Senescence-Driven Inflammatory and Trophic Microenvironment Imprints Mesenchymal Stromal/Stem Cells in Osteoarthritic Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Human MSCs and In Vitro Treatments
2.2. MSC Self-Renewal Capacities
2.3. Senescence-Associated β-Galactosidase Activity Quantification
2.4. Gene Expression Analysis by RT-qPCR
| ARF | For | 5′CCCTCGTGCTGATGCTACTG3′ |
| Rev | 5′ACCTGGTCTTCTAGGAAGCGG3′ | |
| CDKN2B | For | 5′GACCGGGAATAACCTTCCAT3′ |
| Rev | 5′CACCAGGTCCAGTCAAGGAT3′ | |
| CDKN2A | For | 5′GCTGCCCAACGCACCGAATA3′ |
| Rev | 5′ACCACCAGCGTGTCCAGGAA3′ | |
| CDKN1A | For | 5′ACCGAGGCACTCAGAGGAG-3′ |
| Rev | 5′CAGGTCCACATGGTCTTCCT3′ | |
| CDKN1B | For | 5′CGGCTAACTCTGAGGACACG3′ |
| Rev | 5′CTTCTGAGGCCAGGCTTCTT-3′ | |
| CDKN1C | For | 5′GCGGCGATCAAGAAGCTGT-3′ |
| Rev | 5′GCTTGGCGAAGAAATCGGAGA-3′ | |
| RPS9 | For | 5′ATGAAGGACGGGATGTTCAC-3′ |
| Rev | 5′GATTACATCCTGGGCCTGAA-3′ |
2.5. Microarray Analysis of Gene Expression
2.6. Gene Ontology Analysis, Gene Set Enrichment Analysis (GSEA) and Venn Diagram Representation
2.7. Statistical Analysis
3. Results and Discussion
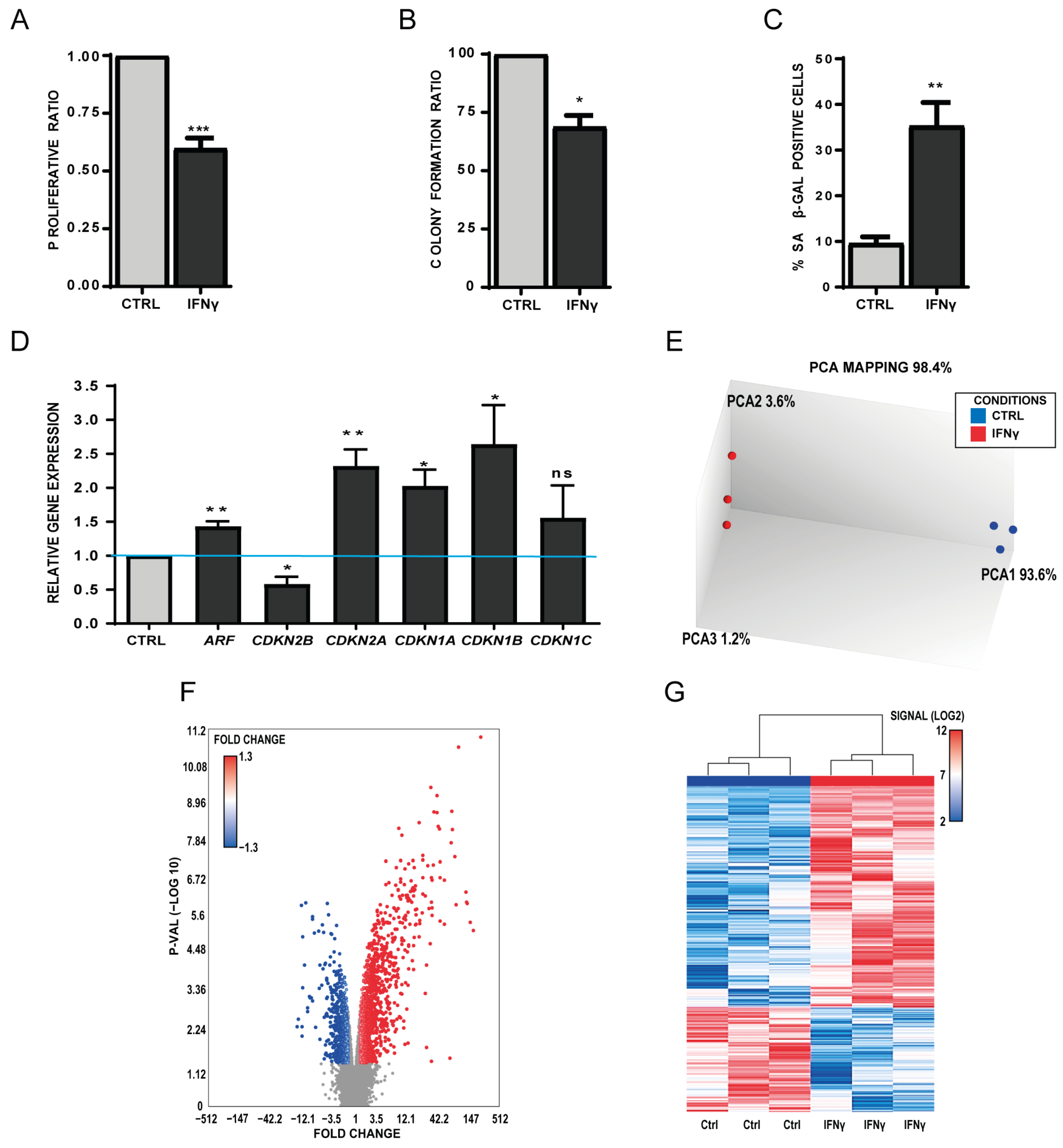
3.1. The Senescence-Driven Microenvironment Mediated by Interferon-Gamma (IFN-γ) Reduces Self-Renewal and Triggers Senescence Features in Healthy Human MSCs
3.2. The Senescence-Driven Microenvironment Mediated by TGFβ1 Triggers a Senescence-Like Phenotype in Healthy MSCs
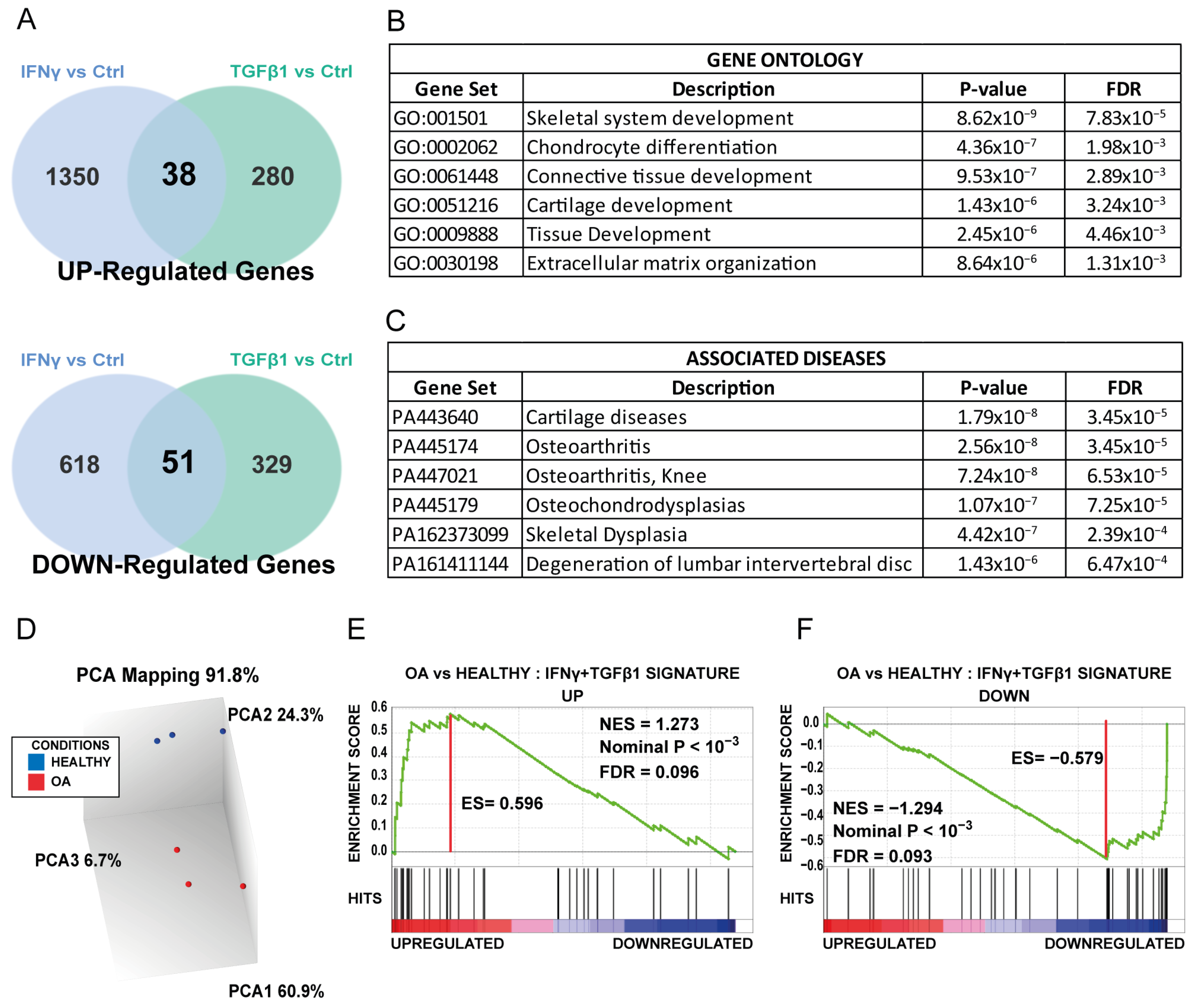
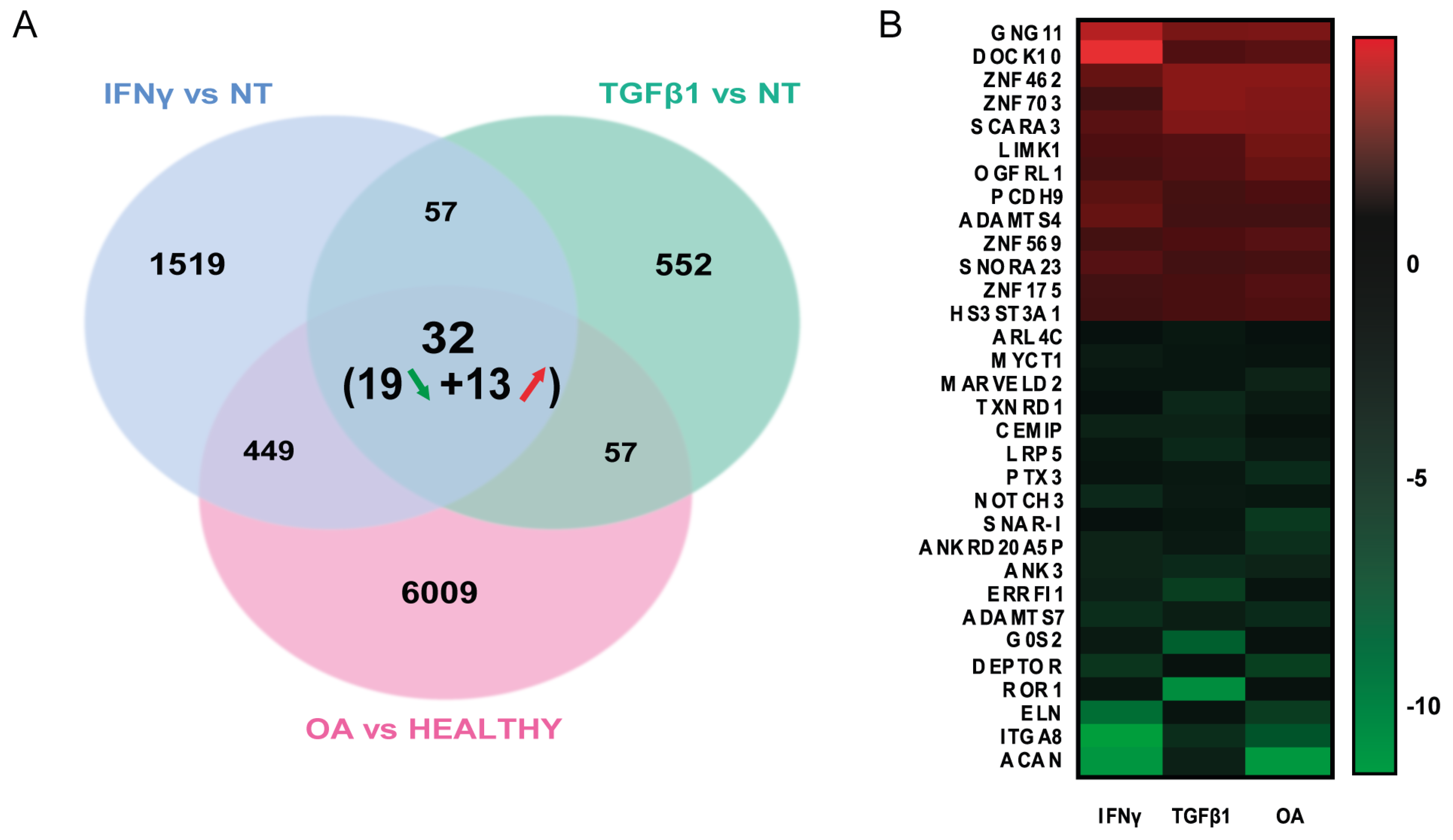
3.3. Common Gene Set of Both Treated MSCs Is Enriched in MSCs Isolated from OA Patients
