4. Sequencing 324 Cancer-Associated Genes
3.5. Mutated Genes Identification Related to Temozolomide Resistance Mechanisms
3.6. Bioinformatic Analysis of Mutations Effect on the Protein Domains
3.7. Prediction of Pathogenic SNPs
3.8. Prediction of Deleterious SNPs for the Protein
3.9. Predicting Protein Stability for Functionally Deleterious SNPs
3.10. Prediction of Protein Structural Alteration and Loss of Activity
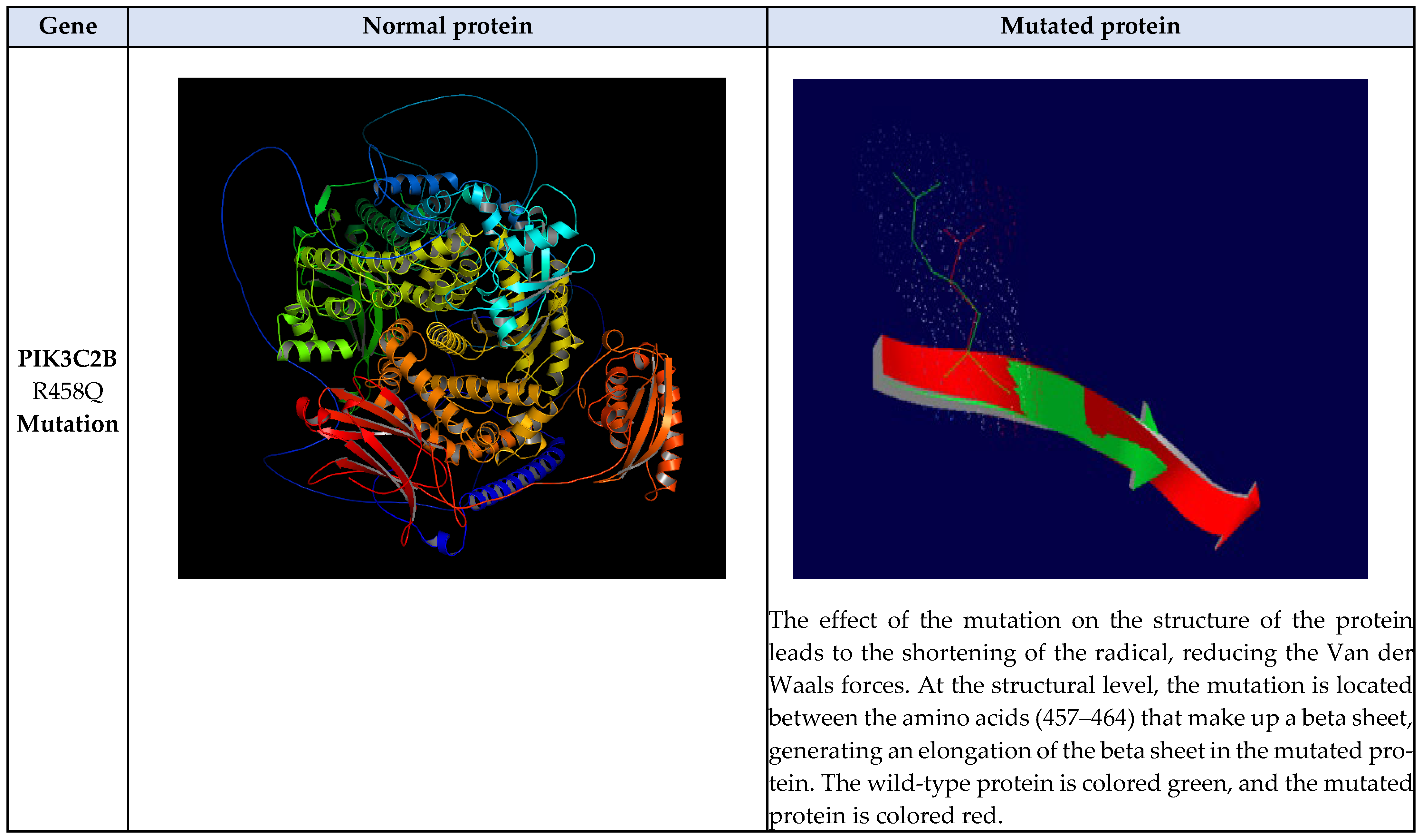
3.11. Structural Comparison Between Normal and Mutated Residues
3.12. Mutations Group
3.13. Observation of Amino Acid Change
3.14. Protein–Protein Interaction
| Chromosome | Position | RS | Gene | Amino Acid Alteration | Classification According to ClinVar | PolyPhen2 Score | Prediction PolyPhen | SIFT Score | Prediction SIFT | PhD SNP | SNP and GO |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | - | PIK3C2B | p.Arg485Gln | New | 0.979 | Probably damaging | 0.45 | Tolerated | Neutral | Neutral |
| 12 | 56086452 | rs560422339 | ERBB3 | p.Arg164Lys | Likely benign | 0.017 | Benign | 0.57 | Tolerated | Neutral | Neutral |
| MUpro | I-Mutant 3.0 | ||||||||||
| Gene | Alteration | Value DDG (Kcal/mol) | Protein stability | Value DDG (Kcal/mol) | Reliability index | Protein stability | |||||
| PIK3C2B | R458Q | −1.0766127 | Decrease | −1.08 | 9 | Decrease | |||||
| ERBB3 | R164K | −1.6580712 | Decrease | −1.47 | 7 | Decrease | |||||
| MuPred2 | |||||||||||
| ID | Substitution | MuPred2 Score | Remarks | Affected PROSITE and ELM motifs | |||||||
| ERBB2 | R164K | 0.128 | - | - | |||||||
| PIK3C2B | R458Q | 0.259 | - | - | |||||||
| Gene | nsSNP | TM-aling | SWISS-MODEL | ||||||||
| TM-score | Value RMSD | Ramachandran graphic score | |||||||||
| PIK3C2B | R458Q | 0.999 | 0.00 | 89.33% | |||||||
| ERBB3 | R164K | 0.980 | 2.26 | 98.18% | |||||||
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gómez Vega, J.C.; Ocampo Navia, M.I.; De Vries, E.; Feo Lee, O. Sobrevida de los tumores cerebrales primarios en Colombia. Univ. Méd. 2020, 61. [Google Scholar] [CrossRef]
- Low, J.T.; Ostrom, Q.T.; Cioffi, G.; Neff, C.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. Primary brain and other central nervous system tumors in the United States (2014–2018): A summary of the CBTRUS statistical report for clinicians. Neuro-Oncol. Pract. 2022, 9, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Jeans, A.; Esiri, M. Brain histology. Pr. Neurol. 2008, 8, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Goryaynov, S.A.; Potapov, A.A.; Ignatenko, M.A.; Zhukov, V.Y.; Protskiy, S.V.; Zakharova, N.A.; Okhlopkov, V.A.; Shishkina, L.V. Glioblastoma metastases: A literature review and a description of six clinical observations. Zhurnal Voprosy Neirokhirurgii Imeni NN Burdenko 2015, 79, 33. [Google Scholar] [CrossRef]
