RUNX1-Regulated Signaling Pathways in Ovarian Cancer
Abstract
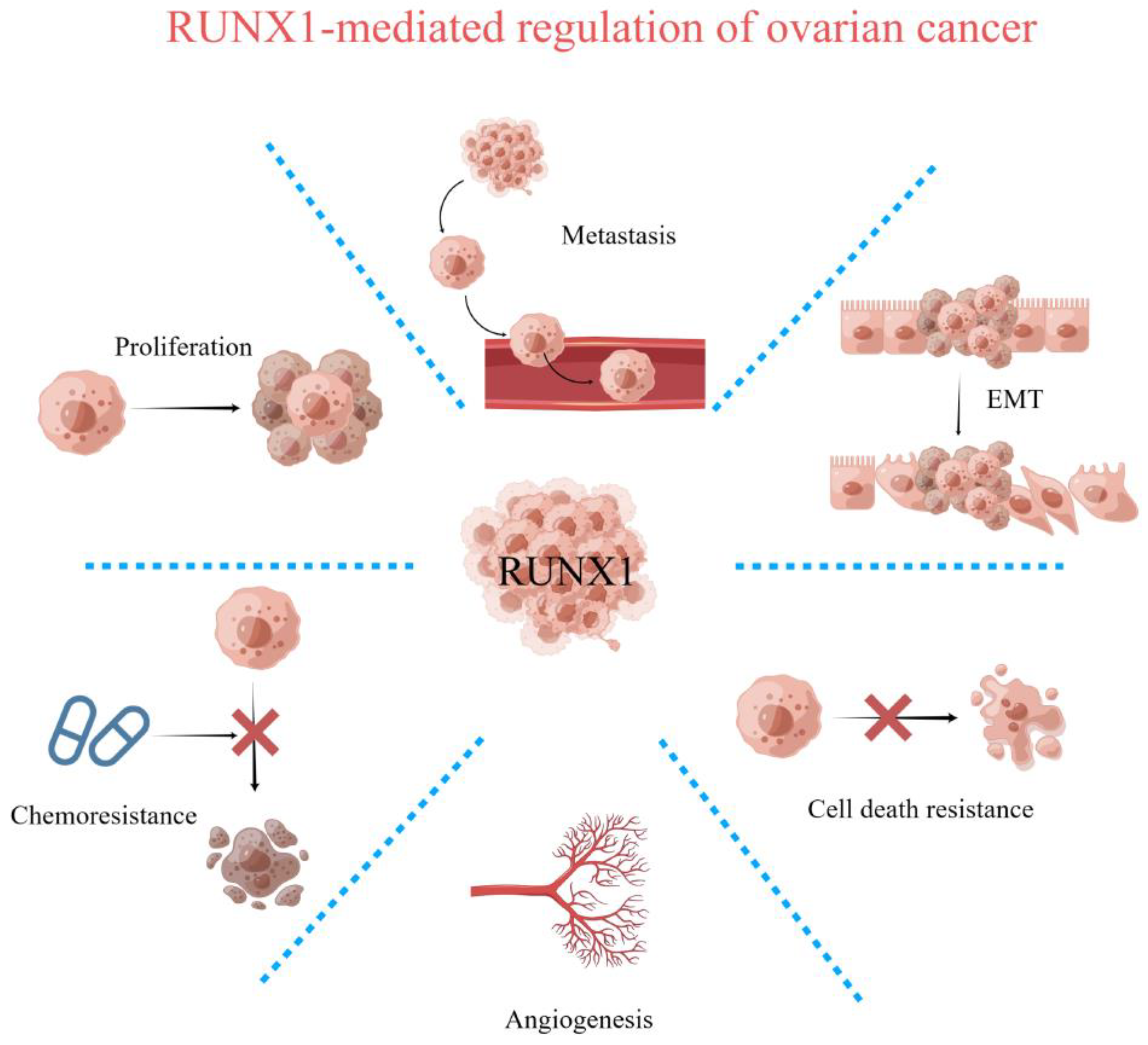
:1. Introduction
2. Expression and Regulation of RUNX1 in Ovarian Cancer
3. Interaction between RUNX1 and Its Cofactors
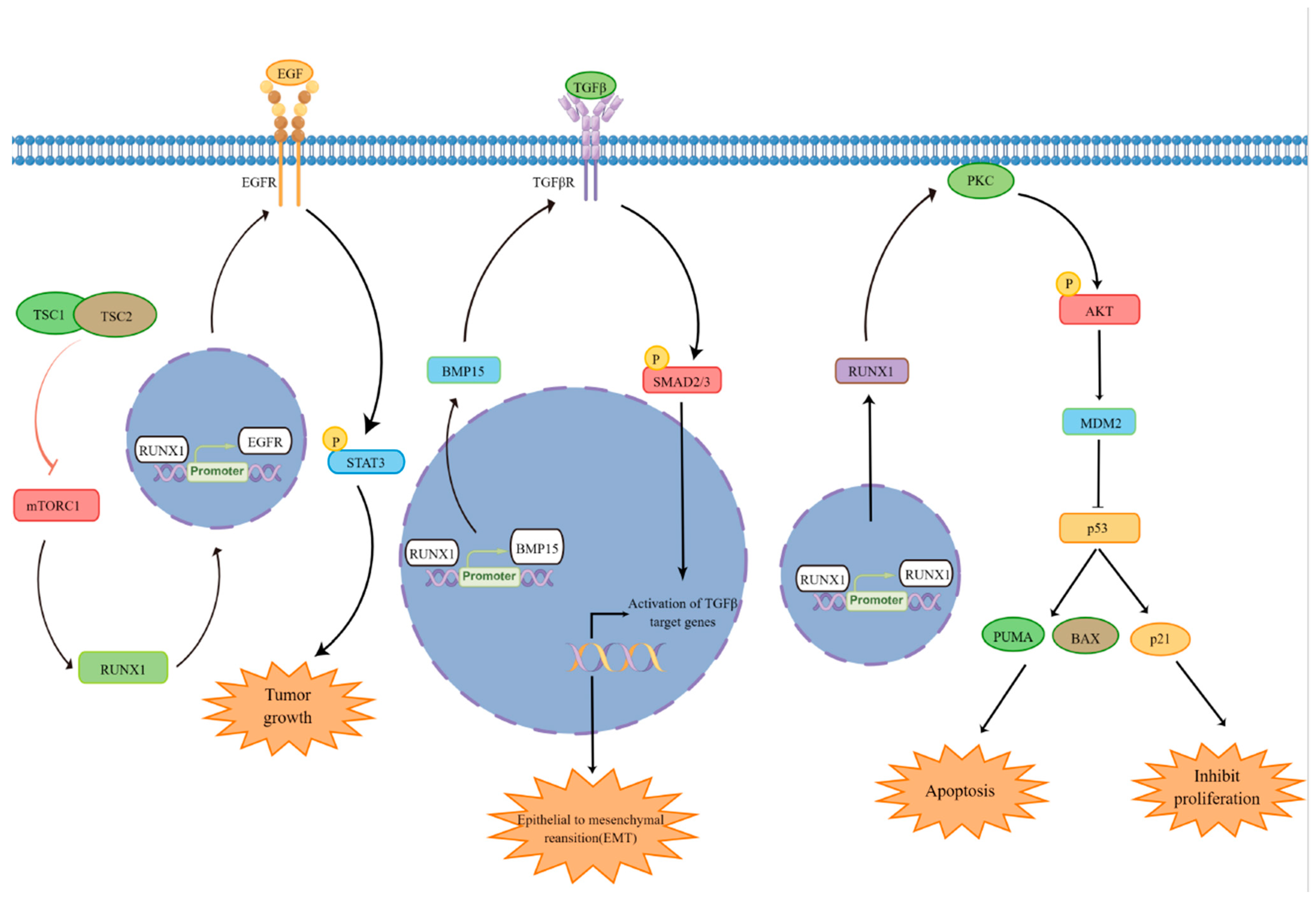
4. Regulation of Cell Proliferation and Survival by RUNX1
5. Regulation of P53 by RUNX1
6. Regulation of Epithelial–Mesenchymal Transition (EMT) by RUNX1
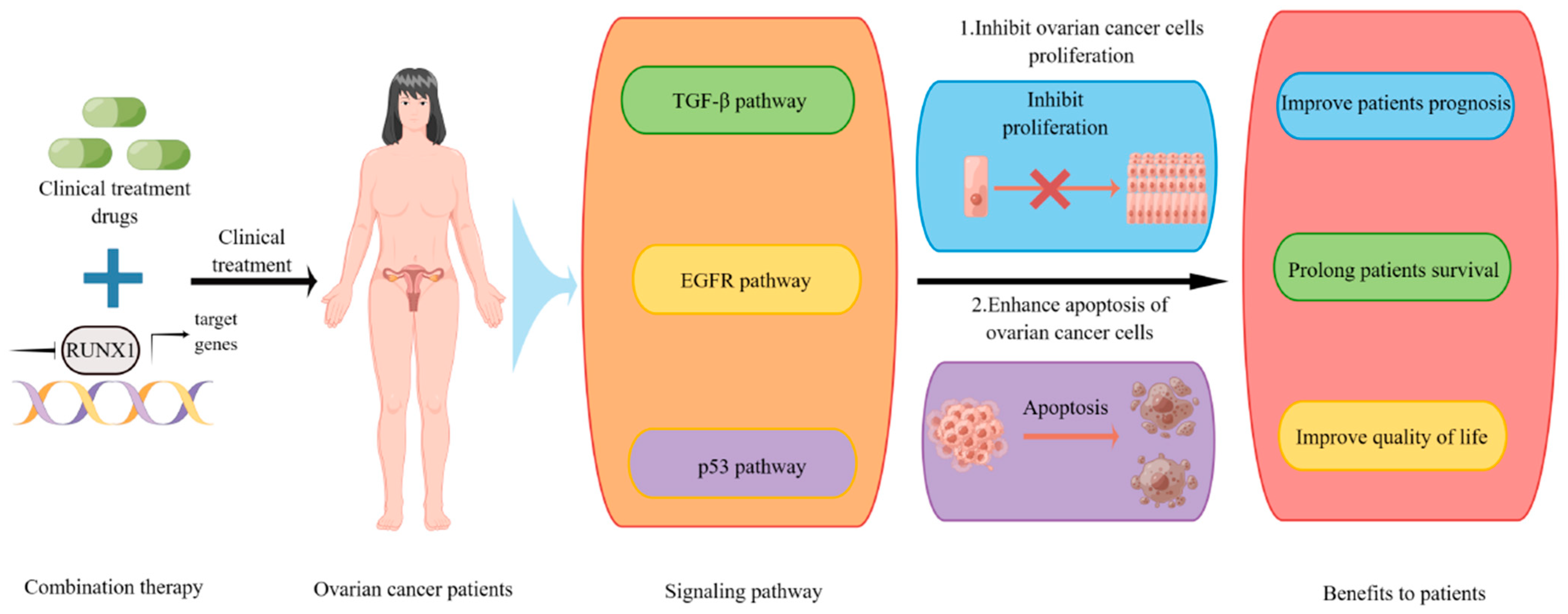
7. Potential of RUNX1 in Clinical Therapy
8. Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.C. Tour d’Horizon of Recent Advances in RUNX Family Gene Research. Mol. Cells 2020, 43, 97–98. [Google Scholar]
- Mevel, R.; Draper, J.E.; Lie, A.L.M.; Kouskoff, V.; Lacaud, G. RUNX transcription factors: Orchestrators of development. Development 2019, 146, dev148296. [Google Scholar] [CrossRef] [PubMed]
- De Braekeleer, E.; Ferec, C.; De Braekeleer, M. RUNX1 translocations in malignant hemopathies. Anticancer Res. 2009, 29, 1031–1037. [Google Scholar] [PubMed]
- Dowdy, C.R.; Xie, R.; Frederick, D.; Hussain, S.; Zaidi, S.K.; Vradii, D.; Javed, A.; Li, X.; Jones, S.N.; Lian, J.B.; et al. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum. Mol. Genet. 2010, 19, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef]
- Lin, T.C. RUNX1 and cancer. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188715. [Google Scholar] [CrossRef]
- Riggio, A.I.; Blyth, K. The enigmatic role of RUNX1 in female-related cancers–current knowledge & future perspectives. FEBS J. 2017, 284, 2345–2362. [Google Scholar]
- Keita, M.; Bachvarova, M.; Morin, C.; Plante, M.; Gregoire, J.; Renaud, M.C.; Sebastianelli, A.; Trinh, X.B.; Bachvarov, D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle 2013, 12, 972–986. [Google Scholar] [CrossRef]
- Nicol, B.; Grimm, S.A.; Chalmel, F.; Lecluze, E.; Pannetier, M.; Pailhoux, E.; Dupin-De-Beyssat, E.; Guiguen, Y.; Capel, B.; Yao, H.H. RUNX1 maintains the identity of the fetal ovary through an interplay with FOXL2. Nat. Commun. 2019, 10, 5116. [Google Scholar] [CrossRef] [PubMed]
- Keita, M.; Wang, Z.Q.; Pelletier, J.F.; Bachvarova, M.; Plante, M.; Gregoire, J.; Renaud, M.C.; Mes-Masson, A.M.; Paquet, E.R.; Bachvarov, D. Global methylation profiling in serous ovarian cancer is indicative for distinct aberrant DNA methylation signatures associated with tumor aggressiveness and disease progression. Gynecol. Oncol. 2013, 128, 356–363. [Google Scholar] [CrossRef]
