An Analysis of JADE2 in Non-Small Cell Lung Cancer (NSCLC)
Abstract
1. Introduction
2. Materials and Methods
2.1. Nomenclature
2.2. Cell Culture
2.3. Primary Tumor Samples
2.4. Formalin Fixed Paraffin Embedded Samples
2.5. Immunohistochemistry
2.6. RNA Isolation, RT-PCR and qPCR Amplification
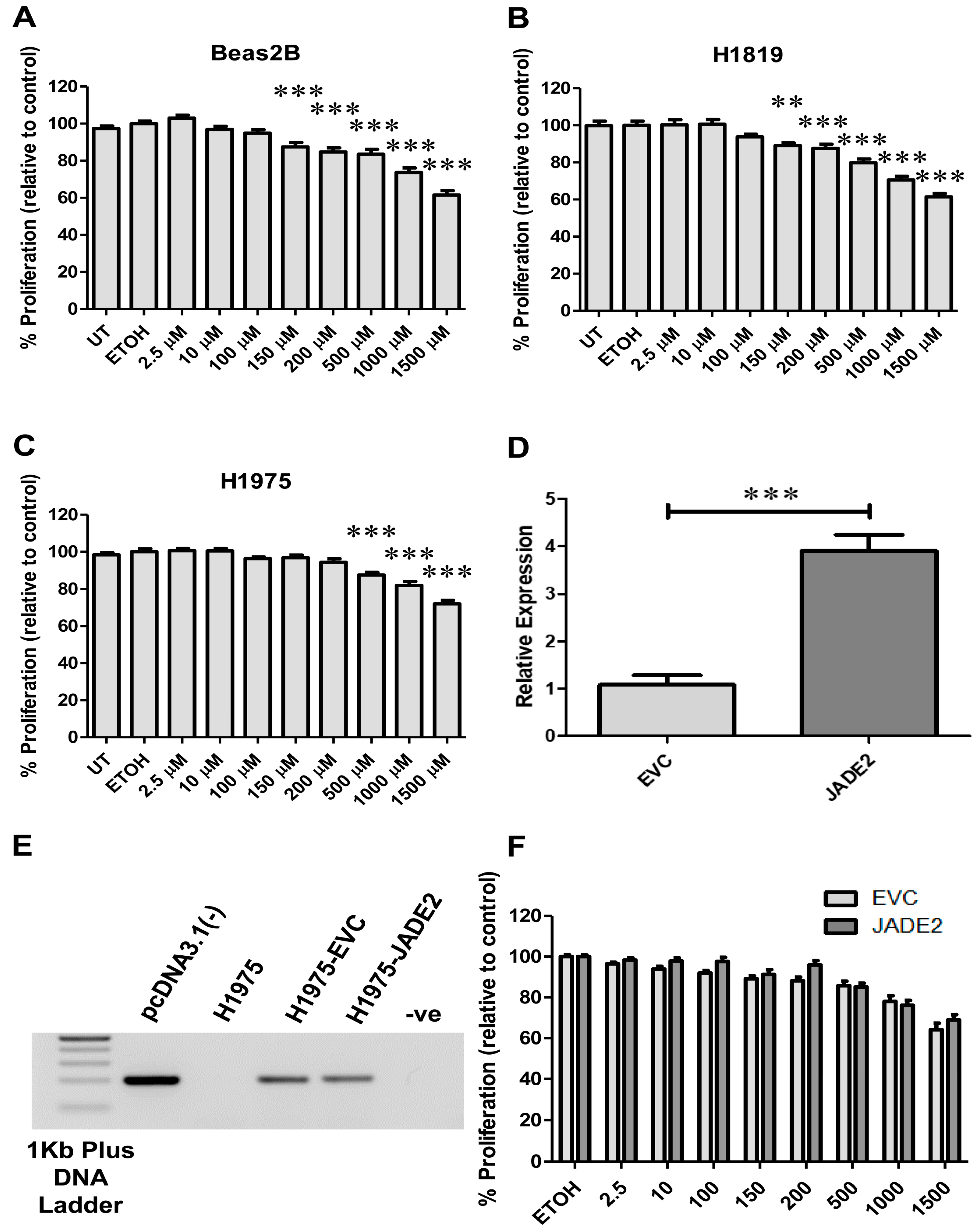
2.7. JADE2 Overexpression Studies
2.8. Validations of Expression Differences for JADE2 in NSCLC
2.9. Survival Analysis for JADE2
2.10. Associations with Key NSCLC Genes, Oncogenic Driver Mutations and Novel Mutated Genes
2.11. Correlations between JADE2 Expression, Tumor Mutational Burden and Immune Infiltrations in NSCLC
2.12. Correlations between JADE2 Expression and Anti-Cancer Drug Sensitivity in NSCLC Cell Lines
2.13. Drug Treatment and Cellular Viability Assays
2.14. Statistical Analysis
- TIMER: https://cistrome.shinyapps.io/timer/ (accessed on 19 August 2022)
- TIMER2.0: http://timer.cistrome.org/ (accessed on 24 August 2022)
- GEPIA2.0: http://gepia2.cancer-pku.cn/#index (accessed on 29 July 2022)
- LCE: http://lce.biohpc.swmed.edu/lungcancer/ (accessed on 22 August 2022)
- KM-PLOT: https://kmplot.com/analysis/index.php?p=background (accessed on 30 August 2022)
- cBioPortal: https://www.cbioportal.org/ (accessed on 25 August 2022)
- muTarget: https://www.mutarget.com/ (accessed on 23 August 2022)
- DepMap: https://depmap.org/portal/ (accessed on 23 August 2022)
- cProSite: https://cprosite.ccr.cancer.gov/#/ (accessed on 13 July 2022)
- PROGgeneV2: http://www.progtools.net/gene/ (accessed on 29 August 2022)
3. Results
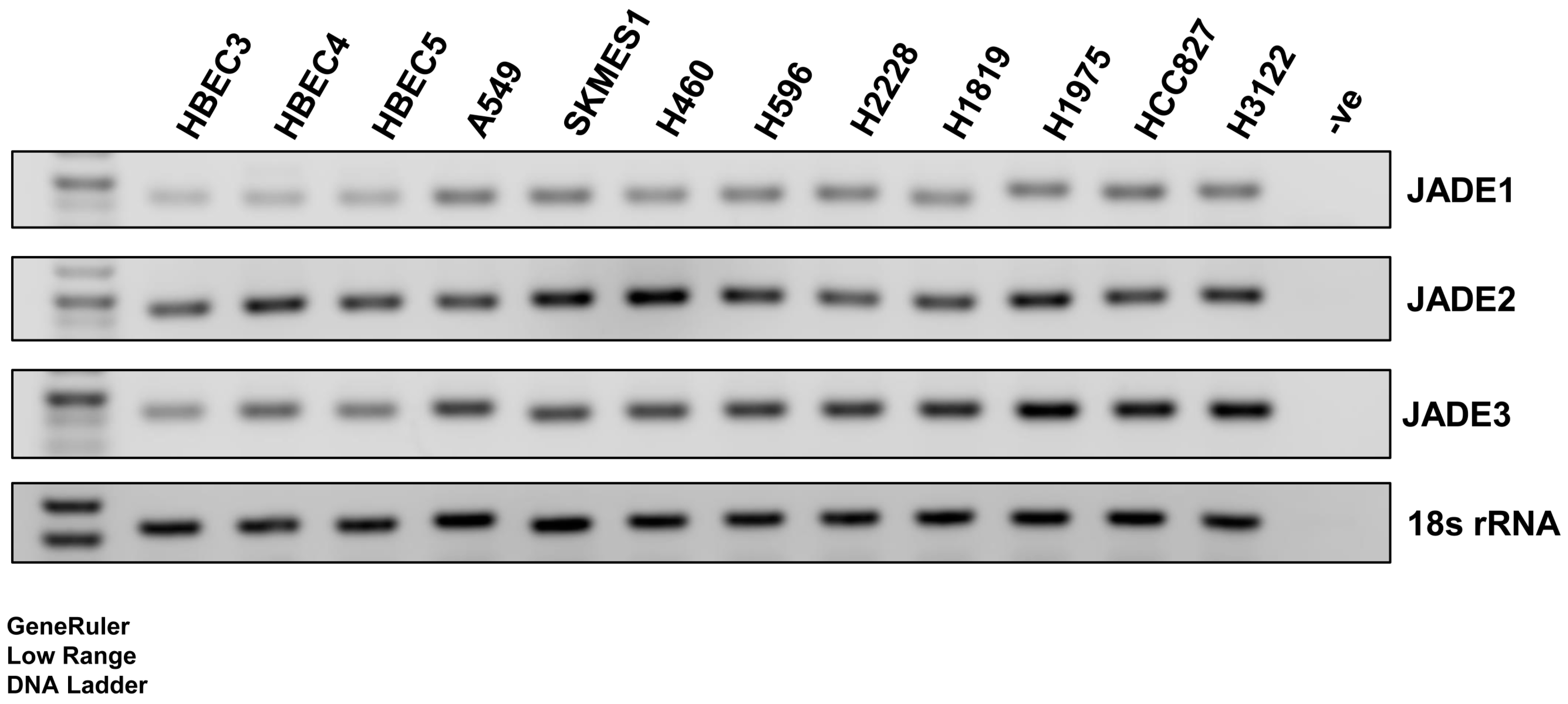
3.1. Expression of JADE2 in a Panel of Normal Lung and NSCLC Cell Lines
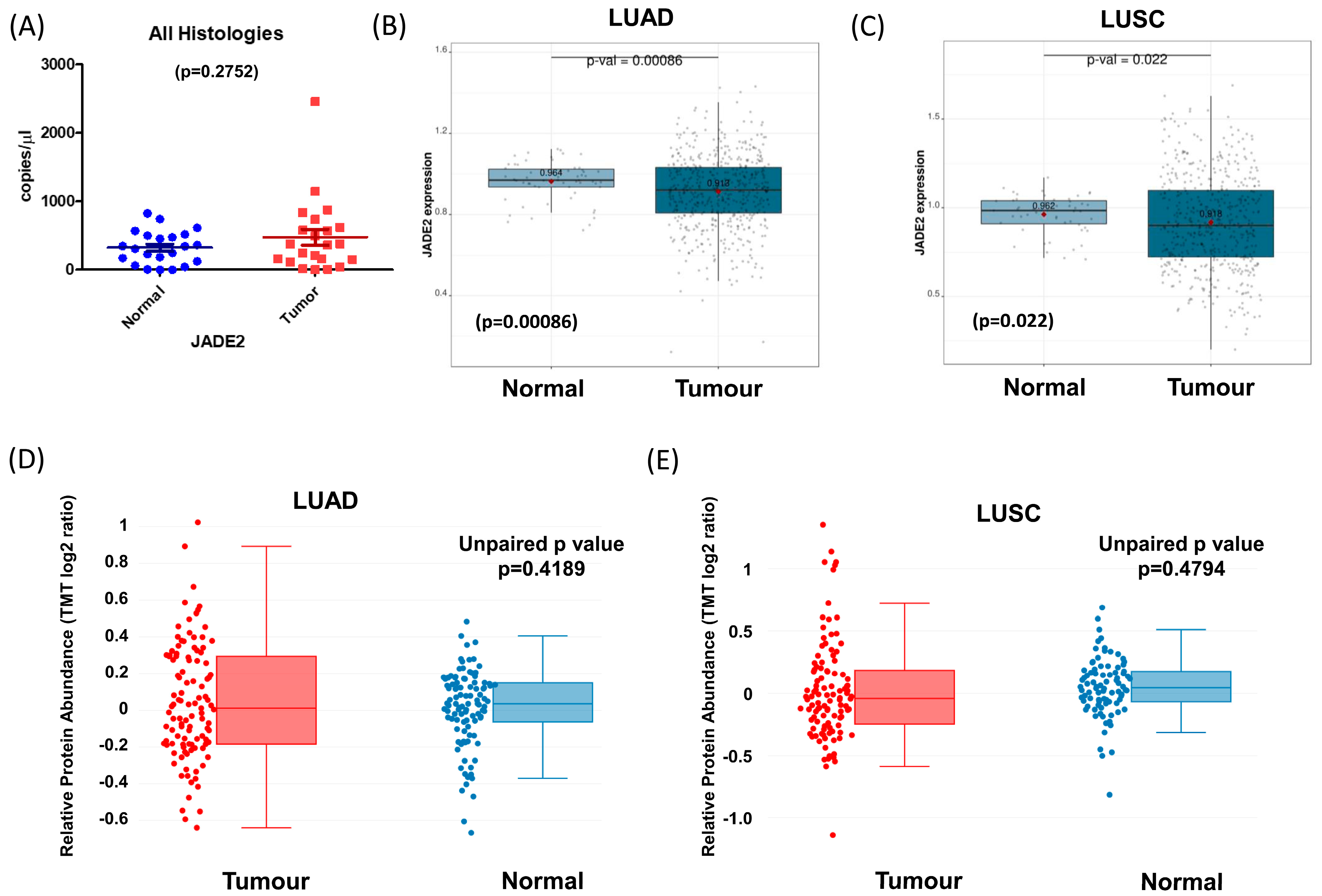
3.2. Expression of JADE2 in Primary NSCLC
3.3. Potential Prognostic Value of JADE2 Protein in NSCLC
3.4. Correlations between JADE2 Expression and Key Genes Associated with NSCLC
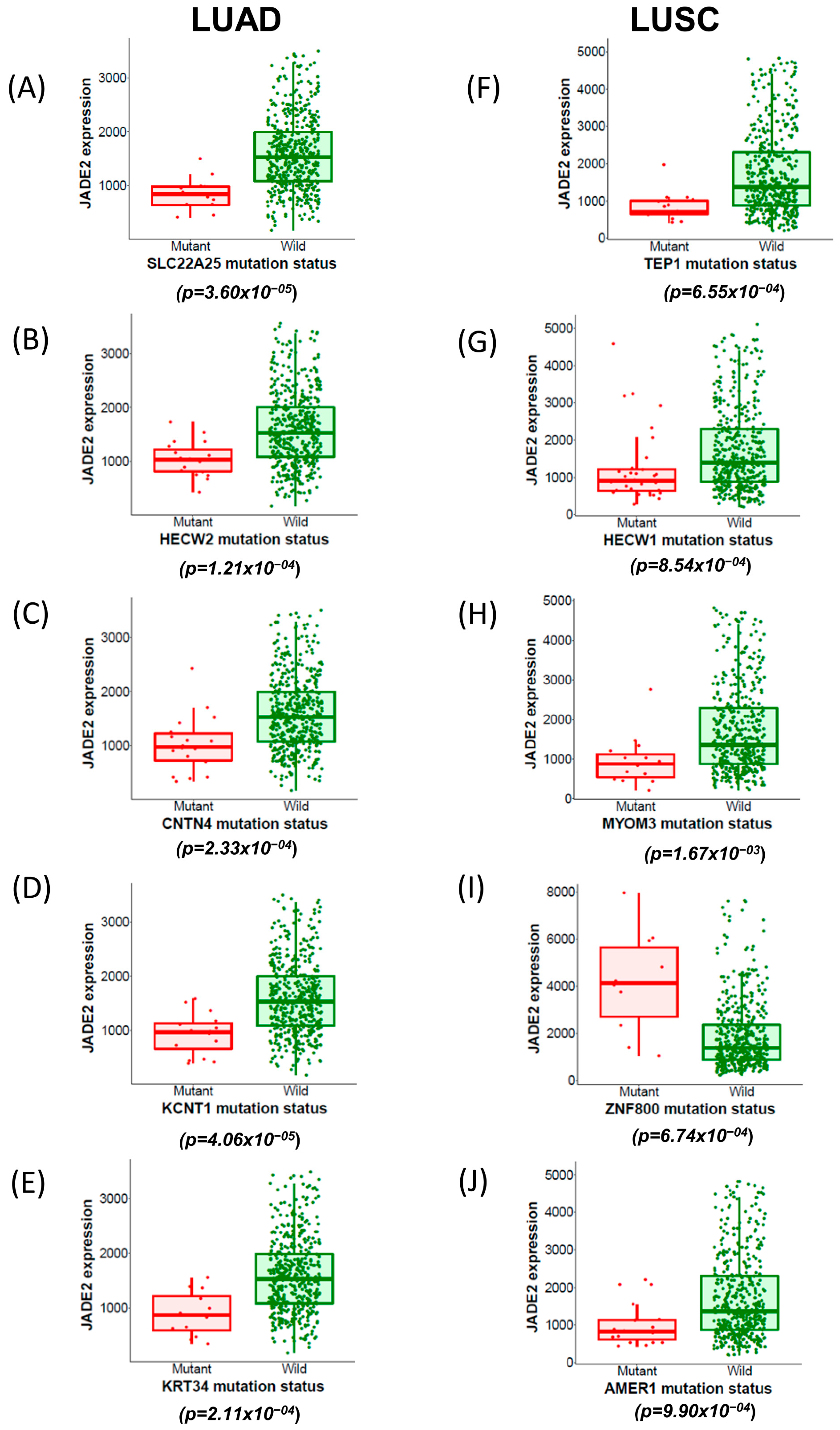
3.5. Correlations between Novel Mutated Genes and JADE2 mRNA Expression
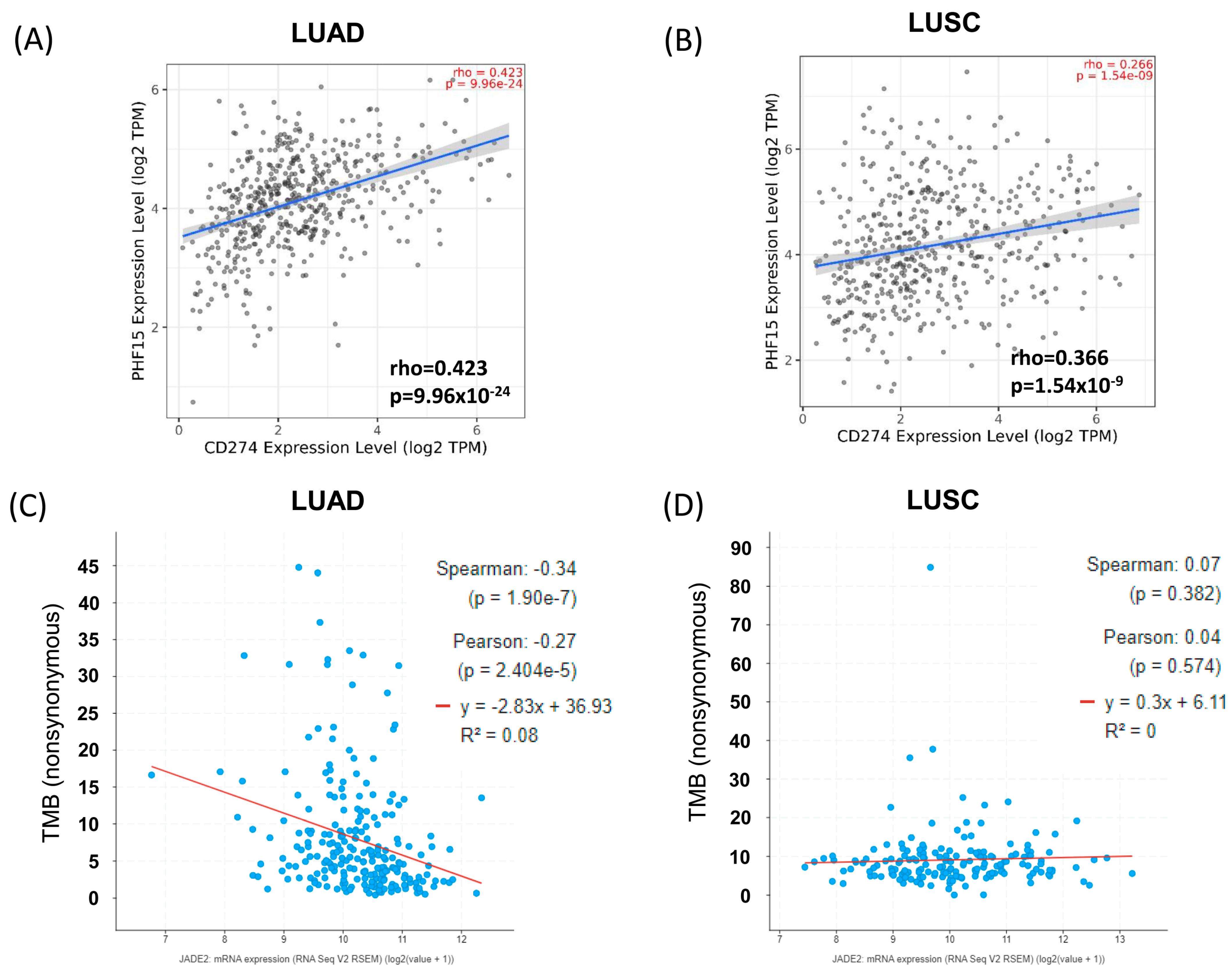
3.6. Correlation Analysis between JADE2 Expression and Tumor Mutational Burden
3.7. Effects of JADE2 mRNA Expression on Immune Cell Infiltration
3.8. Correlation Analysis between JADE2 Expression and Immune Cell Exhaustion
3.9. Identifying Compounds That Can Potentially Target JADE2 in NSCLC
4. Discussion
5. Conclusions
- Knockdown or CRIPSR-based editing of JADE2 expression. In addition, it is potentially conceivable that redundancy between other JADE family members could substitute for JADE2, so studies on JADE1 or JADE3 will also be necessary.
- In vivo animal studies will be required to assess if overexpression of JADE2 affects NSCLC tumourigenicity/aggressiveness.
- Given the limited effects of ornidazole when tested as a “stand-alone” agent, combinatorial treatments of standard NSCLC therapies with ornidazole are warranted to assess if this agent can either enhance therapy or resensitize resistant cells to therapy, or affect the expression of checkpoint inhibitor targets.
- Studies to assess whether ornidazole can affect cancer stem cell populations in NSCLC are warranted given the data emerging from melanoma [71]. In this regard, it may be possible to test this in a panel of isogenic parent/cisplatin-resistant cell lines which we have previously generated and shown to be enriched for CD133+ cells [76].
