Abstract
Small cell lung cancer (SCLC) is an aggressive malignancy characterized by rapid proliferation, early dissemination, acquired therapy resistance, and poor prognosis. Early diagnosis of SCLC is crucial since most patients present with advanced/metastatic disease, limiting the potential for curative treatment. While SCLC exhibits initial responsiveness to chemotherapy and radiotherapy, treatment resistance commonly emerges, leading to a five-year overall survival rate of up to 10%. New effective biomarkers, early detection, and advancements in therapeutic strategies are crucial for improving survival rates and reducing the impact of this devastating disease. This review aims to comprehensively summarize current knowledge on diagnostic options, well-known and emerging biomarkers, and SCLC treatment strategies and discuss future perspectives on this aggressive malignancy.
1. Introduction
Lung cancer remains a significant global health concern, with staggering mortality rates. According to GLOBOCAN, it accounted for 2.1 million new cases and 1.8 million deaths in 2018, making it the leading cause of cancer-related deaths worldwide [1]. Lung cancer is categorized into two main histological types: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC comprises approximately 85% of cases, while SCLC represents around 15% [2]. SCLC is an aggressive neoplasm characterized by rapid proliferation, early dissemination, metastases, acquired therapy resistance, and poor outcomes [3]. Each year, approximately 250,000 new cases of SCLC are reported, resulting in at least 200,000 deaths worldwide [1]. While historically more common in men, the prevalence of SCLC among women has risen due to global smoking trends. Exposure to tobacco carcinogens (polycyclic aromatic hydrocarbons and tobacco-specific nitrosamines) is considered a key risk factor for SCLC, as only 2% of all SCLC cases are among never-smokers [4]. Early diagnosis of SCLC is crucial as most patients present with metastatic disease, limiting the potential for curative treatment. While SCLC exhibits initial responsiveness to chemotherapy and radiotherapy, treatment resistance often emerges, leading to a five-year overall survival rate of only 10% [5]. Poor prognosis is associated with factors such as male gender, poor performance status, and age over 70 [5,6]. Diagnostic procedures for SCLC typically involve physical examination, performance status evaluation, laboratory tests, and imaging techniques, including contrast-enhanced CT scans of the chest and abdomen, brain MRI or CT, and optional FDG PET/CT for limited-stage disease. Pathological examination following bronchoscopy, lymph node biopsy, and metastatic lesion biopsy is essential for accurately classifying SCLC [5]. To combat the high mortality rates associated with lung cancer, smoking cessation, and prevention remain the most critical interventions in reducing lung cancer mortality [5,6].
Common clinical manifestations of SCLC at diagnosis include central tumor masses, mediastinal involvement, and extrathoracic spread in 75–80% of patients [6]. Symptoms may include cough, wheezing, dyspnea, hemoptysis, weight loss, pain, fatigue, and paraneoplastic syndromes. Metastasis frequently occurs in the brain, liver, adrenal glands, bone, and bone marrow, often resulting in neurological deficits and paraneoplastic syndromes [7].
With the addition of programmed cell death protein-1 (PD-1) and programmed death-ligand 1 (PD-L1) inhibitors to chemotherapy in the first line of extensive small cell lung cancer (SCLC), a step forward has been made in improving overall treatment outcomes for patients with SCLC [8]. In routine clinical practice, there are currently no available predictive biomarkers for immunotherapy response, and the use of programmed death-ligand 1 (PD-L1) and tumor mutational burden (TMB) testing is not recommended [5]. The need for biomarkers to predict treatment response in patients with SCLC is urgent. Potential biomarkers such as PD-L1 expression, high TMB (TMB-H), and microsatellite instability (MSI-H) need further investigation for applicability in SCLC. Effective biomarkers, early detection, and advancements in therapeutic strategies are crucial for improving survival rates and reducing the impact of this devastating disease. Therefore, this review aims to comprehensively summarize current diagnostic options, well-known and emerging biomarkers, and treatment options of SCLC and discuss future perspectives of this distinct oncological challenge.
2. Pathology of SCLC
SCLC belongs to the spectrum of neuroendocrine pulmonary neoplasms that share some common morphologic, ultrastructural, immunohistochemical, and molecular genomic characteristics [9,10]. Four major neuroendocrine pulmonary neoplasms are carcinoids (typical and atypical) and neuroendocrine carcinomas (SCLC and large cell neuroendocrine carcinomas; LCNEC). A typical carcinoid is a low-grade neoplasm, and atypical carcinoid is intermediate-grade, whereas both neuroendocrine carcinomas are, per definition, high-grade neoplasms. The current evidence suggests that carcinoids (typical and atypical) are closely related and etiologically different from SCLC and LCNEC [9,10]. Carcinoids are not precursor lesions of neuroendocrine carcinomas (SCLC and LCNEC) and may be seen more frequently among non-smokers [9,10]. A small subset of carcinoids can be seen in patients with multiple endocrine neoplasia 1 (MEN1) syndrome (OMIM#131100), while somatic MEN1 gene mutations are commonly observed in carcinoids [9,10]. Rare cases of histologic transformation of epidermal growth factor receptor (EGFR)—or anaplastic large kinase (ALK)-altered pulmonary adenocarcinomas have also been well-documented [11]. It is widely accepted that SCLC has the same endodermal origins as other major subtypes of lung carcinoma (e.g., adenocarcinoma or squamous cell carcinoma), arising from multipotent precursor cells [9,10,12,13].
Morphologically, SCLC is composed of densely packed, small neoplastic cells with scanty cytoplasm and finely granular nuclear chromatin but without prominent nucleoli; nuclear molding and smudging are commonly present (Figure 1A,B). The cells are round or oval, although spindle cells (fusiform pattern of cancer cells) are frequently seen. Mitotic figures are numerous, while the tumor necrosis and crush artifacts may be extensive.
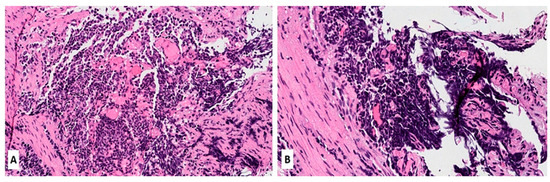
Figure 1.
(A,B) Hematoxylin and Eosin (H&E) stain of a lung biopsy showing a small cell carcinoma with sheet-like diffuse growth pattern and basophilic appearance (A, magnification 10×); Image 1B reveals a prominent nuclear molding of neoplastic cells (magnification 20×).
SCLC expresses neuroendocrine markers, such as synaptophysin, chromogranin-A, and CD56/NCAM, which should be used as a panel [14,15]. CD56 is the most sensitive as it stains 90–100% of all SCLC, while synaptophysin and chromogranin-A can be negative in >50% of cases [16,17,18]. Neuron-specific enolase (NSE) is frequently positive in SCLC but is considered non-specific due to its widespread expression in non-neuroendocrine neoplasms (both pulmonary and extrapulmonary) [19]. Thyroid transcription factor 1 (TTF-1) is positive in ~80–90% of cases [10]. Other pulmonary biomarkers, including Napsin-A (positive in adenocarcinomas), p63, and p40 (positive in squamous cell carcinomas), are not immunoreactive in SCLC and can help in differential diagnosis, particularly on small biopsies. Other challenging cases (metastatic neuroendocrine tumors from other anatomic locations, e.g., mammary, gastrointestinal, or Merkel cell carcinoma from the skin) can be resolved using clinical history and other specific immunohistochemical biomarkers.
