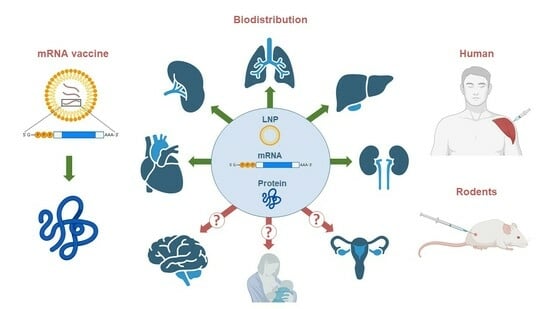
Biodistribution of RNA Vaccines and of Their Products: Evidence from Human and Animal Studies
Abstract
:1. Introduction
2. The Search Strategy
3. Biodistribution of an mRNA-Encoded Protein
| mRNA-LNP | Immunization Details | Species | Results (Antigens) | Ref. |
|---|---|---|---|---|
| mRNA(FLuc)-LNP | A single intramuscular injection of 2 µg of mRNA | BALB/c mice | FLuc was detected at the injection site and liver (max concentration at 6 h) | [17] |
| mRNA(FLuc)-LNP | A single intramuscular injection of 0.08, 0.4, 2.0, or 10 µg of mRNA | BALB/c mice | FLuc was detected at the injection site and liver (max concentration at 6 h) | [18] |
| mRNA(Luc)-LNP | A single intramuscular injection of 5 µg of mRNA | BALB/c mice | +++ injection site + liver (3 h after administration) | [19] |
| mRNA(Luc)-LNP | A single intramuscular injection of 10 µg of mRNA | BALB/c mice | +++ injection site ++ liver + spleen (6 h after administration) | [20] |
| mRNA(FLuc)-LNP | A single intramuscular injection of 5 µg of mRNA | BALB/c mice | + injection site (max concentration at 6 h) | [22] |
| mRNA(Luc)-LNP | A single IV | C57Bl/6J mice | +++ liver ++ spleen + lungs, lymph nodes (max concentration at 6 h) | [26] |
| or SC injection | + lymph nodes (max concentration at 24 h) | |||
| mRNA-1273 | 13 individuals (first/second dose) | Humans | S1 antigen: plasma (max concentration at 5 days) Spike protein: plasma (max concentration at 14 days) | [8] |
| mRNA-1273 or BNT162b2 | Seven individuals (first/second dose) | Humans | Spike protein: ipsilateral axillary lymph nodes (max concentration at 16 days) | [31] |
| mRNA-1273 or BNT162b2 | ~300 individuals (first/second dose) | Humans | Spike protein: plasma (max concentration at 2 days) | [31] |
| BNT162b2 | Eight individuals (first/second dose) | Humans | Spike protein: plasma (max concentration at 14 days) | [9] |
| mRNA-1273 or BNT162b2 | 20 individuals (first/second dose) | Humans | Spike protein: plasma (in 50% of subjects, up to 187 days after vaccination) | [34] |
| mRNA-1273 or BNT162b2 | 22 individuals (first/second dose) | Humans | Spike protein: skin | [11] |
4. Biodistribution of Vaccine mRNA and LNPs
| mRNA-LNP | Immunization Details | Species | Results (mRNA) | Ref. |
|---|---|---|---|---|
| mRNA-1647 | A single intramuscular injection of 100 µg of mRNA-1647 | Sprague–Dawley rats (only males) | +++ injection site, lymph nodes, liver, spleen, eyes ++ blood + heart, lungs, testes, and brain − kidneys (max concentrations at 24 h) | [10] |
| mRNA vaccine against Influenza Viruses | A single injection of 6 mg of formulated mRNA, either intramuscularly or intradermally | CD-1 mice (only males) | ++++ injection site +++ lymph nodes ++ spleen, liver + blood plasma and other tissues (max concentrations at 24 h) | [37] |
| E6- or E7-encoding mRNA (mixed with TriMix) | Two doses intravenously (1 week apart) of 100 µg of mRNA in total | non-human primates | +++ spleen ++ liver, bone marrow + lung, lymph node − heart, brain, kidney, adrenal gland (after the 2nd administration) | [38] |
| mRNA vaccine against yellow fever | A single intramuscular injection of 200 µg | Cynomolgus monkeys | +++ injection site ++ inguinal LNs + iliac and para-aortic LNs (up to 28 h after vaccination) | [40] |
| BNT162b2 | 16 individuals (first/second/boost dose) | Humans | + blood plasma (up to 15 days after vaccination) | [41] |