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Punekar, S.R.; Shum, E.; Grello, C.M.; Lau, S.C.; Velcheti, V. Immunotherapy in non-small cell lung cancer: Past, present, and future directions. Front. Oncol. 2022, 12, 877594. [Google Scholar] [CrossRef] [PubMed]
- Michelotti, A.; de Scordilli, M.; Bertoli, E.; De Carlo, E.; Del Conte, A.; Bearz, A. NSCLC as the Paradigm of Precision Medicine at Its Finest: The Rise of New Druggable Molecular Targets for Advanced Disease. Int. J. Mol. Sci. 2022, 23, 6748. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Tan, D.S.W. Targeted Therapies for Lung Cancer Patients With Oncogenic Driver Molecular Alterations. J. Clin. Oncol. 2022, 40, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Ansari, J.; Shackelford, R.E.; El-Osta, H. Epigenetics in non-small cell lung cancer: From basics to therapeutics. Transl. Lung Cancer Res. 2016, 5, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Darwiche, N. Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. Am. J. Cancer Res. 2020, 10, 1954–1978. [Google Scholar]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Zhao, S.; Allis, C.D.; Wang, G.G. The language of chromatin modification in human cancers. Nat. Rev. Cancer 2021, 21, 413–430. [Google Scholar] [CrossRef]
- Jain, K.; Fraser, C.S.; Marunde, M.R.; Parker, M.M.; Sagum, C.; Burg, J.M.; Hall, N.; Popova, I.K.; Rodriguez, K.L.; Vaidya, A.; et al. Characterization of the plant homeodomain (PHD) reader family for their histone tail interactions. Epigenetics. Chromatin. 2020, 13, 3. [Google Scholar] [CrossRef]
- Shah, M.A.; Denton, E.L.; Liu, L.; Schapira, M. ChromoHub V2: Cancer genomics. Bioinformatics 2014, 30, 590–592. [Google Scholar] [CrossRef][Green Version]
- Panchenko, M.V. Structure, function and regulation of jade family PHD finger 1 (JADE1). Gene 2016, 589, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.I.; Wang, H.; Ross, J.J.; Kuzmin, I.; Xu, C.; Cohen, H.T. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J. Biol. Chem. 2002, 277, 39887–39898. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Panchenko, M.V.; Zhou, M.I.; Cohen, H.T. von Hippel-Lindau partner Jade-1 is a transcriptional co-activator associated with histone acetyltransferase activity. J. Biol. Chem. 2004, 279, 56032–56041. [Google Scholar] [CrossRef]
- Foy, R.L.; Song, I.Y.; Chitalia, V.C.; Cohen, H.T.; Saksouk, N.; Cayrou, C.; Vaziri, C.; Côté, J.; Panchenko, M.V. Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J. Biol. Chem. 2008, 283, 28817–28826. [Google Scholar] [CrossRef]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2006, 21, 51–64. [Google Scholar] [CrossRef]
- Han, J.; Lachance, C.; Ricketts, M.D.; McCullough, C.E.; Gerace, M.; Black, B.E.; Côté, J.; Marmorstein, R. The scaffolding protein JADE1 physically links the acetyltransferase subunit HBO1 with its histone H3-H4 substrate. J. Biol. Chem. 2018, 293, 4498–4509. [Google Scholar] [CrossRef]
- Lan, R.; Wang, Q. Deciphering structure, function and mechanism of lysine acetyltransferase HBO1 in protein acetylation, transcription regulation, DNA replication and its oncogenic properties in cancer. Cell Mol. Life Sci. 2020, 77, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Siriwardana, N.S.; Meyer, R.; Havasi, A.; Dominguez, I.; Panchenko, M.V. Cell cycle-dependent chromatin shuttling of HBO1-JADE1 histone acetyl transferase (HAT) complex. Cell Cycle 2014, 13, 1885–1901. [Google Scholar] [CrossRef]