3.4. Identifying Molecular Pathways Altered in OA MSCs by the Senescence-Driven Microenvironment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pignolo, R.J.; Passos, J.F.; Khosla, S.; Tchkonia, T.; Kirkland, J.L. Reducing Senescent Cell Burden in Aging and Disease. Trends Mol. Med. 2020, 26, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Khosla, S.; Farr, J.N.; Tchkonia, T.; Kirkland, J.L. The Role of Cellular Senescence in Ageing and Endocrine Disease. Nat. Rev. Endocrinol. 2020, 16, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Liu, R.-X.; Huan, S.-W.; Tang, W.; Zeng, Y.-K.; Zhang, J.-C.; Yang, J.; Li, Z.-Y.; Zhou, Y.; Zha, Z.-G.; et al. Senescent Skeletal Cells Cross-Talk with Synovial Cells Plays a Key Role in the Pathogenesis of Osteoarthritis. Arthritis Res. Ther. 2022, 24, 59. [Google Scholar] [CrossRef]
- Tachikart, Y.; Malaise, O.; Mumme, M.; Jorgensen, C.; Brondello, J.-M. Seno-Suppressive Molecules as New Therapeutic Perspectives in Rheumatic Diseases. Biochem. Pharmacol. 2019, 165, 126–133. [Google Scholar] [CrossRef]
- Berenbaum, F.; Walker, C. Osteoarthritis and Inflammation: A Serious Disease with Overlapping Phenotypic Patterns. Postgrad. Med. 2020, 132, 377–384. [Google Scholar] [CrossRef]
- Yang, Z.; Li, H.; Yuan, Z.; Fu, L.; Jiang, S.; Gao, C.; Wang, F.; Zha, K.; Tian, G.; Sun, Z.; et al. Endogenous Cell Recruitment Strategy for Articular Cartilage Regeneration. Acta Biomater. 2020, 114, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Barry, F.; Murphy, M. Mesenchymal Stem Cells in Joint Disease and Repair. Nat. Rev. Rheumatol. 2013, 9, 584–594. [Google Scholar] [CrossRef]
- Fernandes, A.M.; Herlofsen, S.R.; Karlsen, T.A.; Küchler, A.M.; Fløisand, Y.; Brinchmann, J.E. Similar Properties of Chondrocytes from Osteoarthritis Joints and Mesenchymal Stem Cells from Healthy Donors for Tissue Engineering of Articular Cartilage. PLoS ONE 2013, 8, e62994. [Google Scholar] [CrossRef]
