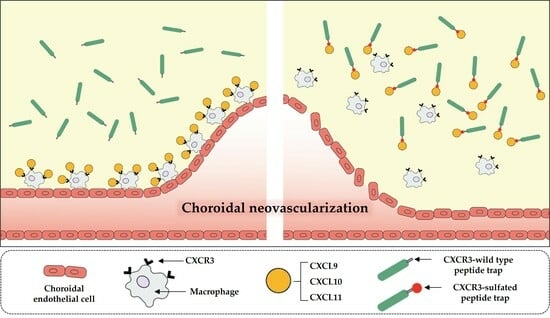
Sulfated CXCR3 Peptide Trap Use as a Promising Therapeutic Approach for Age-Related Macular Degeneration
Abstract
1. Introduction
2. Materials and Methods
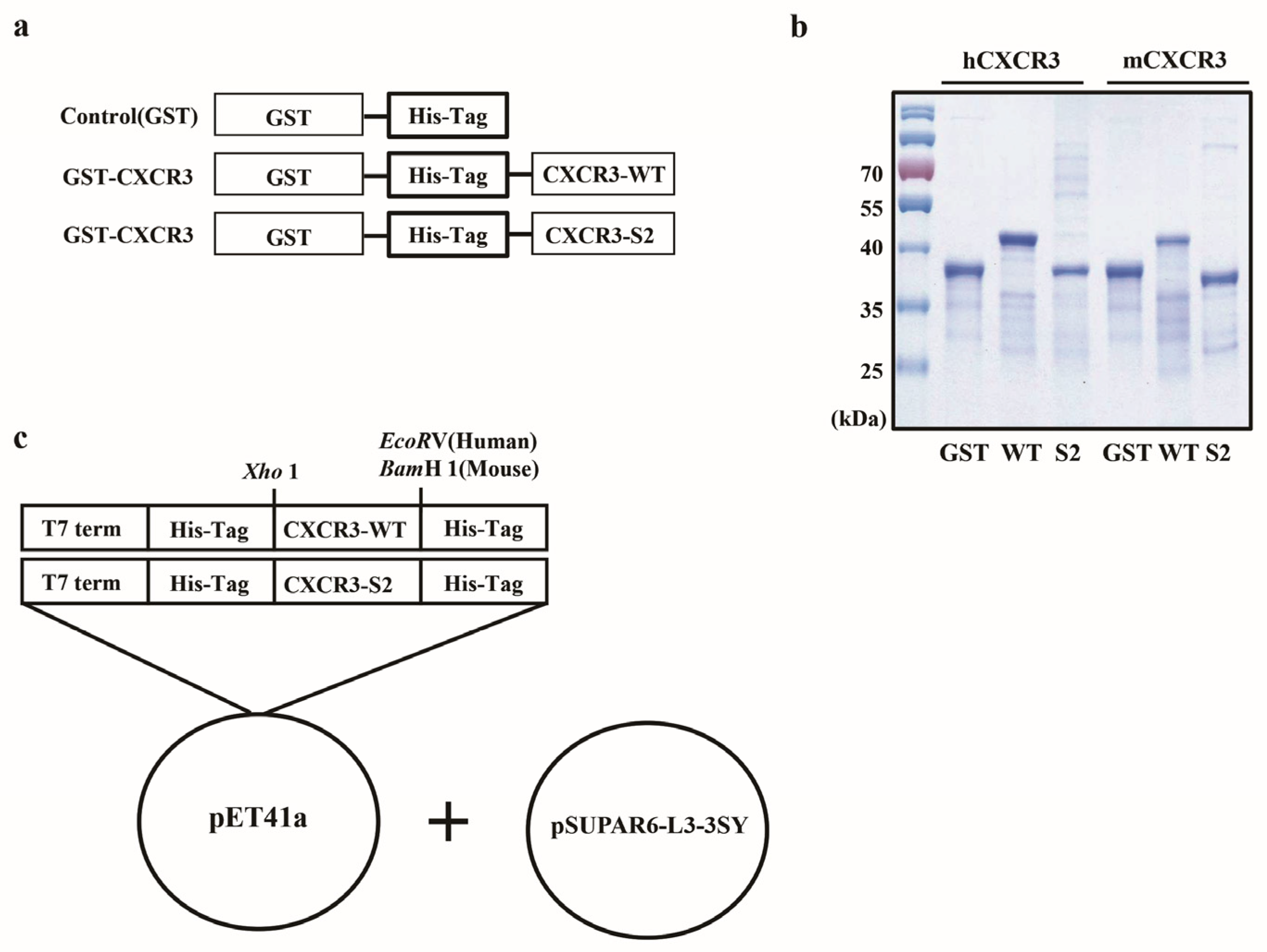
2.1. Plasmid Construction
2.2. Protein Purification
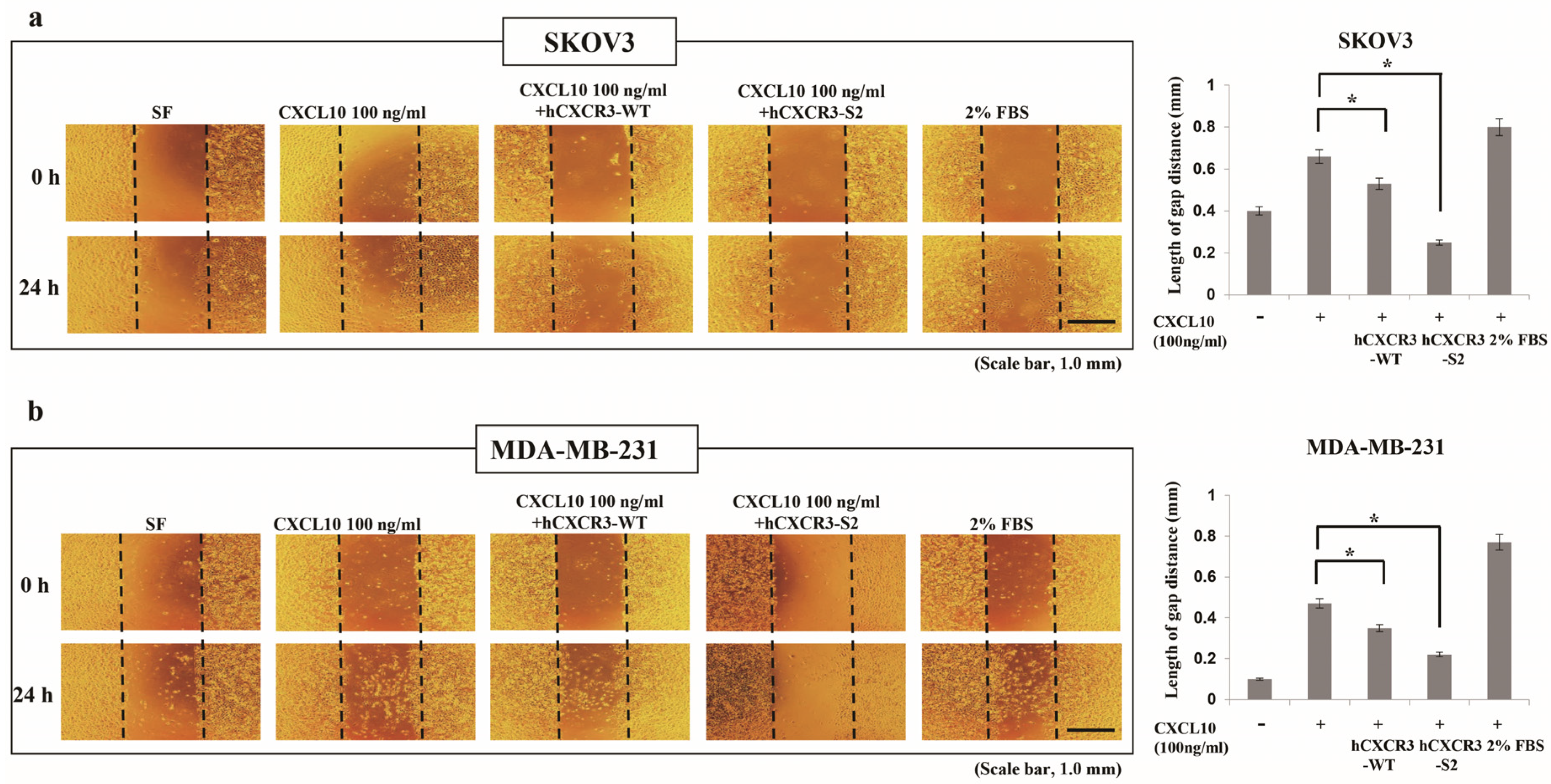
2.3. Wound Healing Assay
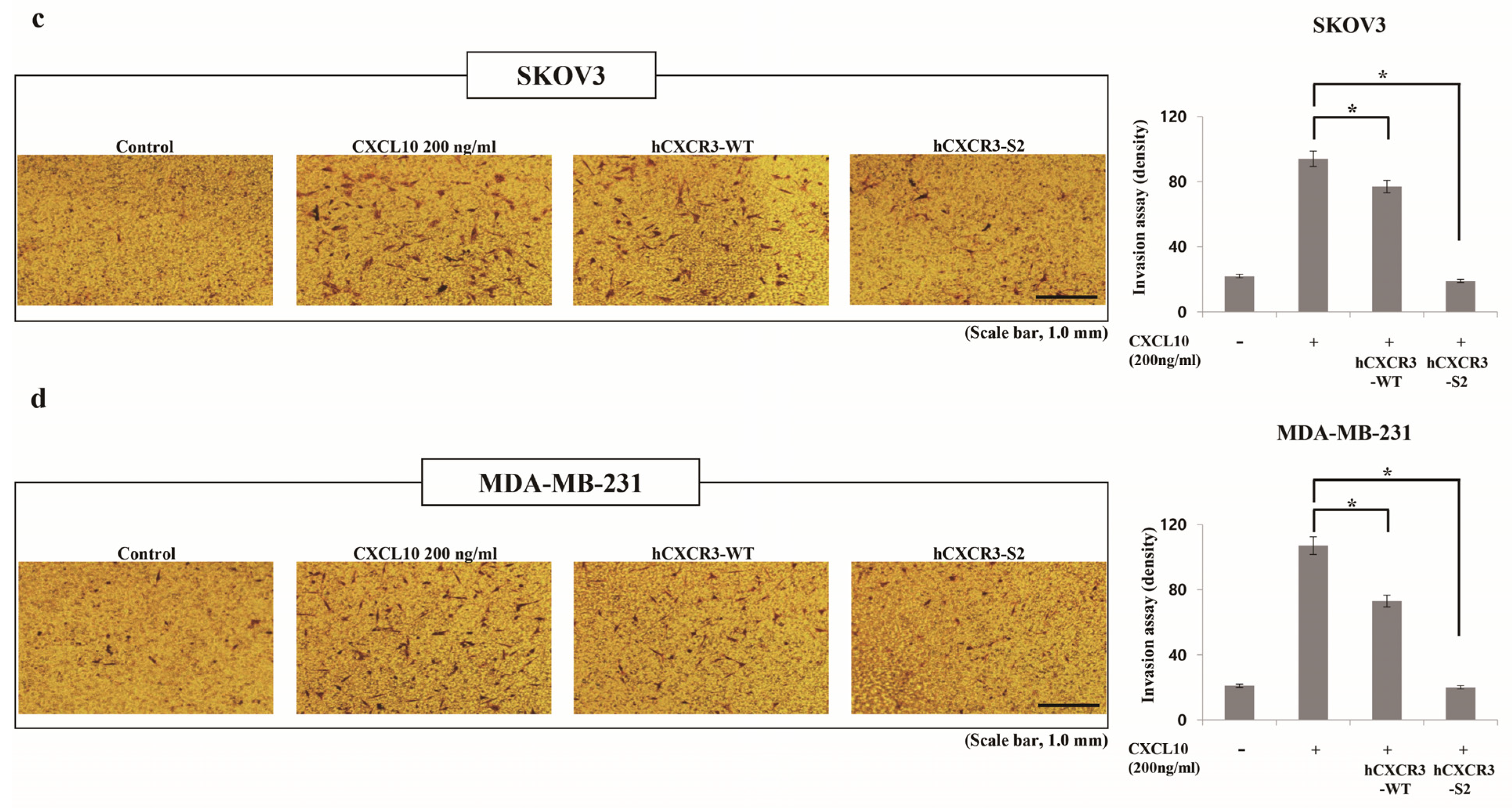
2.4. Invasion Assay
2.5. RAW 264.7 Cell Culture
2.6. Experimental Animals
2.7. Laser-Induced CNV Mouse Models
2.8. Quantitative Reverse Transcriptase-Mediated Real-Time PCR (qPCR)
2.9. Quantitation of CNV in the Mouse Model of Laser-Induced CNV
2.10. Immunofluorescence Staining
2.11. Statistical Analysis
3. Results
3.1. Cloning and Purification of mCXCR3-S2 and hCXCR3-S2
3.2. hCXCR3 Sulfation Attenuated CXCL10-Induced Cell Migration and Invasion
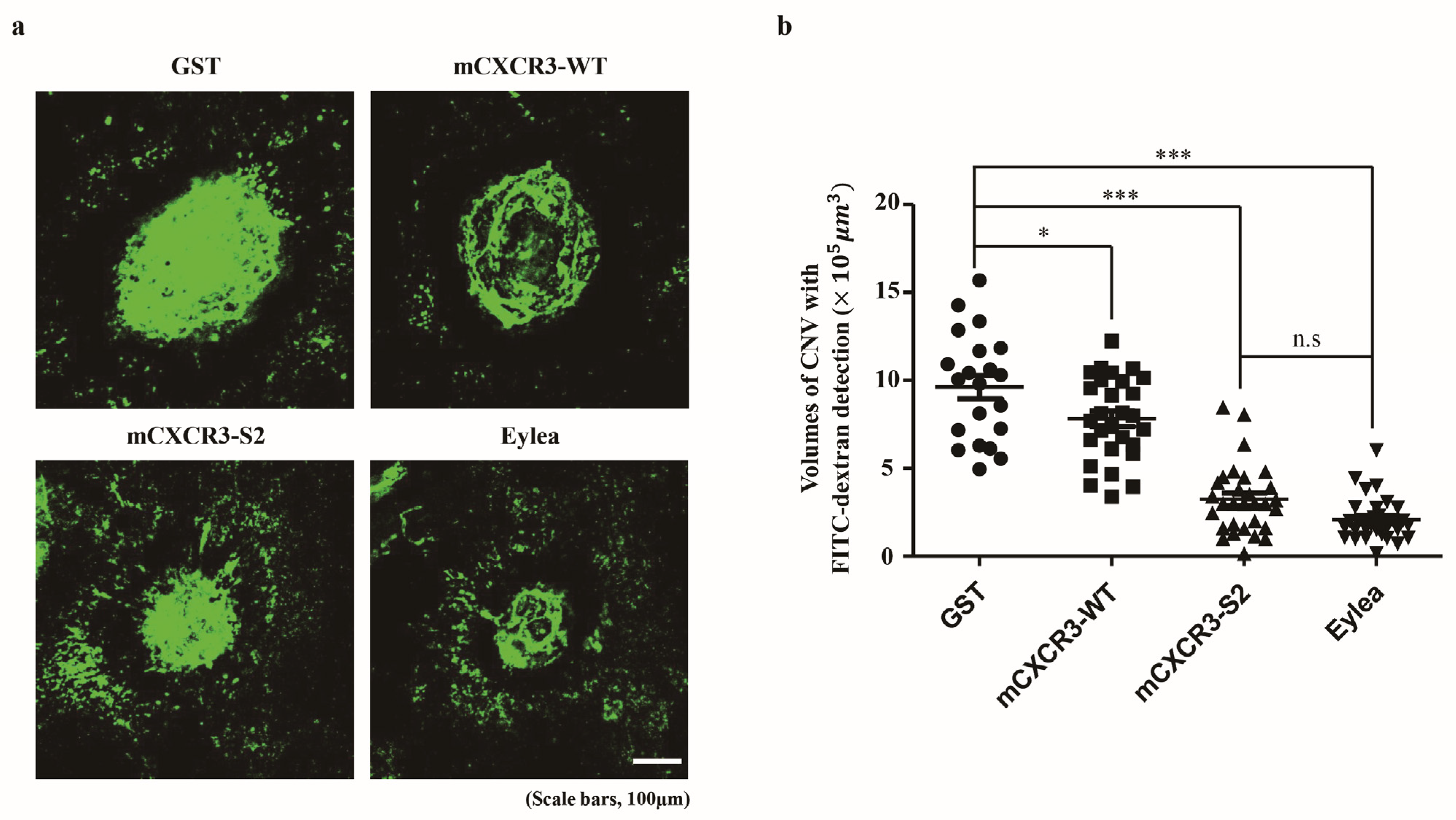
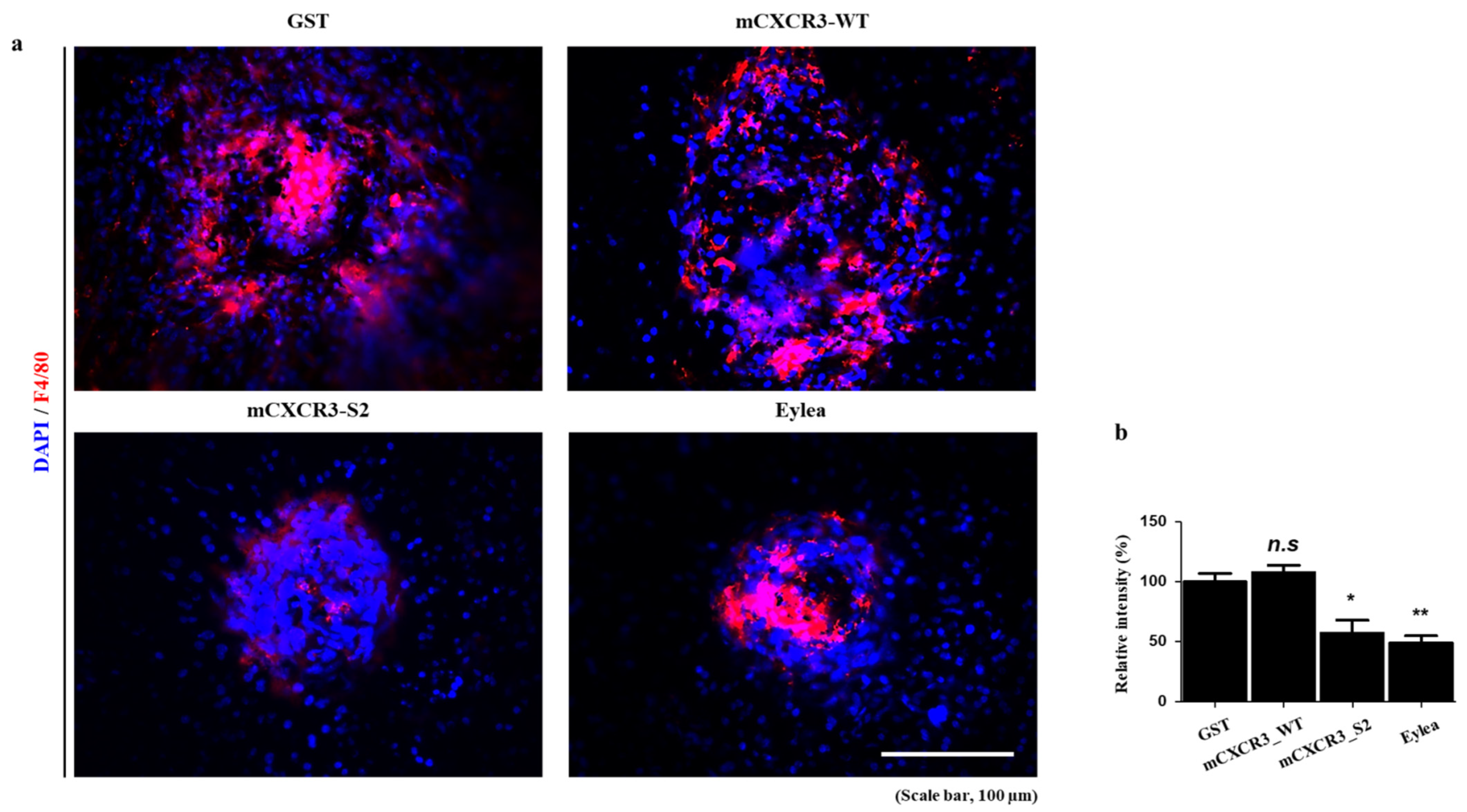
3.3. mCXCR3-S2 Prevented CNV and Macrophage Infiltration in the Laser-Induced CNV Mouse Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ambati, J.; Anand, A.; Fernandez, S.; Sakurai, E.; Lynn, B.C.; Kuziel, W.A.; Rollins, B.J.; Ambati, B.K. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat. Med. 2003, 11, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Ambati, J.; Atkinson, J.; Gelfand, G. Immunology of age-related macular degeneration. Nat. Rev. Immunol. 2013, 13, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Liew, G.; Mitchell, P. Clinical update: New treatments for age-related macular degeneration. Lancet 2007, 9583, s0140–s6736. [Google Scholar] [CrossRef] [PubMed]