| BNT162b2 or mRNA 1273 | HCV-positive patients (first/second dose) | Humans | + blood plasma (up to 28 days after vaccination) | [42] |
| BNT162b2 or mRNA 1273 | Two doses of one of the vaccines | Humans | + lymph nodes (up to 60 days after vaccination) | [31] |
| BNT162b2 or mRNA 1273 | 20 individuals (first/second/boost dose) | Humans | ++ axillary lymph nodes + heart − spleen, liver (up to 26 days after vaccination) | [44] |
| BNT162b2 or mRNA 1273 | Healthy lactating individuals (first/second dose) | Humans | + breast milk (up to 48 h after vaccination, only in ~50% of subjects) | [45,46] |
| BNT162b2 or mRNA 1273 | Healthy lactating individuals (first/second dose) | Humans | − breast milk | [47] |
| BNT162b2 | Healthy lactating individuals (first/second dose) | Humans | + breast milk (up to 3 days after vaccination, only in ~10% of subjects) | [48] |
| rabies saRNA vaccine | A single intramuscular or intradermal injection at 0.15 μg | BALB/c mice | + injection site (max concentration at 4 h) | [49] |
| saRNA vaccine | A single intramuscular injection | C57Bl/6J mice | +++ injection site, lymph nodes ++ liver, spleen + lungs (up to 21 days after vaccination) | [50] |
| rabies saRNA vaccine | A single intramuscular injection at 15 µg | Sprague–Dawley rats | +++ injection site, lymph nodes ++ liver, spleen, lungs + blood plasma – brain, kidneys, heart, reproductive organs | [51] |
| saRNA vaccine against SARS-CoV-2 | A single intramuscular injection at 6 µg | Sprague–Dawley rats | +++ injection site, lymph nodes, spleen ++ liver, blood + heart, lungs, kidneys, reproductive organs − brain | [52] |
| saRNA vaccine against SARS-CoV-2 | A single oral administration at 10 µg | BALB/c mice | ++ small intestine + large intestine, liver | [53] |
5. Can Vaccine mRNA Cross Blood–Breast Milk and Blood–Placental Barriers?
6. Biodistribution of a Self-Amplifying mRNA (saRNA) Vaccine
7. Concluding Remarks and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Self, W.H.; Tenforde, M.W.; Rhoads, J.P.; Gaglani, M.; Ginde, A.A.; Douin, D.J.; Olson, S.M.; Talbot, H.K.; Casey, J.D.; Mohr, N.M.; et al. Comparative Effectiveness of Moderna, Pfizer-BioNTech, and Janssen (Johnson & Johnson) Vaccines in Preventing COVID-19 Hospitalizations Among Adults without Immunocompromising Conditions-United States, March–August 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 1337–1343. [Google Scholar] [PubMed]
- Pang, A.P.S.; Higgins-Chen, A.T.; Comite, F.; Raica, I.; Arboleda, C.; Went, H.; Mendez, T.; Schotsaert, M.; Dwaraka, V.; Smith, R.; et al. Longitudinal Study of DNA Methylation and Epigenetic Clocks Prior to and Following Test-Confirmed COVID-19 and mRNA Vaccination. Front. Genet. 2022, 13, 819749. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, P.S.; Scott, M.K.D.; Hagan, T.; Li, C.; Feng, Y.; Wimmers, F.; Grigoryan, L.; Trisal, M.; Edara, V.V.; Lai, L.; et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature 2021, 596, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Sureshchandra, S.; Lewis, S.A.; Doratt, B.M.; Jankeel, A.; Coimbra Ibraim, I.; Messaoudi, I. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight 2021, 6, e153201. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Zhou, J.Q.; Horvath, S.C.; Schmitz, A.J.; Sturtz, A.J.; Lei, T.; Liu, Z.; Kalaidina, E.; Thapa, M.; Alsoussi, W.B.; et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature 2022, 604, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Kato, Y.; Edahiro, R.; Sondergaard, J.N.; Murakami, T.; Amiya, S.; Nameki, S.; Yoshimine, Y.; Morita, T.; Takeshima, Y.; et al. Consecutive BNT162b2 mRNA vaccination induces short-term epigenetic memory in innate immune cells. JCI Insight 2022, 7, e163347. [Google Scholar] [CrossRef]