- Calvi, B.R. HBO1:JADE1 at the cell cycle chromatin crossroads. Cell Cycle 2014, 13, 2322. [Google Scholar] [CrossRef][Green Version]
- Han, X.; Gui, B.; Xiong, C.; Zhao, L.; Liang, J.; Sun, L.; Yang, X.; Yu, W.; Si, W.; Yan, R.; et al. Destabilizing LSD1 by Jade-2 promotes neurogenesis: An antibraking system in neural development. Mol. Cell 2014, 55, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Liu, Y.; Shang, Y.; Xue, Y.; Liang, J.; Huang, Z. JADE2 Is Essential for Hippocampal Synaptic Plasticity and Cognitive Functions in Mice. Biol. Psychiatry 2022, 92, 800–814. [Google Scholar] [CrossRef] [PubMed]
- Anan, K.; Hino, S.; Shimizu, N.; Sakamoto, A.; Nagaoka, K.; Takase, R.; Kohrogi, K.; Araki, H.; Hino, Y.; Usuki, S.; et al. LSD1 mediates metabolic reprogramming by glucocorticoids during myogenic differentiation. Nucleic Acids Res. 2018, 46, 5441–5454. [Google Scholar] [CrossRef] [PubMed]
- Saksouk, N.; Avvakumov, N.; Champagne, K.S.; Hung, T.; Doyon, Y.; Cayrou, C.; Paquet, E.; Ullah, M.; Landry, A.J.; Côté, V.; et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 2009, 33, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Duan, X.; Wu, X.; Li, C.; Chen, S.; Wang, S.; Cai, Y.; Weng, Z. Expression and clinical significance of von Hippel-Lindau downstream genes: Jade-1 and β-catenin related to renal cell carcinoma. Urology 2012, 80, 485.e7–485.e13. [Google Scholar] [CrossRef]
- Shafique, S.; Rashid, S. Structural basis for renal cancer by the dynamics of pVHL-dependent JADE1 stabilization and β-catenin regulation. Prog. Biophys Mol. Biol. 2019, 145, 65–77. [Google Scholar] [CrossRef]
- Zeng, L.; Bai, M.; Mittal, A.K.; El-Jouni, W.; Zhou, J.; Cohen, D.M.; Zhou, M.I.; Cohen, H.T. Candidate tumor suppressor and pVHL partner Jade-1 binds and inhibits AKT in renal cell carcinoma. Cancer Res. 2013, 73, 5371–5380. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Du, W.Q.; Zhang, R.Y.; Zheng, J.N.; Pei, D.S. Jade-1: Its structure, regulation and functions in the renal cancer. Curr. Mol. Med. 2016, 16, 63–69. [Google Scholar] [CrossRef]
- Zhou, M.I.; Foy, R.L.; Chitalia, V.C.; Zhao, J.; Panchenko, M.V.; Wang, H.; Cohen, H.T. Jade-1, a candidate renal tumor suppressor that promotes apoptosis. Proc. Natl. Acad. Sci. USA 2005, 102, 11035–11040. [Google Scholar] [CrossRef]
- Quintela, M.; Sieglaff, D.H.; Gazze, A.S.; Zhang, A.; Gonzalez, D.; Francis, L.; Webb, P.; Conlan, R.S. HBO1 directs histone H4 specific acetylation, potentiating mechano-transduction pathways and membrane elasticity in ovarian cancer cells. Nanomedicine 2019, 17, 254–265. [Google Scholar] [CrossRef]
- Cheng, C.K.; Chan, H.Y.; Yung, Y.L.; Wan, T.S.K.; Leung, A.W.K.; Li, C.K.; Tian, K.; Chan, N.P.H.; Cheung, J.S.; Ng, M.H.L. A novel NUP98-JADE2 fusion in a patient with acute myeloid leukemia resembling acute promyelocytic leukemia. Blood Adv. 2022, 6, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Wang, M.; Zhang, Y.; Ou, R.; Zhu, Z.; Ou, Y.; Chen, X.; Liang, X.; Ding, Y.; Song, L.; et al. Jade family PHD finger 3 (JADE3) increases cancer stem cell-like properties and tumorigenicity in colon cancer. Cancer Lett. 2018, 428, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liu, H.; Zhang, R.; Freedman, J.A.; Han, Y.; Hung, R.J.; Brhane, Y.; McLaughlin, J.; Brennan, P.; Bickeboeller, H.; et al. Deciphering associations between three RNA splicing-related genetic variants and lung cancer risk. NPJ Precis. Oncol. 