- Prasad, P.S.; Schwartz, S.D.; Hubschman, J.P. Age-related macular degeneration: Current and novel therapies. Maturitas 2010, 1, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Igl, I.; Bailey, J.N.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 2, 134–143. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 2, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Webers, A.; Heneka, M.T.; Gleeson, P.A. The role of innate immune responses and neuroinflammation in amyloid accumulation and progression of Alzheimer’s disease. Immunol. Cell Biol. 2020, 1, 28–41. [Google Scholar] [CrossRef]
- Anand, A.; Sharma, N.K.; Gupta, A.; Prabhakar, S.; Sharma, S.K.; Singh, R.; Gupta, P.K. Single nucleotide polymorphisms in MCP-1 and its receptor are associated with the risk of age related macular degeneration. PLoS ONE 2012, 7, e49905. [Google Scholar] [CrossRef]
- Vandercappellen, J.; Van Damme, J.; Struyf, S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008, 2, 226–244. [Google Scholar] [CrossRef]
- Satarkar, D.; Patra, C. Evolution, Expression and Functional Analysis of CXCR3 in Neuronal and Cardiovascular Diseases: A Narrative Review. Front. Cell Dev. Biol. 2022, 20, 882017. [Google Scholar] [CrossRef]
- Loetscher, M.; Gerber, B.; Loetscher, P.; Jones, S.A.; Piali, L.; Clark, L.I. Chemokine Receptor Specific for IP10 and Mig: Structure, Function, and Expression in Activated T-Lymphocytes. J. Exp. Med. 1996, 184, 963–969. [Google Scholar] [CrossRef]