- Kramer, K.J.; Wilfong, E.M.; Voss, K.; Barone, S.M.; Shiakolas, A.R.; Raju, N.; Roe, C.E.; Suryadevara, N.; Walker, L.M.; Wall, S.C.; et al. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat. Commun. 2022, 13, 3466. [Google Scholar] [CrossRef]
- Ogata, A.F.; Cheng, C.A.; Desjardins, M.; Senussi, Y.; Sherman, A.C.; Powell, M.; Novack, L.; Von, S.; Li, X.; Baden, L.R.; et al. Circulating Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vaccine Antigen Detected in the Plasma of mRNA-1273 Vaccine Recipients. Clin. Infect. Dis. 2022, 74, 715–718. [Google Scholar] [CrossRef]
- Bansal, S.; Perincheri, S.; Fleming, T.; Poulson, C.; Tiffany, B.; Bremner, R.M.; Mohanakumar, T. Cutting Edge: Circulating Exosomes with COVID Spike Protein Are Induced by BNT162b2 (Pfizer-BioNTech) Vaccination prior to Development of Antibodies: A Novel Mechanism for Immune Activation by mRNA Vaccines. J. Immunol. 2021, 207, 2405–2410. [Google Scholar] [CrossRef]
- European Medicines Agency. Moderna Assessment Report COVID-19 Vaccine Moderna; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Magro, C.; Crowson, A.N.; Franks, L.; Schaffer, P.R.; Whelan, P.; Nuovo, G. The histologic and molecular correlates of COVID-19 vaccine-induced changes in the skin. Clin. Dermatol. 2021, 39, 966–984. [Google Scholar] [CrossRef]
- Kirshina, A.K.A.; Kolosova, E.; Imasheva, E.; Vasileva, O.; Zaborova, O.; Terenin, I.; Muslimov, A.; Reshetnikov, V. Effects of various mRNA-LNP vaccine doses on neuroinflammation in BALB/c mice. Bull. Russ. State Med. Univ. 2022, 119–125. [Google Scholar] [CrossRef]
- Ndeupen, S.; Qin, Z.; Jacobsen, S.; Bouteau, A.; Estanbouli, H.; Igyártó, B.Z. The mRNA-LNP platform’s lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience 2021, 24, 103479. [Google Scholar] [CrossRef] [PubMed]
- Shook, L.L.; Edlow, A.G. Safety and Efficacy of Coronavirus Disease 2019 (COVID-19) mRNA Vaccines during Lactation. Obstet. Gynecol. 2023, 141, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Swingle, K.L.; Safford, H.C.; Geisler, H.C.; Hamilton, A.G.; Thatte, A.S.; Billingsley, M.M.; Joseph, R.A.; Mrksich, K.; Padilla, M.S.; Ghalsasi, A.A.; et al. Ionizable Lipid Nanoparticles for In Vivo mRNA Delivery to the Placenta during Pregnancy. J. Am. Chem. Soc. 2023, 145, 4691–4706. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, V.J.; Li, L.; Buarpung, S.; Prahl, M.; Robinson, J.F.; Gaw, S.L. Minimal mRNA uptake and inflammatory response to COVID-19 mRNA vaccine exposure in human placental explants. iScience 2023, 26, 107549. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Assessment Report: Comirnaty; European Medicines Agency: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, Ö.; Thompson, J.; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017, 25, 1316–1327. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, R.; Guo, J.; Li, M.; Ma, L.; Dai, J.; Shi, Y.; Dai, J.; Huang, Y.; Dai, C.; et al. Immunization with a Prefusion SARS-CoV-2 Spike Protein Vaccine (RBMRNA-176) Protects against Viral Challenge in Mice and Nonhuman Primates. Vaccines 2022, 10, 1698. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Deng, Y.; Huang, B.; Huang, L.; Lin, A.; Li, Y.; Wang, W.; Liu, J.; Lu, S.; Zhan, Z.; et al. A core-shell structured COVID-19 mRNA vaccine with favorable biodistribution pattern and promising immunity. Signal Transduct. Target. Ther. 2021, 6, 213. [Google Scholar] [CrossRef]