2022, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Bruford, E.A.; Braschi, B.; Denny, P.; Jones, T.E.M.; Seal, R.L.; Tweedie, S. Guidelines for human gene nomenclature. Nat. Genet. 2020, 52, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, R.D.; Sheridan, S.; Girard, L.; Sato, M.; Kim, Y.; Pollack, J.; Peyton, M.; Zou, Y.; Kurie, J.M.; Dimaio, J.M.; et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004, 64, 9027–9034. [Google Scholar] [CrossRef]
- Young, L.; Sung, J.; Stacey, G.; Masters, J.R. Detection of Mycoplasma in cell cultures. Nat. Protoc. 2010, 5, 929–934. [Google Scholar] [CrossRef]
- Chansky, K.; Detterbeck, F.C.; Nicholson, A.G.; Rusch, V.W.; Vallières, E.; Groome, P.; Kennedy, C.; Krasnik, M.; Peake, M.; Shemanski, L.; et al. The IASLC Lung Cancer Staging Project: External Validation of the Revision of the TNM Stage Groupings in the Eighth Edition of the TNM Classification of Lung Cancer. J. Thorac. Oncol. 2017, 12, 1109–1121. [Google Scholar] [CrossRef]
- Detterbeck, F.C.; Chansky, K.; Groome, P.; Bolejack, V.; Crowley, J.; Shemanski, L.; Kennedy, C.; Krasnik, M.; Peake, M.; Rami-Porta, R. The IASLC Lung Cancer Staging Project: Methodology and Validation Used in the Development of Proposals for Revision of the Stage Classification of NSCLC in the Forthcoming (Eighth) Edition of the TNM Classification of Lung Cancer. J. Thorac. Oncol. 2016, 11, 1433–1446. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J. Thorac. Oncol. 2015, 10, 1240–1242. [Google Scholar] [CrossRef]
- Keogh, A.; Ryan, L.; Nur, M.M.; Baird, A.M.; Nicholson, S.; Cuffe, S.; Fitzmaurice, G.J.; Ryan, R.; Young, V.K.; Finn, S.P.; et al. USO1 expression is dysregulated in non-small cell lung cancer. Transl. Lung Cancer Res. 2022, 11, 1877–1895. [Google Scholar] [CrossRef]
- Sui, J.S.Y.; Martin, P.; Keogh, A.; Murchan, P.; Ryan, L.; Nicholson, S.; Cuffe, S.; Broin, P.; Finn, S.P.; Fitzmaurice, G.J.; et al. Altered expression of ACOX2 in non-small cell lung cancer. BMC Pulm. Med. 2022, 22, 321. [Google Scholar] [CrossRef]
- Baird, A.M.; Gray, S.G.; O’Byrne, K.J. IL-20 is epigenetically regulated in NSCLC and down regulates the expression of VEGF. Eur. J. Cancer 2011, 47, 1908–1918. [Google Scholar] [CrossRef]
- Cregan, S.; McDonagh, L.; Gao, Y.; Barr, M.P.; O’Byrne, K.J.; Finn, S.P.; Cuffe, S.; Gray, S.G. KAT5 (Tip60) is a potential therapeutic target in malignant pleural mesothelioma. Int. J. Oncol. 2016, 48, 1290–1296. [Google Scholar] [CrossRef]
- Singh, A.S.; Heery, R.; Gray, S.G. In Silico and In Vitro Analyses of LncRNAs as Potential Regulators in the Transition from the Epithelioid to Sarcomatoid Histotype of Malignant Pleural Mesothelioma (MPM). Int. J. Mol. Sci. 2018, 19, 1297. [Google Scholar] [CrossRef] [PubMed]