- Lasagni, L.; Francalanci, M.; Annunziato, F.; Lazzeri, E.; Giannini, S.; Cosmi, L. An Alternatively Spliced Variant of CXCR3 Mediates the Inhibition of Endothelial Cell Growth Induced by IP-10, Mig, and I-TAC, and Acts as Functional Receptor for Platelet Factor 4. J. Exp. Med. 2003, 197, 1537–1549. [Google Scholar] [CrossRef]
- Colvin, R.A.; Campanella, G.S.V.; Manice, L.A.; Luster, A.D. CXCR3 Requires Tyrosine Sulfation for Ligand Binding and a Second Extracellular Loop Arginine Residue for Ligand-Induced Chemotaxis. Mol. Cell. Biol. 2006, 26, 5838–5849. [Google Scholar] [CrossRef]
- Mo, F.M.; Proia, A.D.; Johnson, W.H.; Cyr, D.; Lashkari, K. Interferon gamma-inducible protein-10 (IP-10) and eotaxin as biomarkers in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2010, 8, 4226–4236. [Google Scholar] [CrossRef] [PubMed]
- Sakurada, Y.; Nakamura, Y.; Yoneyama, S.; Mabuchi, F.; Gotoh, T.; Tateno, Y.; Sugiyama, A.; Kubota, T.; Iijima, H. Aqueous humor cytokine levels in patients with polypoidal choroidal vasculopathy and neovascular age-related macular degeneration. Ophthalmic Res. 2015, 1, 2–7. [Google Scholar] [CrossRef]
- Fujimura, S.; Takahashi, H.; Yuda, K.; Ueta, T.; Iriyama, A.; Inoue, T.; Kaburaki, T.; Tamaki, Y.; Matsushima, K.; Yanagi, Y. Angiostatic effect of CXCR3 expressed on choroidal neovascularization. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1999–2006. [Google Scholar] [CrossRef]
- Liu, M.; Guo, S.; Hibbert, J.M.; Jain, V.; Singh, N.; Wilson, N.O.; Stiles, J.K. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor. Rev. 2011, 22, 121–130. [Google Scholar] [CrossRef]