3. Genomic Features of SCLC
Recent research has focused on understanding the genetic basis of SCLC to identify new therapeutic targets and develop more effective treatments [20]. Genetic alterations contribute significantly to the development and progression of SCLC. Concomitant inactivation of two tumor suppressor genes, TP53 and RB1, is found in most SCLC cases [21,22] and is found in up to 90% and 50–90% of SCLC cases, respectively. These molecular features are strikingly different from those seen in NSCLC, in which various oncogenic driver mutations/fusions prevail (e.g., EGFR, KRAS, ALK, BRAF, RET, ROS1, MET, NTRK1-3, HER2/ERBB2) [22,23]. Additionally, genetic alterations contributing to SCLC’s development include amplifying the MYC family of oncogenes (MYC, MYCL, and MYCN), inactivation of the phosphatase and tensin homolog (PTEN) tumor suppressor gene, and mutations in the Notch signaling pathway. Genomic alterations of MYC family members are seen in SCLC and represent biomarkers of poor prognosis. In particular, MYCN alterations are related to SCLC cases with immunotherapy failure. The most important genes altered in SCLC in humans are summarized in Table 1.
Different studies have identified recurrent mutations in chromatin remodeling genes, such as ARID1A, ARID1B, and SMARCA4, which regulate gene expression. These mutations may contribute to the dysregulation of critical genes involved in cell proliferation and survival, leading to the development of SCLC, characterized by a high frequency of mutations in genes that regulate cell cycle and DNA damage response pathways, such as TP53, RB1, and PTEN. Additionally, SCLC often exhibits widespread chromosomal instability, with frequent amplifications and deletions of large genome regions. In addition to these genetic alterations, SCLC is characterized by a high frequency of copy number alterations, including amplification of MYC family members and deletion of the tumor suppressor gene cyclin-dependent kinase inhibitor 2A (CDKN2A) [24]. In addition, the changes in the stroma and immune microenvironment are additional factors involved in the pathogenesis of SCLC [25].
Overall, the genetic landscape of SCLC is complex and heterogeneous, with multiple genetic alterations contributing to its aggressive phenotype. Understanding the underlying genetic mechanisms of SCLC is crucial for developing effective targeted therapies and personalized treatment strategies for patients with this aggressive cancer. SCLC mutational characteristics reveal a clear causal connection with smoking. Direct scientific evidence confirms that carcinogens from tobacco are responsible for initiating SCLC [26].
Genomic profiling in patients with SCLC has not revealed mutationally defined subtypes of SCLC. However, due to the lack of larger studies, this may be a consequence of the insufficient number of tumor samples included in analyses. Therefore, there is a substantial need for clinical trials that include the analyses of tumor tissue to identify vital genomic triggers. However, there is an accentuated difficulty in tumor material collection. Ethnicity or smoking status did not affect the consistency of mutational differences; however, the prevalence of oncogenic triggers is considered higher in never-smokers with SCLC compared to tobacco users [27].
In addition, genetically modified mice have provided critical genetic lessons and contributed to the knowledge of molecular mechanisms of SCLC etiopathogenesis, metastasis, and response to treatment. It has been shown that tumors in mice show genetic alterations and histological features like those in humans. Ferone et al. provided a comprehensive review of lung cancers and lessons from mouse studies, showing an enormous contribution of animal studies in pulmooncology [28].

Table 1.
Most important genes altered in SCLC (mostly according to memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets—MSK-IMPACT sequencing of SCLC tumors)—data adopted from Cheng et al. [29], Rudin et al. [23], and Liu et al. [30].
Table 1.
Most important genes altered in SCLC (mostly according to memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets—MSK-IMPACT sequencing of SCLC tumors)—data adopted from Cheng et al. [29], Rudin et al. [23], and Liu et al. [30].
| Gene | Aliases | Gene Location on Human Chromosome and Number of Amino Acids | Gene Alteration in SCLC | Known Function and Features | Frequency of Mutation in SCLC (% in Various Cohorts) | Refs. |
|---|---|---|---|---|---|---|
| TP53 | Tumor protein 53; p53; Phosphoprotein P53; Antigen NY-CO-13; Transformation-Related Protein 53; BCC7, LFS1, TRP53, tumor protein BMFS5 | Chromosome 17 at position 17p13.1.; 375 amino acids | Inactivating mutation; deletion | Nuclear phosphoprotein involved in the regulation of cell proliferation; tumor suppressor; transcription regulation | 77–89 | Chang et al. [31] Rudin et al. [23] |
| RB1 | RB1, pRb, RB, retinoblastoma 1, OSRC, PPP1R130, p105-Rb, pp110, Retinoblastoma protein, RB transcriptional corepressor 1, p110-RB1 | Chromosome 13 at position 13q14.1-q14.2.; 928 amino acids | Inactivating mutation; deletion; loss or inactivation of both copies of the gene | Tumor suppressor protein that is dysfunctional in several major cancers. Prevents excessive cell growth by inhibiting cell cycle progression -key regulator of the G1/S transition of the cell cycle | 50–90 | George et al. [21] Febres-Aldana et al. [32] |
| KMT2D | KMT2D, ALR, KABUK1, MLL2, MLL4, lysine methyltransferase 2D, histone-lysine methyltransferase 2D, TNRC21, AAD10, KMS, CAGL114 | Chromosome 12 at position 12q13.12.; 5316 amino acids | Inactivating mutation; deletion; gene fusion; truncating nonsense/frameshift/splice site mutations | Key regulator of transcriptional enhancer function; major enhancer regulator in mammalian cells, including regulation of development, differentiation, metabolism, and tumor suppression. | 5–13 | Wu et al. [33] Simbolo et al. [34] Augert et al. [35] |
| CREBBP | AW558298, CBP, CBP/p300, KAT3A, p300/CBP, RSTS, CREB binding protein, RSTS1, MKHK1 | Chromosome 16 at position 16p13.3. 2414 amino acids. | Inactivating mutation, deletion | Crucial role in transcriptional regulation and chromatin remodeling. Interacts with various transcription factors and coactivators, influencing the expression of target genes involved in cell growth, differentiation, and development. | 4–10 | Carazo et al. [36] Jia et al. [37] |
| PTEN | PTEN, 10q23del, BZS, CWS1, DEC, GLM2, MHAM, MMAC1, PTEN1, TEP1, phosphatase and tensin homolog, Phosphatase and tensin homolog, PTENbeta | Chromosome 10 at position 10q23.3. 403 amino acids | inactivating mutations, deletions, or loss of expression | Tumor suppressor involved in the regulation of the PI3K/AKT/mTOR pathway, which plays a critical role in cell survival and proliferation. PTEN’s protein phosphatase activity may be involved in the regulation of the Cell cycle, preventing cells from growing and dividing too rapidly. | 3–10 | Sivakumar et al. [38] Zhang et al. [39] |
| FAT1 | CDHF7, CDHR8, FAT, ME5, hFat1, FAT atypical cadherin 1 | Chromosome 4 at position 4q35.2. 4410 amino acids | Inactivation mutation; deletion | Cell-cell adhesion, migration and communication, regulation of tissue growth, cell polarity, and migration; tumor suppressor gene | 2–10 | JiaXin et al. [40] Pop-Bica et al. [41] |