- Cregan, S.; Breslin, M.; Roche, G.; Wennstedt, S.; MacDonagh, L.; Albadri, C.; Gao, Y.; O’Byrne, K.J.; Cuffe, S.; Finn, S.P.; et al. Kdm6a and Kdm6b: Altered expression in malignant pleural mesothelioma. Int. J. Oncol. 2017, 50, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Lin, S.; Girard, L.; Zhou, Y.; Yang, L.; Ci, B.; Zhou, Q.; Luo, D.; Yao, B.; Tang, H.; et al. LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2019, 38, 2551–2564. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Qian, X.; Du, Y.-C.N.; Sanchez-Solana, B.; Chen, K.; Park, B.; Chen, B.; Jenkins, L.; Luo, J.; Tripathi, B.K.; et al. Abstract 3912: cProSite: A web based interactive platform for on-line proteomics and phosphoproteomics data analysis. Cancer Res. 2022, 82, 3912. [Google Scholar] [CrossRef]
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200–225.e235. [Google Scholar] [CrossRef]
- Satpathy, S.; Krug, K.; Jean Beltran, P.M.; Savage, S.R.; Petralia, F.; Kumar-Sinha, C.; Dou, Y.; Reva, B.; Kane, M.H.; Avanessian, S.C.; et al. A proteogenomic portrait of lung squamous cell carcinoma. Cell 2021, 184, 4348–4371.e4340. [Google Scholar] [CrossRef]
- Győrffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic. Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Á.; Győrffy, B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer 2021, 148, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Corsello, S.M.; Nagari, R.T.; Spangler, R.D.; Rossen, J.; Kocak, M.; Bryan, J.G.; Humeidi, R.; Peck, D.; Wu, X.; Tang, A.A.; et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 2020, 1, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Liu, X.; Ma, T.; Lv, D.; Sun, G. Predictive value of tumor mutational burden for immunotherapy in non-small cell lung cancer: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0263629. [Google Scholar] [CrossRef]
- Feng, H.; Shen, W. ACAA1 Is a Predictive Factor of Survival and Is Correlated With T Cell Infiltration in Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 564796. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic. Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Ricciuti, B.; Wang, X.; Alessi, J.V.; Rizvi, H.; Mahadevan, N.R.; Li, Y.Y.; Polio, A.; Lindsay, J.; Umeton, R.; Sinha, R.; et al. Association of High Tumor Mutation Burden in Non-Small Cell Lung Cancers With Increased Immune Infiltration and Improved Clinical Outcomes of PD-L1 Blockade Across PD-L1 Expression Levels. JAMA Oncol. 2022, 8, 1160–1168. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Arrowsmith, C.H.; Schapira, M. Targeting non-bromodomain chromatin readers. Nat. Struct. Mol. Biol. 2019, 26, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.I.; Wang, H.; Foy, R.L.; Ross, J.J.; Cohen, H.T. Tumor suppressor von Hippel-Lindau (VHL) stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res. 2004, 64, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ning, G.; Si, P.; Zhang, C.; Liu, W.; Ge, W.; Cui, K.; Zhang, R.; Ge, S. E3 ubiquitin ligase HECW1 promotes the metastasis of non-small cell lung cancer cells through mediating the ubiquitination of Smad4. Biochem. Cell Biol. 2021, 99, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, E.; Cai, C.; Liu, W.; Li, K.; Zhao, W. Downregulated microRNA-140-5p expression regulates apoptosis, migration and invasion of lung cancer cells by targeting zinc finger protein 800. Oncol. Lett. 2020, 20, 390. [Google Scholar] [CrossRef]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]