- Scheitz, C.J.; Lee, T.S.; McDermitt, D.J.; Tumbar, T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012, 31, 4124–4139. [Google Scholar] [CrossRef]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.; Garcia, R.; Turkson, J.; Jove, R. STATs in oncogenesis. Oncogene 2000, 19, 2474–2488. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Speck, N.A.; Gilliland, D.G. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2002, 2, 502–513. [Google Scholar] [CrossRef]
- Ito, Y.; Bae, S.C.; Chuang, L.S. The RUNX family: Developmental regulators in cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef]
- Malik, N.; Yan, H.; Moshkovich, N.; Palangat, M.; Yang, H.; Sanchez, V.; Cai, Z.; Peat, T.J.; Jiang, S.; Liu, C.; et al. The transcription factor CBFB suppresses breast cancer through orchestrating translation and transcription. Nat. Commun. 2019, 10, 2071. [Google Scholar] [CrossRef]
- Amann, J.M.; Nip, J.; Strom, D.K.; Lutterbach, B.; Harada, H.; Lenny, N.; Downing, J.R.; Meyers, S.; Hiebert, S.W. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 2001, 21, 6470–6483. [Google Scholar] [CrossRef] [PubMed]
- Gelmetti, V.; Zhang, J.; Fanelli, M.; Minucci, S.; Pelicci, P.G.; Lazar, M.A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 1998, 18, 7185–7191. [Google Scholar] [CrossRef]
- Maiques-Diaz, A.; Spencer, G.J.; Lynch, J.T.; Ciceri, F.; Williams, E.L.; Amaral, F.M.R.; Wiseman, D.H.; Harris, W.J.; Li, Y.; Sahoo, S.; et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Rep. 2018, 22, 3641–3659. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, W.; Feng, F.; Cao, Q.; Li, Y.; Hou, Y.; Zhang, L.; Fan, J. Upregulated lncRNA CASC2 May Inhibit Malignant Melanoma Development Through Regulating miR-18a-5p/RUNX1. Oncol. Res. 2019, 27, 371–377. [Google Scholar] [CrossRef]
- Zhao, K.; Cui, X.; Wang, Q.; Fang, C.; Tan, Y.; Wang, Y.; Yi, K.; Yang, C.; You, H.; Shang, R.; et al. RUNX1 contributes to the mesenchymal subtype of glioblastoma in a TGFbeta pathway-dependent manner. Cell Death Dis. 2019, 10, 877. [Google Scholar] [CrossRef]
- Liu, S.; Yang, J.; Sun, G.; Zhang, Y.; Cheng, C.; Xu, J.; Yen, K.; Lu, T. RUNX1 Upregulates CENPE to Promote Leukemic Cell Proliferation. Front. Mol. Biosci. 2021, 8, 692880. [Google Scholar] [CrossRef]