| PIK3CA | PIK3CA, CLOVE, CWS5, MCAP, MCM, MCMTC, PI3K, p110-alpha, PI3K-alpha, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, CLAPO, CCM4 | Chromosome 3 at position 3q26.3.; 1068 amino acids | Activating mutation; mutations in specific regions | The PIK3CA gene for synthesis of the catalytic subunit alpha of the enzyme phosphatidylinositol 3-kinase, having crucial role in cell growth, proliferation, and survival | 1–7 | Hung et al. [42] Pop-Bica et al. [41] |
| NOTCH1 | NOTCH1, Notch1, 9930111A19Rik, Mis6, N1, Tan1, lin-12, AOS5, AOVD1, hN1 | Chromosome 9 at position 9q34.3. 2527 amino acids | Inactivating mutation | Tumor suppressor; involved in cell signalling processes | 1–6 | Li et al. [43] Roper et al. [44] Herbreteau et al. [45] |
| NF1 | NFNS, VRNF, WSS, neurofibromin 1 | Chromosome 17 at position 17q11.2. 2818 amino acids | Inactivating mutation, deletion | Tumor suppressor. Neurofibromin 1 plays a role in regulating cell growth and proliferation by negatively regulating the activity of Ras, associated with uncontrolled cell growth. | 3–4 | Ross et al. [46] Shimizu et al. [47] |
| APC | BTPS2, DP2, DP2.5, DP3, GS, PPP1R46, adenomatous polyposis coli, WNT signaling pathway regulator | Chromosome 5 at position 5q22.2. 2843 amino acids | Inactivating mutation, deletion | Crucial role in regulating the Wnt signaling pathway and controlling cell proliferation, growth, differentiation, and migration. | 3–4 | Jin et al. [48] Grote et al. [49] |
| EGFR | ERBB, ERBB1, HER1, NISBD2, PIG61, mENA, epidermal growth factor receptor, erbB-1, ERRP | Chromosome 7 at position 7p12.1. 1210 amino acids | Activating mutation | Oncogene; a receptor tyrosine kinase that plays a critical role in cell growth, proliferation, and survival; involved in RAS signaling pathway. | 3–4 | Ding et al. [50] Hao et al. [51] |
| KRAS | C-K-RAS, CFC2, K-RAS2A, K-RAS2B, K-RAS4A, K-RAS4B, KI-RAS, KRAS1, KRAS2, NS, NS3, RALD, RASK2, K-ras, KRAS proto-oncogene, GTPase, c-Ki-ras2, OES, c-Ki-ras, K-Ras 2, K-Ras, Kirsten Rat Sarcoma virus | Chromosome 12 at position 12p12.1. 189 amino acids | Activating mutation | A GTPase involved in cell signalingpathways that regulate cell growth and proliferation (RAS/MAPK). KRAS mutations can lead to the constitutive activation of the KRAS protein, resulting in dysregulated cell signaling and increased cell proliferation. | 1–3 | Otegui et al. [52] Li et al. [53] |
| NOTCH3 | CADASIL, CASIL, IMF2, LMNS, CADASIL1, notch 3, notch receptor 3 | Chromosome 19 at position 19p13.2. 2345 amino acids | Inactivating mutation, deletion | Involved in cell signaling pathways. Notch signaling plays a critical role in cellular processes, such as cell fate determination, differentiation, and development. | <3 | Herbreteau et al. [45] Du et al. [54] |
| ARID1A | B120, BAF250, BAF250a, BM029, C1orf4, ELD, MRD14, OSA1, P270, SMARCF1, hELD, hOSA1, CSS2, AT-rich interaction domain 1A | Chromosome 1 at position 1p36.11. 2254 amino acids | Inactivating mutation, deletion | Tumor suppressor gene; plays a crucial role in regulating chromatin remodeling and gene expression; involved in various cellular processes, including DNA repair, cell cycle regulation, and differentiation. | <3 | Du et al. [54] Devarakonda et al. [55] |
| PTPRD | HPTP, HPTPD, HPTPDELTA, PTPD, RPTPDELTA, protein tyrosine phosphatase, receptor type D, protein tyrosine phosphatase receptor type D, R-PTP-delta | Chromosome 9 at position 9p23.3. 1840 amino acids | Inactivating mutation, deletion | Protein tyrosine phosphatase receptor that plays a role in regulating cell signaling pathways, including those involved in cell growth, differentiation, and migration. | <3 | Sato et al. [56] |
| ATRX | ATR2, JMS, MRXHF1, RAD54, RAD54L, SFM1, SHS, XH2, XNP, ZNF-HX, MRX52, alpha thalassemia/mental retardation syndrome X-linked, chromatin remodeler, ATRX chromatin remodeler | X chromosome at position Xq21.1. | Inactivating mutation, deletion | Tumor suppressor; plays a critical role in chromatin remodeling and the regulation of gene expression. ATRX is involved in maintaining the stability and structure of telomeres and in cell signaling | <2 | Du et al. [54] |
4. Biomarkers in SCLC
In contrast to NSCLC, the discovery of therapeutic targets in SCLC has not been easy, partly because driver mutations are in first-line loss of function or untargetable, e.g., MYC family members [23]. The recent division of SCLC into molecular subtypes based on the expression of transcription factors has provided an essential step in searching for new therapeutic targets for the disease. This classification system identifies four distinct subtypes of SCLC: achaete-scute homolog 1 (ASCL1), neurogenic differentiation factor 1 (NEUROD1), yes-associated protein 1 (YAP1), and POU class 2 homeobox 3 (POU2F3) [57].
New blood-based biomarkers for the early detection of lung cancer have been developed and evaluated, with several showing promising results. “Liquid biopsy”—biomarkers such as tumor-derived extracellular vesicles, circulating tumor cells (CTC), and circulating tumor DNA (ctDNA) seem to be promising tools in cancer monitoring. For example, SCLC cells express different tumor-specific markers, including Delta-like protein 3 (DLL-3), which may be associated with a worse prognosis in patients with SCLC [58]. However, whether these biomarkers listed in Table 2 will impact cancer control in the population, especially in cancer with aggressive biologic behavior such as SCLC, remains unknown. To date, it seems that patients with SCLC have the greatest number of CTC, which was suggested to be a prognostic biomarker for clinically evaluating therapy efficacy [59]. Likewise, CTC-derived DNA and plasma cell-free DNA, along with their genomic alterations, have been recognized as potential non-invasive biomarkers that could provide insights into treatment efficacy and the occurrence of SCLC relapse [60].
Furthermore, the characterization of extracellular vesicles, such as exosomes, appears to be a promising tool and alternative source for various analytes in liquid biopsies [61]. This approach has the potential to significantly contribute to the identification of new biomarkers for the diagnosis and monitoring of SCLC patients, as well as the development of promising prognostic models. Emerging predictive and prognostic biomarkers are crucial and indispensable for selecting the most suitable therapeutic option for patients with SCLC.
To date, genetic alterations of MYC were noticed in about 20% of patients with SCLC, representing the third most common genetic abnormality following TP53 and RB1 and a potential biomarker of targeted therapy [62]. PD-L1, TMB, and MSI-H have been studied as potential predictive biomarkers for response to immune checkpoint inhibitors (ICIs) in patients with SCLC [62]. Schlafen 11 (SLFN11) is a DNA/RNA helicase that sensitizes cancer cells to DNA-damaging agents. The newest scientific evidence confirms its importance as a promising predictive biomarker for several therapeutics, including platinum and PARP inhibitors [63]. Expression of SLFN11 in CTCs provides a potential biomarker of sensitivity for DNA-damaging chemotherapy drugs and poly (ADP-ribose) polymerase (PARP) inhibition in SCLC patients [62]. Therefore, detecting SLFN11 by liquid biopsy in circulating CTCs may provide a valuable non-invasive alternative to tissue sampling [64].

Table 2.
Potential biomarkers in small cell lung carcinoma.
Table 2.