- Veldkamp, C.T.; Seibert, C.; Peterson, F.C.; De la Cruz, N.B.; Haugner, J.C.; Basnet, H.; Sakmar, T.P.; Volkman, B.F. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci. Signal. 2008, 37, ra4. [Google Scholar] [CrossRef]
- Zhu, J.Z.; Millard, C.J.; Ludeman, J.P.; Simpson, L.S.; Clayton, D.J.; Payne, R.J.; Widlanski, T.S.; Stone, M.J. Tyrosine sulfation influences the chemokine binding selectivity of peptides derived from chemokine receptor CCR3. Biochemistry 2011, 9, 1524–1534. [Google Scholar] [CrossRef]
- Stemmer, W.P.; Crameri, A.; Ha, K.D.; Brennan, T.M.; Heyneker, H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 1995, 164, 49–53. [Google Scholar] [CrossRef]
- Liu, C.C.; Cellitti, S.E.; Geierstanger, B.H.; Schultz, P.G. Efficient expression of tyrosine-sulfated proteins in E. coli using an expanded genetic code. Nat. Protoc. 2009, 12, 1784–1789. [Google Scholar] [CrossRef]
- Choi, J.; Ahn, S.S.; Lim, Y.; Lee, Y.H.; Shin, S.Y. Inhibitory effect of Alisma canaliculatum ethanolic extract on NF-Κb-dependent CXCR3 and CXCL10 expression in TNFα-exposed MDA-MB-231 breast cancer cells. Int. J. Mol. Sci. 2018, 9, 2607. [Google Scholar] [CrossRef]
- Wu, Z.; Han, X.; Yan, J.; Pan, Y.; Gong, J.; Di, J.; Cheng, Z.; Jin, Z.; Wang, Z.; Zheng, Q.; et al. The prognostic significance of chemokine receptor CXCR3 expression in colorectal carcinoma. Biomed. Pharmacother. 2012, 5, 373–377. [Google Scholar] [CrossRef]