- Kobayashi, S.D.; Voyich, J.M.; Whitney, A.R.; DeLeo, F.R. Spontaneous neutrophil apoptosis and regulation of cell survival by granulocyte macrophage-colony stimulating factor. J. Leukoc. Biol. 2005, 78, 1408–1418. [Google Scholar] [CrossRef]
- Wang, M.L.; Tuli, R.; Manner, P.A.; Sharkey, P.F.; Hall, D.J.; Tuan, R.S. Direct and indirect induction of apoptosis in human mesenchymal stem cells in response to titanium particles. J. Orthop. Res. 2003, 21, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Goyama, S.; Schibler, J.; Cunningham, L.; Zhang, Y.; Rao, Y.; Nishimoto, N.; Nakagawa, M.; Olsson, A.; Wunderlich, M.; Link, K.A.; et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Investig. 2013, 123, 3876–3888. [Google Scholar] [CrossRef] [PubMed]
- Klampfer, L.; Zhang, J.; Zelenetz, A.O.; Uchida, H.; Nimer, S.D. The AML1/ETO fusion protein activates transcription of BCL-2. Proc. Natl. Acad. Sci. USA 1996, 93, 14059–14064. [Google Scholar] [CrossRef]
- Xiao, L.; Peng, Z.; Zhu, A.; Xue, R.; Lu, R.; Mi, J.; Xi, S.; Chen, W.; Jiang, S. Inhibition of RUNX1 promotes cisplatin-induced apoptosis in ovarian cancer cells. Biochem. Pharmacol. 2020, 180, 114116. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A.M.; Attardi, L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018, 25, 93–103. [Google Scholar] [CrossRef]
- Leszczynska, K.B.; Foskolou, I.P.; Abraham, A.G.; Anbalagan, S.; Tellier, C.; Haider, S.; Span, P.N.; O’Neill, E.E.; Buffa, F.M.; Hammond, E.M. Hypoxia-induced p53 modulates both apoptosis and radiosensitivity via AKT. J. Clin. Investig. 2015, 125, 2385–2398. [Google Scholar] [CrossRef]
- Maddocks, O.D.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef]
- Jones, R.G.; Plas, D.R.; Kubek, S.; Buzzai, M.; Mu, J.; Xu, Y.; Birnbaum, M.J.; Thompson, C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 2005, 18, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Samarakkody, A.S.; Shin, N.Y.; Cantor, A.B. Role of RUNX Family Transcription Factors in DNA Damage Response. Mol. Cells 2020, 43, 99–106. [Google Scholar] [PubMed]
- Lee, J.W.; van Wijnen, A.; Bae, S.C. RUNX3 and p53: How Two Tumor Suppressors Cooperate Against Oncogenic Ras? Adv. Exp. Med. Biol. 2017, 962, 321–332. [Google Scholar]
- Morita, K.; Noura, M.; Tokushige, C.; Maeda, S.; Kiyose, H.; Kashiwazaki, G.; Taniguchi, J.; Bando, T.; Yoshida, K.; Ozaki, T.; et al. Autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid leukemia cells. Sci. Rep. 2017, 7, 16604. [Google Scholar] [CrossRef] [PubMed]