Potential biomarkers in small cell lung carcinoma.
| Biomarker | Type | Potential Application | References |
|---|---|---|---|
| Delta-like ligand 3 DLL3 | Tumor-specific marker | Biomarker for SCLC prognosis | Chen et al. [58] |
| Circulating tumor cells (CTC) | Liquid biopsy biomarker | Prognostic biomarker for therapy evaluation of therapy efficacy | Roumeliotou et al. [59] |
| Circulating tumor DNA (ctDNA) | Liquid biopsy biomarker | Biomarker for treatment efficacy and relapse detection | Almodovar et al. [60] |
| Exosomes | Extracellular vesicles | Non-invasive biomarkers for prognosis | Zhang et al. [61] |
| MYC proto-oncogene/bHLH transcription factor (MYC) | Genetic alteration | Potential biomarker for targeted therapy | Taniguchi et al. [62] |
| Programmed death-ligand 1 (PD-L1) | Immune checkpoint protein | Potential biomarker for immunotherapy response | Taniguchi et al. [62] |
| Tumor mutational burden (TMB) | Mutation load of a tumor | Potential biomarker for immunotherapy response | Taniguchi et al. [62] and Li et al. [65] |
| Microsatellite instability (MSI-H) | Genetic marker of Microsatellite Instability | Potential biomarker for immunotherapy response | Taniguchi et al. [62] and Chang et al. [66] |
| Schlafen 11 (SLFN11) | Liquid biopsy biomarker | Potential biomarker for the response on DNA damaging chemotherapy and PARP inhibition | Taniguchi et al. [62] and Zhang et al. [63] |
4.1. Biomarkers of Response to Immune Checkpoint Inhibitors in SCLC
Immunotherapy with ICI has markedly improved the treatment of various solid and hematologic malignancies, including lung cancer. ICIs target PD-1 and its ligand (PD-L1). Validated predictive biomarkers associated with a response to ICI include PD-L1 expression (in tumor or immune cells), TMB-H, and MSI-H status. Several ICIs have been approved for NSCLC, while for SCLC, two ICI (nivolumab and atezolizumab) obtained approvals from FDA in 2018 and 2019, respectively [67,68]. In contrast to NSCLC, no predictive biomarkers to ICI response (or resistance) in SCLC have been validated and approved. However, several studies confirmed a substantial therapeutic benefit of adding ICIs to conventional chemotherapy as a first-line treatment for extensive SCLC [69,70]. Facchinetti et al. summarized the results of four randomized trials involving >1500 patients with SCLC treated with combined ICI/chemotherapy vs. chemotherapy alone (platinum–etoposide). They found a slight (10%) but clinically significant improvement in survival outcomes of SCLC patients treated with a combination of chemotherapy and ICI [69]. The authors also highlighted an unmet need for proper predictive biomarkers for ICI. Findings from another systematic review conducted by Zhou et al. also supported the use of the combined treatment with ICI (durvalumab and atezolizumab) and etoposide-based chemotherapy as an optimal first-line treatment approach for patients with extensive-stage SCLC [71]. Chen et al. summarized the results of four clinical trials (>1500 patients with extensive stage SCLC, ES-SCLC), focusing on the efficacy of four different ICIs as a first-line treatment (atezolizumab, pembrolizumab, nivolumab, and durvalumab). They found that none of the ICIs was superior regarding overall and disease-free survival. However, durvalumab was superior to atezolizumab but with higher toxicity (immune-related adverse effects) [72]. Recently designed and ongoing clinical trials appear to be more promising, including predictive tissue-based testing before and after treatment (e.g., Tempus Sculptor Study) [73].
4.1.1. PD-L1 Expression in SCLC
A recently published systematic review with meta-analysis [74] analyzed PD-L1 expression in ~2800 SCLC samples reported in 27 studies. The overall PD-L1 expression was 26% with a favorable prognostic impact, although it did not reach statistical significance [74]. However, the results were heterogeneous, different cutoffs for the definition of PD-L1 positivity were used, and marked variability in subcellular localization of PD-L1 protein in cancer cells was also observed/assessed. A recently published study by Lang et al. revealed a lower (~10%) PD-L1 positivity in cancer cells. The stromal PD-L1 positivity was observed in ~60% of cases with a significant favorable impact on patients’ outcomes. Notably, the authors did not find a significant correlation between PD-L1 expression and molecular subtypes of SCLC [75]. Yu et al. explored PD-L1 expression in SCLC regarding the anatomic location (central vs. peripheral) and TTF-1 expression (positive vs. negative) [76]. They found a more prevalent PD-L1 expression in centrally located, TTF-1-positive SCLC [76].
In contrast to other studies, they found PD-L1 expression as an adverse prognostic factor in SCLC associated with vascular and lymphatic invasion [76]. In summary, variable PD-L1 expression has been reported in SCLC. However, its predictive value has not yet been established, so routine ICI treatment testing is not recommended.
4.1.2. Tumor Mutational Burden (TMB)
TMB is defined as the number of somatic mutations in cancer per megabase of interrogated genomic sequence [77]. TMB has been validated as a predictive biomarker to ICI in multiple studies, leading to the approval of pembrolizumab for all solid tumors with TMB-H, regardless of histotype (tumor-agnostic approach) [78]. One of the largest studies involving less common solid tumors was conducted by Shao et al. [79]. The study included 305 SCLC samples, of which 37% had TMB-H defined as ≥10 mutations/megabase. The study by Hellmann et al. (2018) involved 401 patients with ES-SCLC, out of which 211 (53%) had comprehensive molecular profiling completed. They found that 27% of tested samples had TMB-H with a significant impact on the therapeutic benefit of ICI, particularly the combination of nivolumab and ipilimumab [80]. Zhou et al. explored a small cohort of SCLC patients using next-generation sequencing (NGS). The median TMB was 21.7 mutations/Mb (range 9.3–55.9), and high TMB (defined as >21 mutations/Mb) was a good prognostic factor of OS [81]. Li et al. (2023) explored a small cohort of SCLC (n = 18), revealing TMB-H in 78% of cases using a threshold of seven mutations/megabase [53]. Based on the current evidence, a substantial proportion of SCLC cases may harbor TMB-H (regardless of the threshold for its definition). This feature may be used for treatment with ICIs.
4.1.3. Microsatellite Instability (MSI-H) in SCLC
Microsatellite instability (MSI-H) is a specific molecular genomic alteration with a hyper-mutable phenotype of a cell caused by the impaired DNA mismatch repair machinery (MMR). MSI-H is particularly common in colorectal cancer, a subset of endometrial, gastric, and upper urinary tract carcinomas, while other cancers are rarely affected by the MSI-H/MMR phenotype. From the clinical point of view, MSI-H/dMMR status predicts response to ICIs and is an approved biomarker for pembrolizumab therapy irrespective of histotype (tumor-agnostic approach) [82].
Merlo et al. published one of the earliest papers reporting MSI status in SCLC [83]. The authors found a frequent MSI-H in primary SCLC. However, later studies did not confirm these observations revealing a rare occurrence of MSI-H in lung cancers, but still higher in SCLC compared with NSCLC (~3% vs. 0.1%) [84]. The same study showed that MSI-H SCLC also had TMB-H [84]. A comprehensive molecular analysis of 21 SCLC cell lines revealed that MMR deficiency does not play a prominent role in the pathogenesis of SCLC [85]. Another study conducted by Yanagawa et al. (2021) revealed a lower (1.1%) prevalence of MSI-H in SCLC samples [86]. A large, comprehensive genomic profiling study of Chinese patients with SCLC revealed no MSI-H among 111 SCLC-tested cases [87]. Based on the available data, SCLC is characterized by a low prevalence of MSI-H; however, rare cases of MSI-H SCLC may also harbor concomitant TMB-H and are more likely to benefit from immunotherapeutic approaches.
4.1.4. Delta-Like Ligand 3 (DLL3)
Delta-like ligand 3 (DLL3) represents an inhibitory ligand of the Notch receptor whose overexpression on the surface of neoplastic neuroendocrine cells is associated with tumor progression [88]. The Notch signaling pathway is a highly conserved pathway involved in various developmental processes, including developing pulmonary neuroendocrine cells [89]. DLL3 expression is regulated by achaete-scute homolog 1 (ASCL1), a transcription factor required to develop pulmonary neuroendocrine cells properly. DLL3 is a powerful oncogenic driver in SCLC [90]. More than 80% of SCLC overexpress DLL3 protein in diffuse and homogenous patterns [89,91]. DLL3 expression has also been described in various extrapulmonary neuroendocrine neoplasms (particularly high-grade carcinomas), such as bladder, cervix, anus, prostate, and bile duct neuroendocrine carcinomas [88,89,92,93]. In contrast, its expression in neuroendocrine carcinomas of the breast was low [94]. DLL3 expression in neuroendocrine neoplasms is associated with tumor progression and poor clinical outcomes [88].