- Kirshina, A.; Vasileva, O.; Kunyk, D.; Seregina, K.; Muslimov, A.; Ivanov, R.; Reshetnikov, V. Effects of Combinations of Untranslated-Region Sequences on Translation of mRNA. Biomolecules 2023, 13, 1677. [Google Scholar] [CrossRef]
- Ripoll, M.; Bernard, M.C.; Vaure, C.; Bazin, E.; Commandeur, S.; Perkov, V.; Lemdani, K.; Nicolaï, M.-C.; Bonifassi, P.; Kichler, A.; et al. An imidazole modified lipid confers enhanced mRNA-LNP stability and strong immunization properties in mice and non-human primates. Biomaterials 2022, 286, 121570. [Google Scholar] [CrossRef]
- Troy, T.; Jekic-McMullen, D.; Sambucetti, L.; Rice, B. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol. Imaging 2004, 3, 9–23. [Google Scholar] [CrossRef]
- Yan, X.; Kuipers, F.; Havekes, L.M.; Havinga, R.; Dontje, B.; Poelstra, K.; Scherphof, G.L.; Kalmps, J.A. The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem. Biophys. Res. Commun. 2005, 328, 57–62. [Google Scholar] [CrossRef]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissmaln, D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 2015, 217, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Duan, X.; Mao, M.; Song, Y.; Rao, Y.; Cheng, D.; Feng, L.; Shao, X.; Jiang, C.; Huang, H.; et al. mRNA-LNP vaccination-based immunotherapy augments CD8(+) T cell responses against HPV-positive oropharyngeal cancer. NPJ Vaccines 2023, 8, 144. [Google Scholar] [CrossRef] [PubMed]
- Ignowski, J.M.; Schaffer, D.V. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol. Bioeng. 2004, 86, 827–834. [Google Scholar] [CrossRef]
- Li, W.; Kitsios, G.D.; Bain, W.; Wang, C.; Li, T.; Fanning, K.V.; Deshpande, R.; Qin, X.; Morris, A.; Lee, J.S.; et al. Stability of SARS-CoV-2-Encoded Proteins and Their Antibody Levels Correlate with Interleukin 6 in COVID-19 Patients. mSystems 2022, 7, e0005822. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2-Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.H.; Rissin, D.M.; Kan, C.W.; Fournier, D.R.; Piech, T.; Campbell, T.G.; Meyer, R.E.; Fishburn, M.W.; Cabrera, C.; Patel, P.P.; et al. The Simoa HD-1 Analyzer: A Novel Fully Automated Digital Immunoassay Analyzer with Single-Molecule Sensitivity and Multiplexing. J. Lab. Autom. 2016, 21, 533–547. [Google Scholar] [CrossRef]
- Roltgen, K.; Nielsen, S.C.A.; Silva, O.; Younes, S.F.; Zaslavsky, M.; Costales, C.; Yang, F.; Wirz, O.F.; Solis, D.; Hoh, R.A.; et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 2022, 185, 1025–1040.e14. [Google Scholar] [CrossRef]
- Elschner, M.C.; Laroucau, K.; Singha, H.; Tripathi, B.N.; Saqib, M.; Gardner, I.; Saini, S.; Kumar, S.; El-Adawy, H.; Melzer, F.; et al. Evaluation of the comparative accuracy of the complement fixation test, Western blot and five enzyme-linked immunosorbent assays for serodiagnosis of glanders. PLoS ONE 2019, 14, e0214963. [Google Scholar] [CrossRef]
- Edouard, S.; Jaafar, R.; Orain, N.; Parola, P.; Colson, P.; La Scola, B.; Fournier, P.-E.; Raoult, D.; Drancourt, M. Automated Western immunoblotting detection of anti-SARS-CoV-2 serum antibodies. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Brogna, C.; Cristoni, S.; Marino, G.; Montano, L.; Viduto, V.; Fabrowski, M.; Lettieri, G.; Piscopo, M. Detection of recombinant Spike protein in the blood of individuals vaccinated against SARS-CoV-2: Possible molecular mechanisms. Proteom. Clin. Appl. 2023, 17, e2300048. [Google Scholar] [CrossRef] [PubMed]