- Malaise, O.; Tachikart, Y.; Constantinides, M.; Mumme, M.; Ferreira-Lopez, R.; Noack, S.; Krettek, C.; Noël, D.; Wang, J.; Jorgensen, C.; et al. Mesenchymal Stem Cell Senescence Alleviates Their Intrinsic and Seno-Suppressive Paracrine Properties Contributing to Osteoarthritis Development. Aging 2019, 11, 9128–9146. [Google Scholar] [CrossRef]
- Fu, L.; Hu, Y.; Song, M.; Liu, Z.; Zhang, W.; Yu, F.-X.; Wu, J.; Wang, S.; Izpisua Belmonte, J.C.; Chan, P.; et al. Up-Regulation of FOXD1 by YAP Alleviates Senescence and Osteoarthritis. PLoS Biol. 2019, 17, e3000201. [Google Scholar] [CrossRef]
- Pichard, L.; Brondelo, J.-M.; Becker, F.; Desprat, R.; De Ceuninck, F.; Pastoureau, P.; Noel, D.; Jorgensen, C.; Lemaitre, J.-M. Generation of Human Pluripotent Stem Cell Lines (IPSCs) from Mesenchymal Stem Cells (MSCs) from Three Elderly Patients with Osteoarthritis. Stem Cell Res. 2020, 44, 101721. [Google Scholar] [CrossRef]
- Pichard, L.; Brondello, J.-M.; Becker, F.; Desprat, R.; De Ceuninck, F.; Pastoureau, P.; Noel, D.; Jorgensen, C.; Lemaitre, J.-M. Establishment of a Collection of Human Pluripotent Stem Cell Lines (IPSC) from Mesenchymal Stem Cells (MSC) from Three Healthy Elderly Donors. Stem Cell Res. 2021, 53, 102297. [Google Scholar] [CrossRef] [PubMed]
- Vernes, S.C.; Spiteri, E.; Nicod, J.; Groszer, M.; Taylor, J.M.; Davies, K.E.; Geschwind, D.H.; Fisher, S.E. High-Throughput Analysis of Promoter Occupancy Reveals Direct Neural Targets of FOXP2, a Gene Mutated in Speech and Language Disorders. Am. J. Hum. Genet. 2007, 81, 1232–1250. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-Z.; Lu, H.-D. Transcriptome Analyses Identify Key Genes and Potential Mechanisms in a Rat Model of Osteoarthritis. J. Orthop. Surg. Res. 2018, 13, 319. [Google Scholar] [CrossRef]