Although initial clinical trials (Phase II TRINITY Study; Phase III TAHOE Study) revealed poor to modest therapeutic effects of DLL3 inhibitor rovalpituzumab tesirine in patients with DLL3-positive SCLC [95,96], recent data with novel DLL3 inhibitors such as tarlatamab appear to be promising [97]. Tarlatamab (TMG 757) is a “first-in-class DLL3-targeted bispecific T-Cell engager” that binds to DLL3 and CD3 receptors, activating T-cell mediated tumor cell lysis. In the Phase I study conducted by Paz-Ares et al. (2023), tarlatamab exhibited encouraging therapeutic effects (objective response rate 23%) and an acceptable safety profile in heavily pretreated SCLC patients. Interestingly, DLL3 expression assessed by immunohistochemistry correlated well with the therapeutic responses [97].
5. SCLC Treatment and Approaches to SCLC Therapy
SCLC patient outcomes have not been substantially improved in the era of precision oncology. The treatment algorithm for SCLC is shown in Figure 2. However, systemic therapies, including immunotherapy, show promising results, although many patients do not respond well to the treatment and need alternative or complementary therapeutic approaches. Discrete molecular subcategories of SCLC differ in their responsiveness to a certain therapeutic approach, which opens new questions and directions in the therapies [57]. Landmark studies have been conducted to discover crucial drivers of drug response in cancer cells. One of the largest studies on SCLC cell lines investigated 526 chemical compounds on 63 SCLC cell lines to find promising candidates for new oncological drugs and therapeutic approaches. This extensive study showed that compounds targeting nuclear kinases appear effective in SCLC lines. However, additional investigations, including xenografts, are needed to elucidate their possible therapeutic effectiveness [98]. Many efforts are being invested in screening various compounds to unravel novel targets and biomarkers in SCLC [99]. In addition to synthetic compounds, many phytochemicals and other naturally occurring bioactive compounds like various polyphenols, alkaloids, terpenoids, thiols, and others are intensively investigated to design new anticancer strategies [100]. Different treatment strategies for SCLC, depending on the stage of the disease, are shown in Table 3. Considering the complex treatment algorithm for patients with lung cancer, which encompasses the pathological and molecular phenotype, it is crucial to present each patient to a multidisciplinary team (MDT). The main purpose of the MDT presentation is to facilitate a comprehensive and collaborative approach to the patient’s diagnosis, treatment, and supportive care.
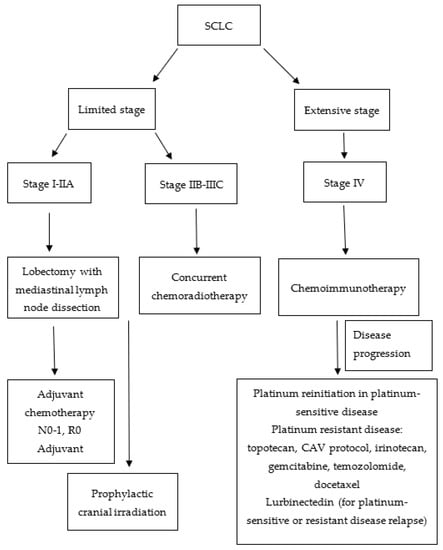
Figure 2.
Flowchart of small cell lung cancer (SCLC) treatment algorithm. CAV: cyclophosphamide, doxorubicin, vincristine.

Table 3.
Treatment strategies for patients with small cell lung carcinoma.
5.1. Limited Stage
Surgical treatment of SCLC for stages I-II due to the aggressiveness of the disease and detection of the disease in most cases at an advanced stage is carried out in a small number of patients. Surgical treatment of lobectomy with mediastinal dissection can be performed after extensive staging, in which stage I or II diseases are confirmed. Several retrospective studies have verified a five-year survival rate of 45–65% for stages I-II after operative treatment. For negative mediastinal lymph nodes and margins, adjuvant chemotherapy with cisplatin and etoposide is advised [5,101]. In the case of positive mediastinal lymph nodes or R1 (microscopic residual tumor) and R2 margins (macroscopic residual tumor), chemoradiotherapy is advised. Prophylactic cranial radiotherapy (PCI) is agreed upon with the patient due to conclusive data from the literature, where it is necessary to disclose the benefit and toxicity of PCI.
SCLC is a very chemosensitive tumor with a response rate of 70–90%, and the basis of treatment for most patients in the limited stage of the disease is chemoradiotherapy. The preferred chemotherapy regimen is cisplatin 75 mg/m2 day one and etoposide 100 to 120 mg/m2 day 1–3 combined with external radiotherapy in a total dose of 45 Gy twice daily (BID). The CONVERT study showed that in combination with chemotherapy, a total radiotherapy dose of 60 Gy once daily is not inferior to 45 Gy BID [70]. By improving disease staging and radiotherapy techniques, the five-year OS of patients treated with chemoradiotherapy was extended from 16% to 34% [102,103]. About 50% of patients have brain metastases, so after the chemoradiotherapy treatment, PCI is performed on the brain area in case of response to the therapy [5]. PCI reduces disease recurrence in the brain and improves three-year overall survival by 5.4% [104]. Due to the greater possibility of cognitive decline, if the patient is older than 70 years or in a worse general condition, ECOG 2 or more PCI is in agreement with the patient.
To improve the treatment of limited SCLC, studies are underway to incorporate immunotherapy with chemoradiotherapy and improve treatment outcomes. For now, immunotherapy and targeted therapy have no role in treating SCLC.
5.2. Extensive Stage
For the last 30 years, the mainstay of treatment for extensive SCLC (eSCLC) has been chemotherapy with cisplatin and etoposide, with a median survival of 7–11 months. The addition of immunotherapy with checkpoint inhibitors (CPI), atezolizumab (PD-L1 inhibitor), and durvalumab (PD-L1 inhibitor) in combination with basic platinum chemotherapy achieves a moderate but statistically significant increase in OS for two months, from 10 to 12 months [105,106]. Based on the above results, the basis of the treatment in the first line of eSCLC is chemoimmunotherapy with atezolizumab or durvalumab. After longer clinical follow-up, immunotherapy with atezolizumab or durvalumab can ensure long-term survival at 18 months, 34.0%, and at 36 months, 17.6% in patients with eSCLC [107,108]. New ICIs, such as serplulimab (PD-1 inhibitor) and adebrelimab (PD-L1 inhibitor) combined with chemotherapy, have shown benefit in the first-line treatment of eSCLC.
In contrast, pembrolizumab (PD-1 inhibitor) and ipilimumab (CTLA-4 inhibitor) have not shown benefit in RCTs [109,110,111,112]. In patients in whom immunotherapy is contraindicated or unavailable, the basis of treatment is carboplatin or cisplatin in combination with etoposide [5,101]. In the case of response to chemotherapy, treatment with consolidation radiotherapy of the lung and PCI or MRI brain surveillance is considered [5,101]. The best symptomatic supportive therapy is advised in patients with poor general conditions. In the second line of treatment, several cytostatics are available, and the choice of cytostatic depends on the patient’s general condition, the previous toxicity of the therapy, and the platinum-free interval. In the case of platinum-sensitive disease, reinitiation of platinum is recommended. In the case of platinum-resistant disease, patients can be treated with chemotherapy with a response rate of 15–30%. Topotecan, CAV (cyclophosphamide, doxorubicin, vincristine) protocol, irinotecan, gemcitabine, temozolomide, and docetaxel are used. Recently, lurbinectedin (inhibitor of DNA synthesis) has been approved, based on a good response rate of 35.2% in a phase II study in patients with platinum-sensitive or resistant disease relapse [113].