- Reichmuth, A.M.; Oberli, M.A.; Jaklenec, A.; Langer, R.; Blankschtein, D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016, 7, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, A.; Zhao, Y.; Huang, Y.F.; Dukler, N.; Rice, E.J.; Chivu, A.G.; Krumholz, K.; Danko, C.G.; Siepel, A. Characterizing RNA stability genome-wide through combined analysis of PRO-seq and RNA-seq data. BMC Biol. 2021, 19, 30. [Google Scholar] [CrossRef]
- Flagella, M.; Bui, S.; Zheng, Z.; Nguyen, C.T.; Zhang, A.; Pastor, L.; Ma, Y.; Yang, W.; Crawford, K.L.; McMaster, G.K.; et al. A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal. Biochem. 2006, 352, 50–60. [Google Scholar] [CrossRef]
- Bevers, S.; Kooijmans, S.A.A.; Van de Velde, E.; Evers, M.J.W.; Seghers, S.; Gitz-Francois, J.J.; van Kronenburg, N.C.H.; Fens, M.H.A.M.; Mastrobattista, E.; Hassler, L.; et al. mRNA-LNP vaccines tuned for systemic immunization induce strong antitumor immunity by engaging splenic immune cells. Mol. Ther. 2022, 30, 3078–3094. [Google Scholar] [CrossRef]
- Bonehill, A.; Tuyaerts, S.; Van Nuffel, A.M.; Heirman, C.; Bos, T.J.; Fostier, K.; Neyns, B.; Thielemans, K. Enhancing the T-cell stimulatory capacity of human dendritic cells by co-electroporation with CD40L, CD70 and constitutively active TLR4 encoding mRNA. Mol. Ther. 2008, 16, 1170–1180. [Google Scholar] [CrossRef]
- Lindsay, K.E.; Bhosle, S.M.; Zurla, C.; Beyersdorf, J.; Rogers, K.A.; Vanover, D.; Xiao, P.; Araínga, M.; Shirreff, L.M.; Pitard, B.; et al. Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat. Biomed. Eng. 2019, 3, 371–380. [Google Scholar] [CrossRef]
- Fertig, T.E.; Chitoiu, L.; Marta, D.S.; Ionescu, V.S.; Cismasiu, V.B.; Radu, E.; Angheluta, G.; Dobre, M.; Serbanescu, A.; Hinescu, M.E.; et al. Vaccine mRNA Can Be Detected in Blood at 15 Days Post-Vaccination. Biomedicines 2022, 10, 1538. [Google Scholar] [CrossRef]
- Castruita, J.A.S.; Schneider, U.V.; Mollerup, S.; Leineweber, T.D.; Weis, N.; Bukh, J.; Pedersen, M.S.; Westh, H. SARS-CoV-2 spike mRNA vaccine sequences circulate in blood up to 28 days after COVID-19 vaccination. APMIS 2023, 131, 128–132. [Google Scholar] [CrossRef]
- Smith, D.A.; Beaumont, K.; Maurer, T.S.; Di, L. Relevance of Half-Life in Drug Design. J. Med. Chem. 2018, 61, 4273–4282. [Google Scholar] [CrossRef] [PubMed]
- Krauson, A.J.; Casimero, F.V.C.; Siddiquee, Z.; Stone, J.R. Duration of SARS-CoV-2 mRNA vaccine persistence and factors associated with cardiac involvement in recently vaccinated patients. NPJ Vaccines 2023, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Hanna, N.; Heffes-Doon, A.; Lin, X.; Manzano De Mejia, C.; Botros, B.; Gurzenda, E.; Nayak, A. Detection of Messenger RNA COVID-19 Vaccines in Human Breast Milk. JAMA Pediatr. 2022, 176, 1268–1270. [Google Scholar] [CrossRef] [PubMed]
- Hanna, N.; De Mejia, C.M.; Heffes-Doon, A.; Lin, X.; Botros, B.; Gurzenda, E.; Clauss-Pascarelli, C.; Nayak, A. Biodistribution of mRNA COVID-19 vaccines in human breast milk. EBioMedicine 2023, 96, 104800. [Google Scholar] [CrossRef] [PubMed]
- Golan, Y.; Prahl, M.; Cassidy, A.; Lin, C.Y.; Ahituv, N.; Flaherman, V.J.; Gaw, S.L. Evaluation of Messenger RNA from COVID-19 BTN162b2 and mRNA-1273 Vaccines in Human Milk. JAMA Pediatr. 2021, 175, 1069–1071. [Google Scholar] [CrossRef] [PubMed]