- Kułach, N.; Pilny, E.; Cichoń, T.; Czapla, J.; Jarosz-Biej, M.; Rusin, M.; Drzyzga, A.; Matuszczak, S.; Szala, S.; Smolarczyk, R. Mesenchymal Stromal Cells as Carriers of IL-12 Reduce Primary and Metastatic Tumors of Murine Melanoma. Sci. Rep. 2021, 11, 18335. [Google Scholar] [CrossRef] [PubMed]
- Brondello, J.-M.; Pers, Y.-M. Taking in Consideration the Bystander Effects of Articular Senescence. Ann. Transl. Med. 2019, 7, S386. [Google Scholar] [CrossRef]
- Cao, X.; Luo, P.; Huang, J.; Liang, C.; He, J.; Wang, Z.; Shan, D.; Peng, C.; Wu, S. Intraarticular Senescent Chondrocytes Impair the Cartilage Regeneration Capacity of Mesenchymal Stem Cells. Stem Cell Res. Ther. 2019, 10, 86. [Google Scholar] [CrossRef]
- Meneghetti, M.C.Z.; Gesteira Ferreira, T.; Tashima, A.K.; Chavante, S.F.; Yates, E.A.; Liu, J.; Nader, H.B.; Lima, M.A. Insights into the Role of 3-O-Sulfotransferase in Heparan Sulfate Biosynthesis. Org. Biomol. Chem. 2017, 15, 6792–6799. [Google Scholar] [CrossRef]
- Shamdani, S.; Chantepie, S.; Flageollet, C.; Henni-Chebra, N.; Jouan, Y.; Eymard, F.; Hay, E.; Cohen-Solal, M.; Papy-Garcia, D.; Chevalier, X.; et al. Heparan Sulfate Functions Are Altered in the Osteoarthritic Cartilage. Arthritis Res. Ther. 2020, 22, 283. [Google Scholar] [CrossRef]
- Plikus, M.V.; Wang, X.; Sinha, S.; Forte, E.; Thompson, S.M.; Herzog, E.L.; Driskell, R.R.; Rosenthal, N.; Biernaskie, J.; Horsley, V. Fibroblasts: Origins, Definitions, and Functions in Health and Disease. Cell 2021, 184, 3852–3872. [Google Scholar] [CrossRef]