- Ou, H.L.; Schumacher, B. DNA damage responses and p53 in the aging process. Blood 2018, 131, 488–495. [Google Scholar] [CrossRef]
- Zhang, D.; Liang, C.; Li, P.; Yang, L.; Hao, Z.; Kong, L.; Tian, X.; Guo, C.; Dong, J.; Zhang, Y.; et al. Runt-related transcription factor 1 (Runx1) aggravates pathological cardiac hypertrophy by promoting p53 expression. J. Cell Mol. Med. 2021, 25, 7867–7877. [Google Scholar] [CrossRef]
- Jung, B.; Staudacher, J.J.; Beauchamp, D. Transforming Growth Factor beta Superfamily Signaling in Development of Colorectal Cancer. Gastroenterology 2017, 152, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Itatani, Y.; Kawada, K.; Sakai, Y. Transforming Growth Factor-beta Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int. J. Mol. Sci. 2019, 20, 5822. [Google Scholar] [CrossRef] [PubMed]
- Bhowmick, N.A.; Ghiassi, M.; Bakin, A.; Aakre, M.; Lundquist, C.A.; Engel, M.E.; Arteaga, C.L.; Moses, H.L. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 2001, 12, 27–36. [Google Scholar] [CrossRef]
- Li, L.; Liu, J.; Xue, H.; Li, C.; Liu, Q.; Zhou, Y.; Wang, T.; Wang, H.; Qian, H.; Wen, T. A TGF-beta-MTA1-SOX4-EZH2 signaling axis drives epithelial-mesenchymal transition in tumor metastasis. Oncogene 2020, 39, 2125–2139. [Google Scholar] [CrossRef]
- Zhou, T.; Luo, M.; Cai, W.; Zhou, S.; Feng, D.; Xu, C.; Wang, H. Runt-Related Transcription Factor 1 (RUNX1) Promotes TGF-beta-Induced Renal Tubular Epithelial-to-Mesenchymal Transition (EMT) and Renal Fibrosis through the PI3K Subunit p110delta. EBioMedicine 2018, 31, 217–225. [Google Scholar] [CrossRef]
- Lu, C.; Yang, Z.; Yu, D.; Lin, J.; Cai, W. RUNX1 regulates TGF-beta induced migration and EMT in colorectal cancer. Pathol. Res. Pract. 2020, 216, 153142. [Google Scholar] [CrossRef]
- Dubey, S.; Dubey, P.K.; Umeshappa, C.S.; Ghebre, Y.T.; Krishnamurthy, P. Inhibition of RUNX1 blocks the differentiation of lung fibroblasts to myofibroblasts. J. Cell Physiol. 2022, 237, 2169–2182. [Google Scholar] [CrossRef]
- Jakobczyk, H.; Jiang, Y.; Debaize, L.; Soubise, B.; Avner, S.; Serandour, A.A.; Rouger-Gaudichon, J.; Rio, A.G.; Carroll, J.S.; Raslova, H.; et al. ETV6-RUNX1 and RUNX1 directly regulate RAG1 expression: One more step in the understanding of childhood B-cell acute lymphoblastic leukemia leukemogenesis. Leukemia 2022, 36, 549–554. [Google Scholar]