- Windmüller, C.; Zech, D.; Avril, S.; Boxberg, M.; Dawidek, T.; Schmalfeldt, B.; Schmitt, M.; Kiechle, M.; Bronger, H. CXCR3 mediates ascites-directed tumor cell migration and predicts poor outcome in ovarian cancer patients. Oncogenesis 2017, 5, e331. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Wang, S.; Ni, H.; Zhao, P.; Chen, G.; Xu, B.; Yuan, L. The role of CXCR3 and its ligands in cancer. Front. Oncol. 2022, 12, 1022688. [Google Scholar] [CrossRef]
- Vincenzo, T.; Chao, C.; Ralf, B.; Jan, P.B.; Astrid, M.S.; Martine, J.S.; Marco, S.; Herman, P.S.; Annemarie, H.M. The CXCR3-CXCL10 signaling axis mediates macrophage recruitment and dis semination of mycobacterial infection. Dis. Model. Mech. 2015, 8, 253–269. [Google Scholar]
- Xin, J.L.; Qiang, C.; Ye, J.R.; Feng, C.; Jiong, C. CXCR3.1 and CXCR3.2 Differentially Contribute to Macrophage Polarization in Teleost Fish. J. Immunol. 2017, 198, 4692–4706. [Google Scholar] [CrossRef]
- Tatsuya, H.; Venkata, S.V.; Yoshio, Y.; Masato, N.; Shigeto, S.; Shigeki, S.; Satoru, Y.; Hideki, K.; Itaru, M.; Yuichiro, N.; et al. Inhibition of the CXCL9-CXCR3 axis suppresses the progression of experimental apical periodontitis by blocking macrophage migration and activation. Sci. Rep. 2021, 11, 2613. [Google Scholar] [CrossRef]
- Monteagudo, C.; Martin, J.M.; Jorda, E.; Llombart-Bosch, A. CXCR3 chemokinereceptor immunoreactivity in primary cutaneous malignant melanoma: Correlationwith clinicopathological prognostic factors. J. Clin. Pathol. 2007, 60, 596–599. [Google Scholar] [CrossRef]