- Lerner, A.; Palmer, K.; Campion, T.; Millner, T.O.; Scott, E.; Lorimer, C.; Paraskevopoulos, D.; McKenna, G.; Marino, S.; Lewis, R.; et al. Gliomas in adults: Guidance on investigations, diagnosis, treatment and surveillance. Clin. Med. 2024, 24, 100240. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- García-Lezama, M.; Carrillo-Ruiz, J.D.; Moreno-Jiménez, S.; Roldán-Valadez, E. WHO CNS5 2021 incluye mutaciones específicas en gliomas que pueden ser identificadas con biomarcadores cuantitativos de resonancia magnética. Gaceta médica de México 2023, 159, 164–171. [Google Scholar] [CrossRef]
- Komori, T. The 2021 WHO classification of tumors, 5th edition, central nervous system tumors: The 10 basic principles. Brain Tumor Pathol. 2022, 39, 47–50. [Google Scholar] [CrossRef]
- Nikitin, P.V.; Musina, G.R.; Pekov, S.I.; Kuzin, A.A.; Popov, I.A.; Belyaev, A.Y.; Kobyakov, G.L.; Usachev, D.Y.; Nikolaev, V.N.; Mikhailov, V.P. Cell-Population Dynamics in Diffuse Gliomas during Gliomagenesis and Its Impact on Patient Survival. Cancers 2022, 15, 145. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, R.; Smith-Cohn, M.; Cohen, A.L.; Colman, H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics 2017, 14, 284–297. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; Van Den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.; Esteller, M. The DNA methylome of glioblastoma multiforme. Neurobiol. Dis. 2010, 39, 40–46. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; De Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Back, M.; Rodriguez, M.; Jayamanne, D.; Khasraw, M.; Lee, A.; Wheeler, H. Understanding the Revised Fourth Edition of the World Health Organization Classification of Tumors of the Central Nervous System (2016) for Clinical Decision-making: A Guide for Oncologists 556 Managing Patients with Glioma. Clin. Oncol. 2018, 30, 556–562. [Google Scholar] [CrossRef]
- Komori, T. Grading of adult diffuse gliomas according to the 2021 WHO Classification of Tumors of the Central Nervous System. Lab. Investig. 2022, 102, 126–133. [Google Scholar] [CrossRef]
- Mikkelsen, V.E.; Solheim, O.; Salvesen, O.; Torp, S.H. The histological representa-tiveness of glioblastoma tissue samples. Acta Neurochir. 2021, 163, 1911–1920. [Google Scholar] [CrossRef]
- Iwanaga, T.; Takahashi, Y.; Fujita, T. Immunohistochemistry of neuron-specific and glia-specific proteins. Arch. Histol. Cytol. 1989, 52, 13–24. [Google Scholar] [CrossRef]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Reviews. Dis. Primers 2015, 1, 15017. [Google Scholar] [CrossRef]
- Zhang, J.; FG Stevens, M.; D Bradshaw, T. Temozolomide: Mechanisms of Action, Repair and Resistance. Curr. Mol. Pharmacol. 2012, 5, 102–114. [Google Scholar] [CrossRef]