- Masuda, T.; Maeda, S.; Shimada, S.; Sakuramoto, N.; Morita, K.; Koyama, A.; Suzuki, K.; Mitsuda, Y.; Matsuo, H.; Kubota, H.; et al. RUNX1 transactivates BCR-ABL1 expression in Philadelphia chromosome positive acute lymphoblastic leukemia. Cancer Sci. 2022, 113, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Mill, C.P.; Fiskus, W.; DiNardo, C.D.; Birdwell, C.; Davis, J.A.; Kadia, T.M.; Takahashi, K.; Short, N.; Daver, N.; Ohanian, M.; et al. Effective therapy for AML with RUNX1 mutation by cotreatment with inhibitors of protein translation and BCL2. Blood 2022, 139, 907–921. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, X.; Zhao, S.; Sun, F.; Ren, W.; Ma, M. RUNX1 promotes liver fibrosis progression through regulating TGF-beta signalling. Int. J. Exp. Pathol. 2023, 104, 188–198. [Google Scholar] [CrossRef]
- Lin, W.; Wan, X.; Sun, A.; Zhou, M.; Chen, X.; Li, Y.; Wang, Z.; Huang, H.; Li, H.; Chen, X.; et al. RUNX1/EGFR pathway contributes to STAT3 activation and tumor growth caused by hyperactivated mTORC1. Mol. Ther. Oncolytics 2021, 23, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lai, Q.; He, C.; Fang, Y.; Yan, Q.; Zhang, Y.; Wang, X.; Gu, C.; Wang, Y.; Ye, L.; et al. RUNX1 promotes tumour metastasis by activating the Wnt/beta-catenin signalling pathway and EMT in colorectal cancer. J. Exp. Clin. Cancer Res. 2019, 38, 334. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, J.; Lin, L.; He, L.; Wu, Y.; Zhang, L.; Yi, Z.; Chen, Y.; Pang, X.; Liu, M. Inhibition of STAT3 signaling pathway by nitidine chloride suppressed the angiogenesis and growth of human gastric cancer. Mol. Cancer Ther. 2012, 11, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef]
- Huynh, J.; Chand, A.; Gough, D.; Ernst, M. Therapeutically exploiting STAT3 activity in cancer–using tissue repair as a road map. Nat. Rev. Cancer 2019, 19, 82–96. [Google Scholar] [CrossRef]
- Chan, K.S.; Carbajal, S.; Kiguchi, K.; Clifford, J.; Sano, S.; DiGiovanni, J. Epidermal growth factor receptor-mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res. 2004, 64, 2382–2389. [Google Scholar] [CrossRef]
- Yu, H.; Jove, R. The STATs of cancer--new molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [Google Scholar] [CrossRef]
- Kim, D.J.; Kataoka, K.; Rao, D.; Kiguchi, K.; Cotsarelis, G.; Digiovanni, J. Targeted disruption of stat3 reveals a major role for follicular stem cells in skin tumor initiation. Cancer Res. 2009, 69, 7587–7594. [Google Scholar] [CrossRef]