- Kawada, K.; Hosogi, H.; Sonoshita, M.; Sakashita, H.; Manabe, T.; Shimahara, Y.; Sakai, Y.; Takabayashi, A.; Oshima, M.; Taketo, M.M. Chemokine receptor CXCR3 promotes colon cancer metastasis to lymphnodes. Oncogene 2007, 26, 4679–4688. [Google Scholar] [CrossRef]
- Ma, X.; Norsworthy, K.; Kundu, N.; Rodgers, W.H.; Gimotty, P.A.; Goloubeva, O.; Lipsky, M.; Li, Y.; Holt, D.; Fulton, A. CXCR3 expression is associated with poor survival in breast cancer andpromotes metastasis in a murine model. Mol. Cancer Ther. 2009, 8, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Seibert, C.; Peterson, F.C.; Sakmar, T.P.; Volkman, B.F. Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1(SDF-1/CXCL12). J. Mol. Biol. 2006, 359, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Duma, L.; Häussinger, D.; Rogowski, M.; Lusso, P.; Grzesiek, S. Recognition of RANTES by extracellular parts of the CCR5 receptor. J. Mol. Biol. 2007, 365, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.S.; Zhu, J.Z.; Widlanski, T.S.; Stone, M.J. Regulation of chemokine recognition by site-specific tyrosine sulfation of receptor peptides. Chem. Biol. 2009, 16, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.; Veldkamp, C.T.; Peterson, F.C.; Chait, B.T.; Volkman, B.F.; Sakmar, T.P. Sequential tyrosine sulfation of CXCR4 by tyrosylprotein sulfotransferases. Biochemistry 2008, 47, 11251–11262. [Google Scholar] [CrossRef]
- Gao, J.M.; Xiang, R.L.; Jiang, L.; Li, W.H.; Feng, Q.P.; Guo, Z.J.; Sun, Q.; Zeng, Z.P.; Fang, F.D. Sulfated tyrosines 27 and 29 in the N-terminus of human CXCR3 participate in binding native IP-10. Acta Pharmacol. Sin. 2009, 30, 193–201. [Google Scholar] [CrossRef]