6. New Drugs in the Second Line or Beyond
New emerging therapeutic strategies for SCLC that are under investigation are Aurora A inhibitor, poly (ADP-ribose) polymerase (PARP) inhibitor, ATR kinase inhibitor, CHK1 inhibitor, DLL3 inhibitor, MYC inhibitor, Ganglioside fucosyl-GM1 and inhibitor of the bromodomain (BRD) and extra-terminal domain (BET) family of proteins [114]. Personalization of therapy in a patient with extensive SCLC and DLL3 ligand expression was attempted with rovalpituzumab tesirine, an antibody-drug conjugate (ADC) that destroys tumor cells by intracellular cytostatic deposition. In the Phase 1 study (protocol number NCT01901653), the activity of rovalpituzumab tesirine was evaluated in 82 patients with SCLC who had progressed to one or more lines of therapy. The key finding of the study indicated that 38% of the patients demonstrated a positive response to treatment with rovalpituzumab tesirine in patients with high DLL3 expression, defined as having ≥50% expression on tumor cells [115]. In the Phase II TRINITY study (protocol number: NCT02674568), rovalpituzumab tesirine demonstrated modest clinical activity among the 339 pretreated patients with SCLC, and the response rate for DLL3-positive patients was 13.2% [95]. In a randomized Phase 3 TAHOE study (protocol number: NCT03061812) involving patients with SCLC undergoing second-line therapy, rovalpituzumab tesirine showed inferior OS when compared to topotecan, with a median OS of 6.3 months versus 8.6 months, respectively [96]. The limitations of the Phase 1 and Phase 2 studies with rovalpituzumab tesirine are primarily attributed to their single-arm study designs, lacking a control comparator arm for direct comparison. Furthermore, when compared to the Phase 2 and Phase 3 studies with rovalpituzumab tesirine, the Phase 1 study’s limitations become apparent due to the selective inclusion of patients who may not be representative of the real-world population. For example, one of the key exclusion criteria in the Phase 1 study was the presence of active central nervous system (CNS) metastases [115]. In a different approach to DLL3 receptors, recent data with novel DLL3 inhibitors such as tarlatamab appear promising [97]. The Phase 1 study (protocol number NCT03319940) assessing the efficacy of tarlatamab in heavily pretreated SCLC patients has demonstrated promising results, with a response rate of 23.4% [97]. The Phase 1/2 study (protocol number NCT02446704) evaluating the combination of the PARP inhibitor olaparib with temozolomide in 50 patients with previously treated SCLC revealed a promising new therapeutic strategy, with an overall response rate of 41.7% [116]. In the Phase 2 study (protocol NCT02038647), the combination of the Aurora A kinase inhibitor alisertib with paclitaxel demonstrated efficacy signals in relapsed or refractory SCLC [117]. Additionally, c-Myc expression and mutations were identified as potential predictive biomarkers of alisertib. The disease control rate, defined as the combination of complete response, partial response, and stable disease lasting at least 8 weeks, was significantly higher with alisertib/paclitaxel compared to placebo/paclitaxel (55% versus 33%) in the subgroup of resistant or refractory patients efficacy [118]. Among c-Myc-positive patients, the median progression-free survival was 4.64 months with alisertib/paclitaxel, whereas it was 2.27 months with placebo/paclitaxel. In the phase II study (protocol number NCT02487095), the combination of M6620 (berzosertib), ataxia telangiectasia, and rad3-related inhibitor (ATR), with topotecan in 25 patients with relapsed SCLC, demonstrated a response rate of 36%, successfully meeting the primary efficacy endpoint. Intensive scientific and clinical efforts are being invested in unraveling new therapeutic options for SCLC to provide better responsiveness to this challenging malignancy.
7. Conclusions
SCLC represents a significant global health burden with high mortality rates. The discovery of therapeutic targets in SCLC has been challenging compared to NSCLC due to the high prevalence of untargetable driver mutations. Immune checkpoint inhibitors (ICI) combined with chemotherapy have shown significant benefits in treating SCLC. PD-L1 expression, tumor mutational burden, and microsatellite instability are validated predictive biomarkers for ICI response in various cancers, but their predictive value in SCLC remains uncertain. Although several emerging targets have been identified (e.g., DLL3), they failed to provide satisfactory therapeutic benefits. Therefore, further studies are needed to provide and validate novel therapeutic targets and biomarkers for this highly aggressive malignancy.
Author Contributions
Conceptualization, K.K., S.V., K.T. and L.B.; writing—original draft preparation, K.K., S.V., K.T., M.S. and L.B.; writing—review and editing, S.V. and L.B.; supervision, S.V., K.T. and L.B.; project administration, K.K.; funding acquisition, K.K., M.S., S.V. and L.B. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by the Croatian Thoracic Society.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.M.; Bubendorf, L.; Edelman, M.J.; Marchetti, A.; Mok, T.; Novello, S.; O’Byrne, K.; Stahel, R.; Peters, S.; Felip, E.; et al. Second ESMO consensus conference on lung cancer: Pathology and molecular biomarkers for non-small-cell lung cancer. Ann. Oncol. 2014, 25, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tang, J.; Sun, T.; Zheng, X.; Li, J.; Sun, H.; Zhou, X.; Zhou, C.; Zhang, H.; Cheng, Z.; et al. Survival changes in patients with small cell lung cancer and disparities between different sexes, socioeconomic statuses and ages. Sci. Rep. 2017, 7, 1339. [Google Scholar] [CrossRef]
- Varghese, A.M.; Zakowski, M.F.; Yu, H.A.; Won, H.H.; Riely, G.J.; Krug, L.M.; Kris, M.G.; Rekhtman, N.; Ladanyi, M.; Wang, L.; et al. Small-cell lung cancers in patients who never smoked cigarettes. J. Thorac. Oncol. 2014, 9, 892–896. [Google Scholar] [CrossRef]
- Dingemans, A.C.; Fruh, M.; Ardizzoni, A.; Besse, B.; Faivre-Finn, C.; Hendriks, L.E.; Lantuejoul, S.; Peters, S.; Reguart, N.; Rudin, C.M.; et al. Small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2021, 32, 839–853. [Google Scholar] [CrossRef]
- Wang, S.; Zimmermann, S.; Parikh, K.; Mansfield, A.S.; Adjei, A.A. Current Diagnosis and Management of Small-Cell Lung Cancer. Mayo Clin. Proc. 2019, 94, 1599–1622. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.D.; Detterbeck, F.C.; Yahalom, J. Clinical practice. Superior vena cava syndrome with malignant causes. N. Engl. J. Med. 2007, 356, 1862–1869. [Google Scholar] [CrossRef]
- Remon, J.; Facchinetti, F.; Besse, B. The efficacy of immune checkpoint inhibitors in thoracic malignancies. Eur. Respir. Rev. 2021, 30, 200387. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart; International Agency for Research on Cancer: Lyon, France, 2015. [Google Scholar]