- Li, N.; Grivennikov, S.I.; Karin, M. The unholy trinity: Inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell 2011, 19, 429–431. [Google Scholar] [CrossRef]
- Osorio, K.M.; Lilja, K.C.; Tumbar, T. Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. J. Cell Biol. 2011, 193, 235–250. [Google Scholar]
- Osorio, K.M.; Lee, S.E.; McDermitt, D.J.; Waghmare, S.K.; Zhang, Y.V.; Woo, H.N.; Tumbar, T. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development 2008, 135, 1059–1068. [Google Scholar] [CrossRef]
- Sarper, S.E.; Kurosaka, H.; Inubushi, T.; Ono Minagi, H.; Kuremoto, K.I.; Sakai, T.; Taniuchi, I.; Yamashiro, T. Runx1-Stat3-Tgfb3 signaling network regulating the anterior palatal development. Sci. Rep. 2018, 8, 11208. [Google Scholar] [CrossRef] [PubMed]
- Sarper, S.E.; Inubushi, T.; Kurosaka, H.; Ono Minagi, H.; Kuremoto, K.I.; Sakai, T.; Taniuchi, I.; Yamashiro, T. Runx1-Stat3 signaling regulates the epithelial stem cells in continuously growing incisors. Sci. Rep. 2018, 8, 10906. [Google Scholar] [CrossRef] [PubMed]
- Jelovac, D.; Armstrong, D.K. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J. Clin. 2011, 61, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Seoud, M.; Lundqvist, E.A.; Fujiwara, K. Targeted therapy in gynecologic cancers: Ready for prime time? Int. J. Gynaecol. Obstet. 2015, 131 (Suppl. S2), S150–S152. [Google Scholar] [CrossRef]
- Huang, M.; Huang, X.; Jiang, B.; Zhang, P.; Guo, L.; Cui, X.; Zhou, S.; Ren, L.; Zhang, M.; Zeng, J.; et al. linc00174-EZH2-ZNF24/Runx1-VEGFA Regulatory Mechanism Modulates Post-burn Wound Healing. Mol. Ther. Nucleic Acids 2020, 21, 824–836. [Google Scholar]
- Abu-Jawdeh, G.M.; Faix, J.D.; Niloff, J.; Tognazzi, K.; Manseau, E.; Dvorak, H.F.; Brown, L.F. Strong expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in ovarian borderline and malignant neoplasms. Lab. Investig. 1996, 74, 1105–1115. [Google Scholar]
- Tossetta, G. Metformin Improves Ovarian Cancer Sensitivity to Paclitaxel and Platinum-Based Drugs: A Review of In Vitro Findings. Int. J. Mol. Sci. 2022, 23, 12893. [Google Scholar]
- Kavallaris, M.; Kuo, D.Y.; Burkhart, C.A.; Regl, D.L.; Norris, M.D.; Haber, M.; Horwitz, S.B. Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific beta-tubulin isotypes. J. Clin. Investig. 1997, 100, 1282–1293. [Google Scholar] [CrossRef]