- Denoyer, A.; Godefroy, D.; Celerier, I.; Frugier, J.; Degardin, J.; Harrison, J.K.; Brignole-Baudouin, F.; Picaud, S.; Baleux, F.; Sahel, J.A.; et al. CXCR3 antagonism of SDF-1(5-67) restores trabecular function and prevents retinal neurodegeneration in a rat model of ocular hypertension. PLoS ONE 2012, 7, e37873. [Google Scholar] [CrossRef]
- Ludeman, J.P.; Stone, M.J. The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 2014, 5, 1167–1179. [Google Scholar] [CrossRef]
- Millard, C.J.; Ludeman, J.P.; Canals, M.; Bridgford, J.L.; Hinds, M.G.; Clayton, D.J.; Christopoulos, A.; Payne, R.J.; Stone, M.J. Structural basis of receptor sulfotyrosine recognition by a CC chemokine: The N-terminal region of CCR3 bound to CCL11/eotaxin-1. Structure 2014, 11, 1571–1581. [Google Scholar] [CrossRef]
- Espinosa-Heidmann, D.G.; Suner, I.J.; Hernandez, E.P.; Monroy, D.; Csaky, K.G.; Cousins, S.W. Macrophage depletion diminishes lesion size and severity in experimental choroidal neovascularization. Investig. Ophthalmol. Vis. Sci. 2003, 8, 3586–3592. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, F.; Tang, M.; Yuan, M.; Hu, A.; Zhan, Z.; Li, Z.; Li, J.; Ding, X.; Lu, L. Macrophage polarization in experimental and clinical choroidal neovascularization. Sci. Rep. 2016, 6, 30933. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Takagi, H.; Takagi, C.; Suzuma, K.; Otani, A.; Ishida, K.; Matsumura, M.; Ogura, Y.; Honda, Y. The potential angiogenic role of macrophages in the formation of choroidal neovascular membranes. Investig. Ophthalmol. Vis. Sci. 1999, 9, 1891–1898. [Google Scholar]