- Colomer, R.; Mondejar, R.; Romero-Laorden, N.; Alfranca, A.; Sanchez-Madrid, F.; Quintela-Fandino, M. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine 2020, 25, 100487. [Google Scholar] [CrossRef]
- Lopez de Rodas, M.; Nagineni, V.; Ravi, A.; Datar, I.J.; Mino-Kenudson, M.; Corredor, G.; Barrera, C.; Behlman, L.; Rimm, D.L.; Herbst, R.S.; et al. Role of tumor infiltrating lymphocytes and spatial immune heterogeneity in sensitivity to PD-1 axis blockers in non-small cell lung cancer. J. Immunother. Cancer 2022, 10, e004440. [Google Scholar] [CrossRef]
- Sanmamed, M.F.; Nie, X.; Desai, S.S.; Villaroel-Espindola, F.; Badri, T.; Zhao, D.; Kim, A.W.; Ji, L.; Zhang, T.; Quinlan, E.; et al. A Burned-Out CD8(+) T-cell Subset Expands in the Tumor Microenvironment and Curbs Cancer Immunotherapy. Cancer Discov. 2021, 11, 1700–1715. [Google Scholar] [CrossRef]
- Okkan, S.; Atkovar, G.; Sahinler, I.; Turkan, S.; Uzel, R. A randomised study of ornidazole as a radiosensitiser in carcinoma of the cervix: Long term results. Br. J. Cancer Suppl. 1996, 27, S282–S286. [Google Scholar]
- Overgaard, J. Clinical evaluation of nitroimidazoles as modifiers of hypoxia in solid tumors. Oncol. Res. 1994, 6, 509–518. [Google Scholar]
- Evyapan, G.; Luleyap, U.; Kaplan, H.M.; Kara, I.O. Ornidazole suppresses CD133+ melanoma stem cells via inhibiting hedgehog signaling pathway and inducing multiple death pathways in a mouse model. Croat. Med. J. 2022, 63, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Qi, X.W.; Yan, G.N.; Zhang, Q.B.; Xu, C.; Bian, X.W. Is CD133 expression a prognostic biomarker of non-small-cell lung cancer? A systematic review and meta-analysis. PLoS ONE 2014, 9, e100168. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhu, H.; Feng, J.; Ni, S.; Huang, J. High CD133 expression in the nucleus and cytoplasm predicts poor prognosis in non-small cell lung cancer. Dis. Markers 2015, 2015, 986095. [Google Scholar] [CrossRef] [PubMed]
- Tirino, V.; Camerlingo, R.; Franco, R.; Malanga, D.; La Rocca, A.; Viglietto, G.; Rocco, G.; Pirozzi, G. The role of CD133 in the identification and characterisation of tumour-initiating cells in non-small-cell lung cancer. Eur. J. Cardiothorac Surg. 2009, 36, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Fakiruddin, K.S.; Lim, M.N.; Nordin, N.; Rosli, R.; Zakaria, Z.; Abdullah, S. Targeting of CD133+ Cancer Stem Cells by Mesenchymal Stem Cell Expressing TRAIL Reveals a Prospective Role of Apoptotic Gene Regulation in Non-Small Cell Lung Cancer. Cancers 2019, 11, 1261. [Google Scholar] [CrossRef] [PubMed]
- Barr, M.P.; Gray, S.G.; Hoffmann, A.C.; Hilger, R.A.; Thomale, J.; O’Flaherty, J.D.; Fennell, D.A.; Richard, D.; O’Leary, J.J.; O’Byrne, K.J. Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS ONE 2013, 8, e54193. [Google Scholar] [CrossRef]



| Sample | Histology | Sex | Age | Stage (7th Edition) | TNM |
|---|---|---|---|---|---|
| 1 | Adenocarcinoma | Female | 75 | IV | pT4 N2 M1a |
| 2 | Adenocarcinoma | Male | 71 | IA | pT1a N0 |
| 3 | Adenocarcinoma | Female | 75 | IIA | pT1a N1 |
| 4 | Adenocarcinoma | Male | 71 | IB | pT2a |
| 5 | Adenocarcinoma | Female | 78 | IB | pT2a |
| 6 | Adenocarcinoma | Female | 67 | IIIB | pT4 N2 |
| 7 | Adenocarcinoma | Female | 66 | IB | pT2a N0 |
| 8 | Adenocarcinoma | Female | 69 | IB | pT2a N0 |
| 9 | Adenocarcinoma | Male | 66 | IIIA | pT2a N0 |