- Wang, J.Y.J.; Edelmann, W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006, 9, 417–418. [Google Scholar] [CrossRef] [PubMed]
- Massoud, T.F.; Paulmurugan, R. (Eds.) Glioblastoma Resistance to Chemotherapy: Molecular Mechanisms and Innovative Reversal Strategies, 1st ed.; Academic Press: San Diego, CA, USA, 2021; Volume 582, p. 24. [Google Scholar]
- Shen, W.; Hu, J.A.; Zheng, J.S. Mechanism of temozolomide-induced antitumour effects on glioma cells. J. Int. Med. Res. 2014, 42, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, F.; Zhou, Y.; Chen, W.; Shao, H.; Zhang, Y. MGMT Promoter Methylation and Glioblastoma Prognosis: A Systematic Review and Meta-analysis. Arch. Med. Res. 2013, 44, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Nabors, L.B.; Portnow, J.; Ahluwalia, M.; Baehring, J.; Brem, H.; Brem, S.; Butowski, N.; Campian, J.L.; Clark, S.W.; Fabiano, A.J.; et al. Central Nervous System Cancers, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 1537–1570. [Google Scholar] [CrossRef]
- Weller, M.; Stupp, R.; Reifenberger, G.; Brandes, A.A.; Van Den Bent, M.J.; Wick, W.; Hegi, M.E. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat. Rev. Neurol. 2010, 6, 39–51. [Google Scholar] [CrossRef]
- Binabaj, M.M.; Bahrami, A.; ShahidSales, S.; Joodi, M.; Joudi Mashhad, M.; Hassanian, S.M.; Anvari, K.; Avan, A. The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. J. Cell. Physiol. 2018, 233, 378–386. [Google Scholar] [CrossRef]
- Knobbe, C.B.; Reifenberger, G. Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3′-kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain Pathol. 2003, 13, 507–518. [Google Scholar] [CrossRef]
- Shepherd, P.R. Mechanisms regulating phosphoinositide 3-kinase signalling in insulin-600 sensitive tissues. Acta Physiol. Scand. 2005, 183, 3–12. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Jaiswal, B.S.; Kljavin, N.M.; Stawiski, E.W.; Chan, E.; Parikh, C.; Durinck, S.; Chaudhuri, S.; Pujara, K.; Guillory, J.; Edgar, K.A.; et al. Oncogenic ERBB3 Mutations in Human Cancers. Cancer Cell 2013, 23, 603–617. [Google Scholar] [CrossRef]
- Roskoski, R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef] [PubMed]
- Vanajothi, R.; Rajamanika, S.; Sudha, A.; Srinivasan, P. Structural and Functional Analysis of KIT Gene Encoding Receptor Tyrosine Kinase and its Interaction with Sunitinib and HDAC Inhibitors: An in-silico Approach. Pak. J. Biol. Sci. 2012, 15, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Pentikäinen, O.T.; Singh, P.K. A holistic view on c-Kit in cancer: Structure, signaling, pathophysiology and its inhibitors. Biochim. Biophys. Acta BBA—Rev. Cancer 2021, 1876, 188631. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Schwartzbaum, J.; Wiklund, F.; Sjöström, S.; Liu, Y.; Tsavachidis, S.; Ahlbom, A.; Auvinen, A.; Collatz-Laier, H.; Feychting, M.; et al. A comprehensive study of the association between the EGFR and ERBB2 genes and glioma risk. Acta Oncol. 2010, 49, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Puputti, M.; Tynninen, O.; Sihto, H.; Blom, T.; Mäenpää, H.; Isola, J.; Paetau, A.; Joensuu, H.; Nupponen, N.N. Amplification of KIT, PDGFRA, VEGFR2, and EGFR in Gliomas. Mol. Cancer Res. 2006, 4, 927–934. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef]
- Ida, J.A.; Dubois-Dalcq, M.; McKinnon, R.D. Expression of the receptor tyrosine kinase c-kit in oligodendrocyte progenitor cells. J. Neurosci. Res. 1993, 36, 596–606. [Google Scholar] [CrossRef]
- Nobusawa, S.; Stawski, R.; Kim, Y.H.; Nakazato, Y.; Ohgaki, H. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: A population-based study: PDGFRA amplification in glioblastoma. Neuropathology 2011, 31, 583–588. [Google Scholar] [CrossRef]
- Yan, Y.; Takayasu, T.; Hines, G.; Dono, A.; Hsu, S.H.; Zhu, J.J.; Riascos-Castaneda, R.F.; Kamali, A.; Bhattacharjee, M.B.; Blanco, A.I.; et al. Landscape of Genomic Alterations in IDH Wild-Type Glioblastoma Identifies PI3K as a Favorable Prognostic Factor. JCO Precis. Oncol. 2020, 4, 575–584. [Google Scholar] [CrossRef]
- Li, X.; Wu, C.; Chen, N.; Gu, H.; Yen, A.; Cao, L.; Wang, E.; Wang, L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016, 7, 33440–33450. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [PubMed]
- De Bacco, F.; Boccaccio, C. ERBB3 as a therapeutic target in glioblastoma: Overexpression can make the difference. Mol. Cell Oncol. 2021, 8, 1990677. [Google Scholar] [CrossRef]
- Ortiz, R.; Perazzoli, G.; Cabeza, L.; Jiménez-Luna, C.; Luque, R.; Prados, J.; Melguizo, C. Temozolomide: An Updated Overview of Resistance Mechanisms, Nanotech-nology Advances and Clinical Applications. Curr. Neuropharmacol. 2021, 19, 513–537. [Google Scholar]
- Indraccolo, S.; Lombardi, G.; Fassan, M.; Pasqualini, L.; Giunco, S.; Marcato, R.; Gasparini, A.; Candiotto, C.; Nalio, S.; Fiduccia, P.; et al. Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clin. Cancer Res. 2019, 25, 1828–1837. [Google Scholar] [CrossRef]
- Felsberg, J.; Thon, N.; Eigenbrod, S.; Hentschel, B.; Sabel, M.C.; Westphal, M.; Schackert, G.; Kreth, F.W.; Pietsch, T.; Löffler, M.; et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glio-blastomas. Int. J. Cancer 2011, 129, 659–670. [Google Scholar] [CrossRef]
- Higuchi, F.; Nagashima, H.; Ning, J.; Koerner, M.V.A.; Wakimoto, H.; Cahill, D.P. Restoration of Temozolomide Sensitivity by PARP Inhibitors in Mismatch Repair Deficient Glioblastoma is Independent of Base Excision Repair. Clin. Cancer Res. 2020, 26, 1690–1699. [Google Scholar] [CrossRef]