- Liu, Y.K.; Jia, Y.J.; Liu, S.H.; Ma, J. FSTL1 increases cisplatin sensitivity in epithelial ovarian cancer cells by inhibition of NF-kappaB pathway. Cancer Chemother. Pharmacol. 2021, 87, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Gao, H.Y.; Liu, J. The role of taurine in improving neural stem cells proliferation and differentiation. Nutr. Neurosci. 2017, 20, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yoon, Y.S.; Kim, J.C.; Kim, Y.M. Assessment of the Applicability of Integrative Tumor Response Assays in Advanced Epithelial Ovarian Cancer. Anticancer Res. 2019, 39, 313–318. [Google Scholar] [CrossRef]
- Vardy, J.L.; Liew, A.; Warby, A.; Elder, A.; Keshet, I.; Devine, R.; Ouliaris, C.; Renton, C.; Tattersall, M.H.N.; Dhillon, H.M. On the receiving end: Have patient perceptions of the side-effects of cancer chemotherapy changed since the twentieth century? Support. Care Cancer 2022, 30, 3503–3512. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y. Oncogenic potential of the RUNX gene family: ‘overview’. Oncogene 2004, 23, 4198–4208. [Google Scholar] [CrossRef]
- Hong, D.; Fritz, A.J.; Finstad, K.H.; Fitzgerald, M.P.; Weinheimer, A.; Viens, A.L.; Ramsey, J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Mol. Cancer Res. 2018, 16, 1952–1964. [Google Scholar] [CrossRef]
- Deltcheva, E.; Nimmo, R. RUNX transcription factors at the interface of stem cells and cancer. Biochem. J. 2017, 474, 1755–1768. [Google Scholar] [CrossRef]
- Dansonka-Mieszkowska, A.; Szafron, L.A.; Kulesza, M.; Stachurska, A.; Leszczynski, P.; Tomczyk-Szatkowska, A.; Sobiczewski, P.; Parada, J.; Kulinczak, M.; Moes-Sosnowska, J.; et al. PROM1, CXCL8, RUNX1, NAV1 and TP73 genes as independent markers predictive of prognosis or response to treatment in two cohorts of high-grade serous ovarian cancer patients. PLoS ONE 2022, 17, e0271539. [Google Scholar]
- Eldholm, V.; Haugen, A.; Zienolddiny, S. CTCF mediates the TERT enhancer-promoter interactions in lung cancer cells: Identification of a novel enhancer region involved in the regulation of TERT gene. Int. J. Cancer 2014, 134, 2305–2313. [Google Scholar] [PubMed]
- Wu, H.; Zheng, J.; Deng, J.; Zhang, L.; Li, N.; Li, W.; Li, F.; Lu, J.; Zhou, Y. LincRNA-uc002yug.2 involves in alternative splicing of RUNX1 and serves as a predictor for esophageal cancer and prognosis. Oncogene 2015, 34, 4723–4734. [Google Scholar] [CrossRef]