| 10 | Adenocarcinoma | Male | 86 | IIIA | pT3 N1 |
| 11 | Adenocarcinoma | Male | 69 | IIIA | pT3 N1 |
| 12 | Squamous Cell Carcinoma | Female | 67 | IB | pT2a N0 IB |
| 13 | Squamous Cell Carcinoma | Male | 71 | IB | pT2a N0 |
| 14 | Squamous Cell Carcinoma | Female | 59 | IIA | pT2a N1 |
| 15 | Squamous Cell Carcinoma | Female | 66 | IIA | pT2a N1 |
| 16 | Squamous Cell Carcinoma | Male | 78 | IIA | pT1b N1 |
| 17 | Squamous Cell Carcinoma | Male | 79 | IIIA | pT3 N2 |
| 18 | Squamous Cell Carcinoma | Male | 70 | IB | T2 N0 |
| 19 | Squamous Cell Carcinoma | Female | 80 | IIA | pT2a N1 |
| 20 | Squamous Cell Carcinoma | Male | 72 | IIB | pT2b N1 |
| 21 | Squamous Cell Carcinoma | Male | 66 | IIIA | pT1b N2 |
| 22 | Squamous Cell Carcinoma | Female | 76 | IA | pT1b N0 |
| n | |
|---|---|
| LUSC | 108 |
| LUAD | 82 |
| Pleomorphic carcinoma | 7 |
| Large cell | 3 |
| Adenosquamous | 4 |
| Female | 79 |
| Male | 125 |
| Age < 65 | 92 |
| Age ≥ 65 | 112 |
| Node positive | 89 |
| Node negative | 115 |
| Tumor size ≥ 5 cm | 82 |
| Tumor size < 5 cm | 122 |
| Grade 1 | 16 |
| Grade 2 | 110 |
| Grade 3 | 78 |
| Stage I | 100 |
| Stage II | 49 |
| Stage III | 54 |
| Stage IV | 1 |
| Smoker | 100 |
| Ex-smoker | 78 |
| Never smoker | 26 |
| LUAD | LUSC | ||||
|---|---|---|---|---|---|
| Parameter | Gene | Partial cor. | Adj. p-Value | Partial cor. | Adj. p-Value |
| JADE2 expression correlated with | TP53 | 0.082710513 | 0.178404525 | 0.081198214 | 0.179881123 |
| KRAS | 0.27347762 | 2.21 × 10−9 | 0.194184439 | 4.33 × 10−5 | |
| EGFR | 0.472416177 | 8.97 × 10−28 | 0.308070729 | 2.01 × 10−11 | |
| ERBB2 | 0.265614636 | 1.19 × 10−8 | 0.030231142 | 0.728719461 | |
| PI3KCA | 0.397114574 | 1.51 × 10−19 | 0.326686247 | 5.30 × 10−13 | |
| ALK | 0.234086121 | 1.46 × 10−6 | 0.119036922 | 0.019498159 | |
| LUAD | LUSC | ||||
|---|---|---|---|---|---|
| Mutated Gene | log2 Fold Changes | Adj. p-Value | log2 Fold Changes | Adj. p-Value | |
| JADE2 | TP53 | −0.051608347 | 0.163694231 | 0.064624702 | 0.163694231 |
| KRAS | 0.043519348 | 0.408863297 | −0.000381679 | 0.891865206 | |
| EGFR | 0.055108336 | 0.541222696 | 0.043119948 | 0.937619343 | |
| ERBB2 | −0.080435586 | 0.796276167 | −0.200601312 | 0.609981468 | |
| PIK3CA | 0.061946812 | 0.648789762 | 0.006501538 | 0.945101486 | |
| ALK | 0.055475718 | 0.728248885 | -0.04583388 | 0.884587181 | |
| ROS1 | −0.068125014 | 0.561008551 | −0.093883364 | 0.468144673 | |
| CDKN2A | 0.052081676 | 0.861629309 | 0.182166956 | 0.002894015 | |
| PTEN | −0.081612099 | 0.71248883 | 0.050782786 | 0.71248883 | |
| BRAF | −0.05377631 | 0.610383827 | −0.073436266 | 0.835224651 | |
| MET | 0.052563434 | 0.868825624 | −0.132008367 | 0.868825624 | |
| NF1 | −0.043139679 | 0.792161892 | −0.028611057 | 0.833802286 | |
| LUAD | LUSC | ||||
|---|---|---|---|---|---|
| Variable | R | p-Value | R | p-Value | |
| DNA damage response (DDR) pathway | BRCA1 | 0.1 | 0.026 * | 0.34 | 1.2 × 10−4 *** |
| ATM | 0.42 | 1.3 × 10−22 *** | 0.18 | 5.2 × 10−5 *** | |
| ATR | 0.15 | 8 × 10−4 *** | 0.18 | 5 × 10−5 *** | |
| CDK1 | −0.17 | 1.8 × 10−3 *** | 0.085 | 0.065 | |
| CHEK1 | −0.078 | 0.087 | 0.083 | 0.069 | |
| CHEK2 | −0.22 | 7.7 × 10−7 *** | 0.012 | 0.79 | |
| TP53 | 0.17 | 1.3 × 10−4 *** | 0.1 | 0.028 * | |
| Combined Signature | 0.057 | 0.21 | 0.21 | 4.7 × 10−6 *** | |