- Quintanal-Villalonga, A.; Chan, J.M.; Yu, H.A.; Pe’er, D.; Sawyers, C.L.; Sen, T.; Rudin, C.M. Lineage plasticity in cancer: A shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 2020, 17, 360–371. [Google Scholar] [CrossRef]
- McFadden, D.G.; Papagiannakopoulos, T.; Taylor-Weiner, A.; Stewart, C.; Carter, S.L.; Cibulskis, K.; Bhutkar, A.; McKenna, A.; Dooley, A.; Vernon, A.; et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 2014, 156, 1298–1311. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, R.; Linn, S.C.; Linnoila, R.I.; Zevenhoven, J.; Mooi, W.J.; Berns, A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003, 4, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Maleki, Z. Diagnostic issues with cytopathologic interpretation of lung neoplasms displaying high-grade basaloid or neuroendocrine morphology. Diagn. Cytopathol. 2011, 39, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, S.A.; Beasley, M.B.; Brambilla, E.; Hasleton, P.S.; Colby, T.V.; Sheppard, M.N.; Falk, R.; Travis, W.D. Small cell lung carcinoma (SCLC): A clinicopathologic study of 100 cases with surgical specimens. Am. J. Surg. Pathol. 2002, 26, 1184–1197. [Google Scholar] [CrossRef] [PubMed]
- Kontogianni, K.; Nicholson, A.G.; Butcher, D.; Sheppard, M.N. CD56: A useful tool for the diagnosis of small cell lung carcinomas on biopsies with extensive crush artefact. J. Clin. Pathol. 2005, 58, 978–980. [Google Scholar] [CrossRef]
- Hiroshima, K.; Iyoda, A.; Shida, T.; Shibuya, K.; Iizasa, T.; Kishi, H.; Tanizawa, T.; Fujisawa, T.; Nakatani, Y. Distinction of pulmonary large cell neuroendocrine carcinoma from small cell lung carcinoma: A morphological, immunohistochemical, and molecular analysis. Mod. Pathol. 2006, 19, 1358–1368. [Google Scholar] [CrossRef]
- Bobos, M.; Hytiroglou, P.; Kostopoulos, I.; Karkavelas, G.; Papadimitriou, C.S. Immunohistochemical distinction between merkel cell carcinoma and small cell carcinoma of the lung. Am. J. Dermatopathol. 2006, 28, 99–104. [Google Scholar] [CrossRef]
- Mjones, P.; Sagatun, L.; Nordrum, I.S.; Waldum, H.L. Neuron-Specific Enolase as an Immunohistochemical Marker Is Better Than Its Reputation. J. Histochem. Cytochem. 2017, 65, 687–703. [Google Scholar] [CrossRef]
- Lee, M.C.; Cai, H.; Murray, C.W.; Li, C.; Shue, Y.T.; Andrejka, L.; He, A.L.; Holzem, A.M.E.; Drainas, A.P.; Ko, J.H.; et al. A multiplexed in vivo approach to identify driver genes in small cell lung cancer. Cell Rep. 2023, 42, 111990. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretic, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef]
- Nong, J.; Gong, Y.; Guan, Y.; Yi, X.; Yi, Y.; Chang, L.; Yang, L.; Lv, J.; Guo, Z.; Jia, H.; et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat. Commun. 2018, 9, 3114. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Wooten, D.J.; Groves, S.M.; Tyson, D.R.; Liu, Q.; Lim, J.S.; Albert, R.; Lopez, C.F.; Sage, J.; Quaranta, V. Systems-level network modeling of Small Cell Lung Cancer subtypes identifies master regulators and destabilizers. PLoS Comput. Biol. 2019, 15, e1007343. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wei, Y.; Hung, R.J.; Liu, G.; Su, L.; Zhang, R.; Zong, X.; Zhang, Z.F.; Morgenstern, H.; Bruske, I.; et al. Associated Links Among Smoking, Chronic Obstructive Pulmonary Disease, and Small Cell Lung Cancer: A Pooled Analysis in the International Lung Cancer Consortium. EBioMedicine 2015, 2, 1677–1685. [Google Scholar] [CrossRef]
- Wang, Q.; Gumus, Z.H.; Colarossi, C.; Memeo, L.; Wang, X.; Kong, C.Y.; Boffetta, P. SCLC: Epidemiology, Risk Factors, Genetic Susceptibility, Molecular Pathology, Screening, and Early Detection. J. Thorac. Oncol. 2023, 18, 31–46. [Google Scholar] [CrossRef]
- Zhou, F.; Zhou, C. Lung cancer in never smokers-the East Asian experience. Transl. Lung Cancer Res. 2018, 7, 450–463. [Google Scholar] [CrossRef]
- Ferone, G.; Lee, M.C.; Sage, J.; Berns, A. Cells of origin of lung cancers: Lessons from mouse studies. Genes Dev. 2020, 34, 1017–1032. [Google Scholar] [CrossRef]
- Cheng, D.T.; Mitchell, T.N.; Zehir, A.; Shah, R.H.; Benayed, R.; Syed, A.; Chandramohan, R.; Liu, Z.Y.; Won, H.H.; Scott, S.N.; et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. 2015, 17, 251–264. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Z.; Wei, S.; Li, B.; Zhao, Z. Genomic features of Chinese small cell lung cancer. BMC Med. Genom. 2022, 15, 117. [Google Scholar] [CrossRef]
- Chang, F.; Syrjanen, S.; Kurvinen, K.; Syrjanen, K. The p53 tumor suppressor gene as a common cellular target in human carcinogenesis. Am. J. Gastroenterol. 1993, 88, 174–186. [Google Scholar]
- Febres-Aldana, C.A.; Chang, J.C.; Ptashkin, R.; Wang, Y.; Gedvilaite, E.; Baine, M.K.; Travis, W.D.; Ventura, K.; Bodd, F.; Yu, H.A.; et al. Rb Tumor Suppressor in Small Cell Lung Cancer: Combined Genomic and IHC Analysis with a Description of a Distinct Rb-Proficient Subset. Clin. Cancer Res. 2022, 28, 4702–4713. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, Y.; Zhang, Y.; Chen, L.H.; Ouyang, H.F.; Xu, X.; Du, Y.; Ti, X.Y. Mutational landscape of homologous recombination-related genes in small-cell lung cancer. Cancer Med. 2023, 12, 4486–4495. [Google Scholar] [CrossRef] [PubMed]
- Simbolo, M.; Mafficini, A.; Sikora, K.O.; Fassan, M.; Barbi, S.; Corbo, V.; Mastracci, L.; Rusev, B.; Grillo, F.; Vicentini, C.; et al. Lung neuroendocrine tumours: Deep sequencing of the four World Health Organization histotypes reveals chromatin-remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. J. Pathol. 2017, 241, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Augert, A.; Zhang, Q.; Bates, B.; Cui, M.; Wang, X.; Wildey, G.; Dowlati, A.; MacPherson, D. Small Cell Lung Cancer Exhibits Frequent Inactivating Mutations in the Histone Methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J. Thorac. Oncol. 2017, 12, 704–713. [Google Scholar] [CrossRef]
- Carazo, F.; Bertolo, C.; Castilla, C.; Cendoya, X.; Campuzano, L.; Serrano, D.; Gimeno, M.; Planes, F.J.; Pio, R.; Montuenga, L.M.; et al. DrugSniper, a Tool to Exploit Loss-Of-Function Screens, Identifies CREBBP as a Predictive Biomarker of VOLASERTIB in Small Cell Lung Carcinoma (SCLC). Cancers 2020, 12, 1824. [Google Scholar] [CrossRef]
- Jia, D.; Augert, A.; Kim, D.W.; Eastwood, E.; Wu, N.; Ibrahim, A.H.; Kim, K.B.; Dunn, C.T.; Pillai, S.P.S.; Gazdar, A.F.; et al. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov. 2018, 8, 1422–1437. [Google Scholar] [CrossRef]
- Sivakumar, S.; Moore, J.A.; Montesion, M.; Sharaf, R.; Lin, D.I.; Colon, C.I.; Fleishmann, Z.; Ebot, E.M.; Newberg, J.Y.; Mills, J.M.; et al. Integrative Analysis of a Large Real-World Cohort of Small Cell Lung Cancer Identifies Distinct Genetic Subtypes and Insights into Histologic Transformation. Cancer Discov. 2023, 13, 1572–1591. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Zhang, B.; Zheng, J.; Singh, P.K.; Bshara, W.; Wang, J.; Gomez, E.C.; Zhang, X.; Wang, Y.; et al. PTEN Loss Expands the Histopathologic Diversity and Lineage Plasticity of Lung Cancers Initiated by Rb1/Trp53 Deletion. J. Thorac. Oncol. 2023, 18, 324–338. [Google Scholar] [CrossRef]