- Shimizu, K.; Yamagata, K.; Kurokawa, M.; Mizutani, S.; Tsunematsu, Y.; Kitabayashi, I. Roles of AML1/RUNX1 in T-cell malignancy induced by loss of p53. Cancer Sci. 2013, 104, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xia, P.; Huang, G.; Zhu, P.; Liu, J.; Ye, B.; Du, Y.; Fan, Z. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nat. Commun. 2016, 7, 11023. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.Y.; Wu, M.; Zhao, D.; Edwards, D.; McVicar, A.; Luo, Y.; Zhu, G.; Wang, Y.; Zhou, H.D.; Chen, W.; et al. Runx1 is a central regulator of osteogenesis for bone homeostasis by orchestrating BMP and WNT signaling pathways. PLoS Genet. 2021, 17, e1009233. [Google Scholar] [CrossRef]
- Deng, X.L.; Feng, L.; Wang, Z.X.; Zhao, Y.E.; Zhan, Q.; Wu, X.M.; Xiao, B.; Shu, Y. The Runx1/Notch1 Signaling Pathway Participates in M1/M2 Microglia Polarization in a Mouse Model of Temporal Lobe Epilepsy and in BV-2 Cells. Neurochem. Res. 2020, 45, 2204–2216. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Jia, Y.; Wang, Y.; Cai, X. Dysregulated MAPK signaling pathway in acute myeloid leukemia with RUNX1 mutations. Expert Rev. Hematol. 2022, 15, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Sun, L.; Wang, G.; Chen, B.; Luo, F. RUNX1: A Regulator of NF-kB Signaling in Pulmonary Diseases. Curr. Protein Pept. Sci. 2018, 19, 172–178. [Google Scholar]
- Christie, E.L.; Bowtell, D.D.L. Acquired chemotherapy resistance in ovarian cancer. Ann. Oncol. 2017, 28 (Suppl. S8), viii13–viii15. [Google Scholar] [CrossRef]
- He, Z.; Lin, J.; Chen, C.; Chen, Y.; Yang, S.; Cai, X.; He, Y.; Liu, S. Identification of BGN and THBS2 as metastasis-specific biomarkers and poor survival key regulators in human colon cancer by integrated analysis. Clin. Transl. Med. 2022, 12, e973. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; He, Y.; Liu, S. RUNX1-Regulated Signaling Pathways in Ovarian Cancer. Biomedicines 2023, 11, 2357. https://doi.org/10.3390/biomedicines11092357
Chen Y, He Y, Liu S. RUNX1-Regulated Signaling Pathways in Ovarian Cancer. Biomedicines. 2023; 11(9):2357. https://doi.org/10.3390/biomedicines11092357
Chicago/Turabian StyleChen, Yuanzhi, Yingying He, and Shubai Liu. 2023. "RUNX1-Regulated Signaling Pathways in Ovarian Cancer" Biomedicines 11, no. 9: 2357. https://doi.org/10.3390/biomedicines11092357
APA StyleChen, Y., He, Y., & Liu, S. (2023). RUNX1-Regulated Signaling Pathways in Ovarian Cancer. Biomedicines, 11(9), 2357. https://doi.org/10.3390/biomedicines11092357