| Mismatch excision repair (MMR) related genes | PMS2 | 0.32 | 1 × 10−12 *** | 0.19 | 3.6 × 10−5 *** |
| MLH1 | 0.39 | 2 × 10−19 *** | 0.14 | 2.1 × 10−3 *** | |
| MSH2 | 0.16 | 3.3 × 10−4 *** | 0.064 | 0.16 | |
| MSH3 | 0.52 | 7.8 × 10−35 *** | 0.21 | 2.2 × 10−6 *** | |
| MSH6 | 0.27 | 8.1 × 10−10 *** | 0.23 | 2.3 × 10−7 *** | |
| PCNA | −0.19 | 0.013 * | 0.053 | 0.24 | |
| Combined Signature | 0.32 | 3 × 10−13 *** | 0.17 | 1.7 × 10−4 *** | |
| (a) Gene Correlations | |||||
| LUAD | LUSC | ||||
| Variable | Partial cor. | p-Value | Partial cor. | p-Value | |
| JADE2 | Purity | −0.180664434 | 5.38 × 10−5 *** | −0.102110571 | 0.025584713 * |
| B cell | 0.267772037 | 2.16 × 10−9 *** | −0.008044225 | 0.86162598 | |
| CD8+ T cell | 0.221168991 | 8.25 × 10−7 *** | 0.076819743 | 0.094463581 | |
| CD4+ T cell | 0.397473929 | 9.08 × 10−20 *** | 0.171171466 | 0.000177919 *** | |
| Macrophage | 0.21693886 | 1.42 × 10−6 *** | −0.014194577 | 0.757156887 | |
| Neutrophil | 0.301271192 | 1.36 × 10−11 *** | 0.086648578 | 0.058887476 | |
| Dendritic cell | 0.397200525 | 6.83 × 10−20 *** | 0.166597378 | 0.000269484 *** | |
| (b) Survival | |||||
| LUAD | LUSC | ||||
| Variable | p-Value | p-Value | |||
| JADE2 | B cell | 0.000268218 *** | 0.778203963 | ||
| CD8+ T cell | 0.345905392 | 0.370701923 | |||
| CD4+ T cell | 0.507773 | 0.142871314 | |||
| Macrophage | 0.110109126 | 0.651047592 | |||
| Neutrophil | 0.081068767 | 0.126999461 | |||
| Dendritic cell | 0.047523634 * | 0.324066598 | |||
| JADE2 | 0.901934355 | 0.298085102 | |||
| LUAD | LUSC | |||||
|---|---|---|---|---|---|---|
| Partial cor. | p-Value | Adj. p-Value | Partial cor. | p-Value | Adj. p-Value | |
| PD-1 (PDCD1) | 0.222296 | 6.17 × 10−7 | 2.06 × 10−6 | 0.259232 | 9.17 × 10−9 | 5.24 × 10−8 |
| CTLA4 | 0.200917 | 6.94 × 10−6 | 2.31 × 10−5 | 0.238258 | 1.39 × 10−7 | 6.97 × 10−7 |
| LAG3 | 0.122737 | 0.00636 | 0.014134 | 0.228991 | 4.29 × 10−7 | 2.45 × 10−6 |
| TIM-3 (HAVCR2) | 0.250338 | 1.76 × 10−8 | 5.86 × 10−8 | 0.228991 | 4.29 × 10−7 | 2.45 × 10−6 |
| GZMB | −0.03006 | 0.50554 | 0.623764 | 0.153332 | 0.00078 | 0.003119 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murphy, C.; Gornés Pons, G.; Keogh, A.; Ryan, L.; McCarra, L.; Jose, C.M.; Kesar, S.; Nicholson, S.; Fitzmaurice, G.J.; Ryan, R.; et al. An Analysis of JADE2 in Non-Small Cell Lung Cancer (NSCLC). Biomedicines 2023, 11, 2576. https://doi.org/10.3390/biomedicines11092576
Murphy C, Gornés Pons G, Keogh A, Ryan L, McCarra L, Jose CM, Kesar S, Nicholson S, Fitzmaurice GJ, Ryan R, et al. An Analysis of JADE2 in Non-Small Cell Lung Cancer (NSCLC). Biomedicines. 2023; 11(9):2576. https://doi.org/10.3390/biomedicines11092576
Chicago/Turabian StyleMurphy, Ciara, Glòria Gornés Pons, Anna Keogh, Lisa Ryan, Lorraine McCarra, Chris Maria Jose, Shagun Kesar, Siobhan Nicholson, Gerard J. Fitzmaurice, Ronan Ryan, and et al. 2023. "An Analysis of JADE2 in Non-Small Cell Lung Cancer (NSCLC)" Biomedicines 11, no. 9: 2576. https://doi.org/10.3390/biomedicines11092576
APA StyleMurphy, C., Gornés Pons, G., Keogh, A., Ryan, L., McCarra, L., Jose, C. M., Kesar, S., Nicholson, S., Fitzmaurice, G. J., Ryan, R., Young, V., Cuffe, S., Finn, S. P., & Gray, S. G. (2023). An Analysis of JADE2 in Non-Small Cell Lung Cancer (NSCLC). Biomedicines, 11(9), 2576. https://doi.org/10.3390/biomedicines11092576