- Yang, X.; Zhao, L.; Campos, M.; Abu-Asab, M.; Ortolan, D.; Hotaling, N.; Bharti, K.; Wong, W.T. Choroid-resident macrophages maintain local vasculature and RPE integrity and spontaneously regenerate following depletion. bioRxiv 2019. [Google Scholar] [CrossRef]
- Xuan, W.; Qu, Q.; Zheng, B.; Xiong, S.; Fan, G.H. The chemotaxis of M1 and M2 macrophages is regulated by different chemokines. J. Leukoc. Biol. 2015, 1, 61–69. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, G.; Chae, J.-B.; Jung, S.-A.; Lyu, J.; Chung, H.; Lee, J.H. Sulfated CXCR3 Peptide Trap Use as a Promising Therapeutic Approach for Age-Related Macular Degeneration. Biomedicines 2024, 12, 241. https://doi.org/10.3390/biomedicines12010241
Jo G, Chae J-B, Jung S-A, Lyu J, Chung H, Lee JH. Sulfated CXCR3 Peptide Trap Use as a Promising Therapeutic Approach for Age-Related Macular Degeneration. Biomedicines. 2024; 12(1):241. https://doi.org/10.3390/biomedicines12010241
Chicago/Turabian StyleJo, Gukheui, Jae-Byoung Chae, Sun-Ah Jung, Jungmook Lyu, Hyewon Chung, and Joon H. Lee. 2024. "Sulfated CXCR3 Peptide Trap Use as a Promising Therapeutic Approach for Age-Related Macular Degeneration" Biomedicines 12, no. 1: 241. https://doi.org/10.3390/biomedicines12010241
APA StyleJo, G., Chae, J.-B., Jung, S.-A., Lyu, J., Chung, H., & Lee, J. H. (2024). Sulfated CXCR3 Peptide Trap Use as a Promising Therapeutic Approach for Age-Related Macular Degeneration. Biomedicines, 12(1), 241. https://doi.org/10.3390/biomedicines12010241