- Niklason, L.E. Understanding the Extracellular Matrix to Enhance Stem Cell-Based Tissue Regeneration. Cell Stem Cell 2018, 22, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Loreti, M.; Sacco, A. The Jam Session between Muscle Stem Cells and the Extracellular Matrix in the Tissue Microenvironment. NPJ Regen. Med. 2022, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, J.; Magli, S.; Rabbachin, L.; Sampaolesi, S.; Nicotra, F.; Russo, L. 3D Extracellular Matrix Mimics: Fundamental Concepts and Role of Materials Chemistry to Influence Stem Cell Fate. Biomacromolecules 2020, 21, 1968–1994. [Google Scholar] [CrossRef]
- Huang, Y.-Z.; Xie, H.-Q.; Silini, A.; Parolini, O.; Zhang, Y.; Deng, L.; Huang, Y.-C. Mesenchymal Stem/Progenitor Cells Derived from Articular Cartilage, Synovial Membrane and Synovial Fluid for Cartilage Regeneration: Current Status and Future Perspectives. Stem Cell Rev. Rep. 2017, 13, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Biroccio, A.; Cherfils-Vicini, J.; Augereau, A.; Pinte, S.; Bauwens, S.; Ye, J.; Simonet, T.; Horard, B.; Jamet, K.; Cervera, L.; et al. TRF2 Inhibits a Cell-Extrinsic Pathway through Which Natural Killer Cells Eliminate Cancer Cells. Nat. Cell Biol. 2013, 15, 818–828. [Google Scholar] [CrossRef]
- Hayat, R.; Manzoor, M.; Hussain, A. Wnt Signaling Pathway: A Comprehensive Review. Cell Biol. Int. 2022, 46, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Goldring, M.B.; Marcu, K.B. Cartilage Homeostasis in Health and Rheumatic Diseases. Arthritis Res. Ther. 2009, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Deroyer, C.; Charlier, E.; Neuville, S.; Malaise, O.; Gillet, P.; Kurth, W.; Chariot, A.; Malaise, M.; de Seny, D. CEMIP (KIAA1199) Induces a Fibrosis-like Process in Osteoarthritic Chondrocytes. Cell Death Dis. 2019, 10, 103. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fusi, G.; Constantinides, M.; Fissoun, C.; Pichard, L.; Pers, Y.-M.; Ferreira-Lopez, R.; Pantesco, V.; Poulet, C.; Malaise, O.; De Seny, D.; et al. Senescence-Driven Inflammatory and Trophic Microenvironment Imprints Mesenchymal Stromal/Stem Cells in Osteoarthritic Patients. Biomedicines 2023, 11, 1994. https://doi.org/10.3390/biomedicines11071994
Fusi G, Constantinides M, Fissoun C, Pichard L, Pers Y-M, Ferreira-Lopez R, Pantesco V, Poulet C, Malaise O, De Seny D, et al. Senescence-Driven Inflammatory and Trophic Microenvironment Imprints Mesenchymal Stromal/Stem Cells in Osteoarthritic Patients. Biomedicines. 2023; 11(7):1994. https://doi.org/10.3390/biomedicines11071994
Chicago/Turabian StyleFusi, Giuseppe, Michael Constantinides, Christina Fissoun, Lydiane Pichard, Yves-Marie Pers, Rosanna Ferreira-Lopez, Veronique Pantesco, Christophe Poulet, Olivier Malaise, Dominique De Seny, and et al. 2023. "Senescence-Driven Inflammatory and Trophic Microenvironment Imprints Mesenchymal Stromal/Stem Cells in Osteoarthritic Patients" Biomedicines 11, no. 7: 1994. https://doi.org/10.3390/biomedicines11071994
APA StyleFusi, G., Constantinides, M., Fissoun, C., Pichard, L., Pers, Y.-M., Ferreira-Lopez, R., Pantesco, V., Poulet, C., Malaise, O., De Seny, D., Lemaitre, J.-M., Jorgensen, C., & Brondello, J.-M. (2023). Senescence-Driven Inflammatory and Trophic Microenvironment Imprints Mesenchymal Stromal/Stem Cells in Osteoarthritic Patients. Biomedicines, 11(7), 1994. https://doi.org/10.3390/biomedicines11071994