- Yang, B.; Han, N.; Sun, J.; Jiang, H.; Xu, H.Y. CtIP contributes to non-homologous end joining formation through interacting with ligase IV and promotion of TMZ resistance in glioma cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2092–2102. [Google Scholar]
- Zhang, P.; Wang, H.; Chen, Y.; Lodhi, A.F.; Sun, C.; Sun, F.; Yan, L.; Deng, Y.; Ma, H. DR5 related autophagy can promote apoptosis in gliomas after irradiation. Biochem. Biophys. Res. Commun. 2020, 522, 910–916. [Google Scholar] [CrossRef]







| Gene | p-Value | Hazard Ratio (CI 95%) |
|---|---|---|
| PIK3C2B | 0.004 | 13.81 (2.258–84.45) |
| NOTCH3 | 0.005 | 6.026 (1.742–20.84) |
| KIT | 0.024 | 3.985 (1.204–13.18) |
| ERBB3 | 0.04 | 3.871 (1.066–14.04) |
| MLH1 | 0.06 | 3.528 (0.950–13.09) |
| Gene | p-Value | HR Raw Value (CI 95%) | p-Value | HR Adjusted Value (CI 95%) |
|---|---|---|---|---|
| PIK3C2B | 0.004 | 13.81 (2.258–84.45) | 0.0001 | 82.37 (8.36–811.67) |
| KIT | 0.024 | 3.985 (1.204–13.18) | 0.002 | 10.24 (2.42–43.34) |
| ERBB3 | 0.04 | 3.871 (1.066–14.04) | 0.001 | 13.20 (2.77–62.77) |
| MLH1 | 0.06 | 3.528 (0.950–13.09) | 0.006 | 8.50 (1.83–39.45) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz Gómez, L.D.; Contreras Martínez, H.J.; Galvis Pareja, D.A.; Vélez Gómez, S.; Salazar Flórez, J.E.; Monroy, F.P.; Peláez Sánchez, R.G. Mutations in the PIK3C2B, ERBB3, KIT, and MLH1 Genes and Their Relationship with Resistance to Temozolomide in Patients with High-Grade Gliomas. Biomedicines 2024, 12, 2777. https://doi.org/10.3390/biomedicines12122777
Ortiz Gómez LD, Contreras Martínez HJ, Galvis Pareja DA, Vélez Gómez S, Salazar Flórez JE, Monroy FP, Peláez Sánchez RG. Mutations in the PIK3C2B, ERBB3, KIT, and MLH1 Genes and Their Relationship with Resistance to Temozolomide in Patients with High-Grade Gliomas. Biomedicines. 2024; 12(12):2777. https://doi.org/10.3390/biomedicines12122777
Chicago/Turabian StyleOrtiz Gómez, León Darío, Heidy Johanna Contreras Martínez, David Andrés Galvis Pareja, Sara Vélez Gómez, Jorge Emilio Salazar Flórez, Fernando P. Monroy, and Ronald Guillermo Peláez Sánchez. 2024. "Mutations in the PIK3C2B, ERBB3, KIT, and MLH1 Genes and Their Relationship with Resistance to Temozolomide in Patients with High-Grade Gliomas" Biomedicines 12, no. 12: 2777. https://doi.org/10.3390/biomedicines12122777
APA StyleOrtiz Gómez, L. D., Contreras Martínez, H. J., Galvis Pareja, D. A., Vélez Gómez, S., Salazar Flórez, J. E., Monroy, F. P., & Peláez Sánchez, R. G. (2024). Mutations in the PIK3C2B, ERBB3, KIT, and MLH1 Genes and Their Relationship with Resistance to Temozolomide in Patients with High-Grade Gliomas. Biomedicines, 12(12), 2777. https://doi.org/10.3390/biomedicines12122777