- Low, J.M.; Gu, Y.; Ng, M.S.F.; Amin, Z.; Lee, L.Y.; Ng, Y.P.M.; Shunmuganathan, B.D.; Niu, Y.; Gupta, R.; Tambyah, P.A.; et al. Codominant IgG and IgA expression with minimal vaccine mRNA in milk of BNT162b2 vaccinees. NPJ Vaccines 2021, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Anderluzzi, G.; Lou, G.; Woods, S.; Schmidt, S.T.; Gallorini, S.; Brazzoli, M.; Johnson, R.; Roberts, C.W.; O’Hagan, D.T.; Baudner, B.C.; et al. The role of nanoparticle format and route of administration on self-amplifying mRNA vaccine potency. J. Control. Release 2022, 342, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Leal, J.M.; Simpson, A.; Warner, N.L.; Berube, B.J.; Archer, J.F.; Park, S.; Kurtz, R.; Hinkley, T.; Nicholes, K.; et al. A localizing nanocarrier formulation enables multi-target immune responses to multivalent replicating RNA with limited systemic inflammation. Mol. Ther. 2023, 31, 2360–2375. [Google Scholar] [CrossRef]
- Stokes, A.; Pion, J.; Binazon, O.; Laffont, B.; Bigras, M.; Dubois, G.; Blouin, K.; Young, J.K.; Ringenberg, M.A.; Ben Abdeljelil, N.; et al. Nonclinical safety assessment of repeated administration and biodistribution of a novel rabies self-amplifying mRNA vaccine in rats. Regul. Toxicol. Pharmacol. 2020, 113, 104648. [Google Scholar] [CrossRef]
- Maruggi, G.; Mallett, C.P.; Westerbeck, J.W.; Chen, T.; Lofano, G.; Friedrich, K.; Qu, L.; Sun, J.T.; McAuliffe, J.; Kanitkar, A.; et al. A self-amplifying mRNA SARS-CoV-2 vaccine candidate induces safe and robust protective immunity in preclinical models. Mol. Ther. 2022, 30, 1897–1912. [Google Scholar] [CrossRef]
- Keikha, R.; Hashemi-Shahri, S.M.; Jebali, A. The evaluation of novel oral vaccines based on self-amplifying RNA lipid nanparticles (saRNA LNPs), saRNA transfected Lactobacillus plantarum LNPs, and saRNA transfected Lactobacillus plantarum to neutralize SARS-CoV-2 variants alpha and delta. Sci. Rep. 2021, 11, 21308. [Google Scholar] [CrossRef] [PubMed]
- Halasa, N.B.; Olson, S.M.; Staat, M.A.; Newhams, M.M.; Price, A.M.; Boom, J.A.; Sahni, L.C.; Cameron, M.A.; Pannaraj, P.S.; Bline, K.E.; et al. Effectiveness of Maternal Vaccination with mRNA COVID-19 Vaccine During Pregnancy Against COVID-19-Associated Hospitalization in Infants Aged <6 Months-17 States, July 2021–January 2022. MMWR Morb. Mortal. Wkly Rep. 2022, 71, 264–270. [Google Scholar] [PubMed]
- Blakney, A.K.; Ip, S.; Geall, A.J. An Update on Self-Amplifying mRNA Vaccine Development. Vaccines 2021, 9, 97. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pateev, I.; Seregina, K.; Ivanov, R.; Reshetnikov, V. Biodistribution of RNA Vaccines and of Their Products: Evidence from Human and Animal Studies. Biomedicines 2024, 12, 59. https://doi.org/10.3390/biomedicines12010059
Pateev I, Seregina K, Ivanov R, Reshetnikov V. Biodistribution of RNA Vaccines and of Their Products: Evidence from Human and Animal Studies. Biomedicines. 2024; 12(1):59. https://doi.org/10.3390/biomedicines12010059
Chicago/Turabian StylePateev, Ildus, Kristina Seregina, Roman Ivanov, and Vasiliy Reshetnikov. 2024. "Biodistribution of RNA Vaccines and of Their Products: Evidence from Human and Animal Studies" Biomedicines 12, no. 1: 59. https://doi.org/10.3390/biomedicines12010059
APA StylePateev, I., Seregina, K., Ivanov, R., & Reshetnikov, V. (2024). Biodistribution of RNA Vaccines and of Their Products: Evidence from Human and Animal Studies. Biomedicines, 12(1), 59. https://doi.org/10.3390/biomedicines12010059