- Yin, J.; Cong, X.; Cui, P.; Zhao, S.; Liu, Z. Repeatedly next-generation sequencing during treatment follow-up of patients with small cell lung cancer. Medicine 2023, 102, e34143. [Google Scholar] [CrossRef]
- Pop-Bica, C.; Ciocan, C.A.; Braicu, C.; Harangus, A.; Simon, M.; Nutu, A.; Pop, L.A.; Slaby, O.; Atanasov, A.G.; Pirlog, R.; et al. Next-Generation Sequencing in Lung Cancer Patients: A Comparative Approach in NSCLC and SCLC Mutational Landscapes. J. Pers. Med. 2022, 12, 453. [Google Scholar] [CrossRef]
- Hung, M.C.; Wang, W.P.; Chi, Y.H. AKT phosphorylation as a predictive biomarker for PI3K/mTOR dual inhibition-induced proteolytic cleavage of mTOR companion proteins in small cell lung cancer. Cell Biosci. 2022, 12, 122. [Google Scholar] [CrossRef]
- Li, W.; Ye, L.; Huang, Y.; Zhou, F.; Wu, C.; Wu, F.; He, Y.; Li, X.; Wang, H.; Xiong, A.; et al. Characteristics of Notch signaling pathway and its correlation with immune microenvironment in SCLC. Lung Cancer 2022, 167, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Roper, N.; Velez, M.J.; Chiappori, A.; Kim, Y.S.; Wei, J.S.; Sindiri, S.; Takahashi, N.; Mulford, D.; Kumar, S.; Ylaya, K.; et al. Notch signaling and efficacy of PD-1/PD-L1 blockade in relapsed small cell lung cancer. Nat. Commun. 2021, 12, 3880. [Google Scholar] [CrossRef] [PubMed]
- Herbreteau, G.; Langlais, A.; Greillier, L.; Audigier-Valette, C.; Uwer, L.; Hureaux, J.; Moro-Sibilot, D.; Guisier, F.; Carmier, D.; Madelaine, J.; et al. Circulating Tumor DNA as a Prognostic Determinant in Small Cell Lung Cancer Patients Receiving Atezolizumab. J. Clin. Med. 2020, 9, 3861. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.S.; Wang, K.; Elkadi, O.R.; Tarasen, A.; Foulke, L.; Sheehan, C.E.; Otto, G.A.; Palmer, G.; Yelensky, R.; Lipson, D.; et al. Next-generation sequencing reveals frequent consistent genomic alterations in small cell undifferentiated lung cancer. J. Clin. Pathol. 2014, 67, 772–776. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, E.; Shinohara, T.; Mori, N.; Yokota, J.; Tani, K.; Izumi, K.; Obashi, A.; Ogura, T. Loss of heterozygosity on chromosome arm 17p in small cell lung carcinomas, but not in neurofibromas, in a patient with von Recklinghausen neurofibromatosis. Cancer 1993, 71, 725–728. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, L.; Sun, X.; Zhou, F.; Zhang, Y.; Zhao, S.; Guo, T.; Sun, Z.; Li, F.; Su, W.; et al. Genetic Mutation Analysis in Small Cell Lung Cancer by a Novel NGS-Based Targeted Resequencing Gene Panel and Relation with Clinical Features. Biomed. Res. Int. 2021, 2021, 3609028. [Google Scholar] [CrossRef]
- Grote, H.J.; Schmiemann, V.; Kiel, S.; Bocking, A.; Kappes, R.; Gabbert, H.E.; Sarbia, M. Aberrant methylation of the adenomatous polyposis coli promoter 1A in bronchial aspirates from patients with suspected lung cancer. Int. J. Cancer 2004, 110, 751–755. [Google Scholar] [CrossRef]
- Ding, J.; Leng, Z.; Gu, H.; Jing, X.; Song, Y. Etoposide/platinum plus anlotinib for patients with transformed small-cell lung cancer from EGFR-mutant lung adenocarcinoma after EGFR-TKI resistance: A retrospective and observational study. Front. Oncol. 2023, 13, 1153131. [Google Scholar] [CrossRef]
- Hao, L.; Chen, H.; Wang, L.; Zhou, H.; Zhang, Z.; Han, J.; Hou, J.; Zhu, Y.; Zhang, H.; Wang, Q. Transformation or tumor heterogeneity: Mutations in EGFR, SOX2, TP53, and RB1 persist in the histological rapid conversion from lung adenocarcinoma to small-cell lung cancer. Thorac. Cancer 2023, 14, 1036–1041. [Google Scholar] [CrossRef]
- Otegui, N.; Houry, M.; Arozarena, I.; Serrano, D.; Redin, E.; Exposito, F.; Leon, S.; Valencia, K.; Montuenga, L.; Calvo, A. Cancer Cell-Intrinsic Alterations Associated with an Immunosuppressive Tumor Microenvironment and Resistance to Immunotherapy in Lung Cancer. Cancers 2023, 15, 3076. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, Y.; Xu, Y.; Li, K.; Song, L.; Xue, Y.; Dong, D. Comparative study of the genomic landscape and tumor microenvironment among large cell carcinoma of the lung, large cell neuroendocrine of the lung, and small cell lung cancer. Medicine 2023, 102, e32781. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Thompson, J.; Fisher, H.; Zhang, P.; Huang, C.C.; Wang, L. Genomic alterations of plasma cell-free DNAs in small cell lung cancer and their clinical relevance. Lung Cancer 2018, 120, 113–121. [Google Scholar] [CrossRef]
- Devarakonda, S.; Sankararaman, S.; Herzog, B.H.; Gold, K.A.; Waqar, S.N.; Ward, J.P.; Raymond, V.M.; Lanman, R.B.; Chaudhuri, A.A.; Owonikoko, T.K.; et al. Circulating Tumor DNA Profiling in Small-Cell Lung Cancer Identifies Potentially Targetable Alterations. Clin. Cancer Res. 2019, 25, 6119–6126. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Takahashi, K.; Nagayama, K.; Arai, Y.; Ito, N.; Okada, M.; Minna, J.D.; Yokota, J.; Kohno, T. Identification of chromosome arm 9p as the most frequent target of homozygous deletions in lung cancer. Genes Chromosomes Cancer 2005, 44, 405–414. [Google Scholar] [CrossRef]
- Rudin, C.M.; Poirier, J.T.; Byers, L.A.; Dive, C.; Dowlati, A.; George, J.; Heymach, J.V.; Johnson, J.E.; Lehman, J.M.; MacPherson, D.; et al. Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 2019, 19, 289–297. [Google Scholar] [CrossRef]
- Chen, B.; Li, H.; Liu, C.; Wang, S.; Zhang, F.; Zhang, L.; Li, M.; Li, G. Potential prognostic value of delta-like protein 3 in small cell lung cancer: A meta-analysis. World J. Surg. Oncol. 2020, 18, 226. [Google Scholar] [CrossRef]
- Roumeliotou, A.; Pantazaka, E.; Xagara, A.; Dimitrakopoulos, F.I.; Koutras, A.; Christopoulou, A.; Kourelis, T.; Aljarba, N.H.; Alkahtani, S.; Koinis, F.; et al. Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients. Cancers 2022, 15, 171. [Google Scholar] [CrossRef]
- Almodovar, K.; Iams, W.T.; Meador, C.B.; Zhao, Z.; York, S.; Horn, L.; Yan, Y.; Hernandez, J.; Chen, H.; Shyr, Y.; et al. Longitudinal Cell-Free DNA Analysis in Patients with Small Cell Lung Cancer Reveals Dynamic Insights into Treatment Efficacy and Disease Relapse. J. Thorac. Oncol. 2018, 13, 112–123. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, C.; Wang, K.; Teng, X.; Chen, M. Identifying diagnostic markers and constructing a prognostic model for small-cell lung cancer based on blood exosome-related genes and machine-learning methods. Front. Oncol. 2022, 12, 1077118. [Google Scholar] [CrossRef]
- Taniguchi, H.; Sen, T.; Rudin, C.M. Targeted Therapies and Biomarkers in Small Cell Lung Cancer. Front. Oncol. 2020, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Stewart, C.A.; Wang, Q.; Cardnell, R.J.; Rocha, P.; Fujimoto, J.; Solis Soto, L.M.; Wang, R.; Novegil, V.; Ansell, P.; et al. Dynamic expression of Schlafen 11 (SLFN11) in circulating tumour cells as a liquid biomarker in small cell lung cancer. Br. J. Cancer 2022, 127, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Keogh, A.; Finn, S.; Radonic, T. Emerging Biomarkers and the Changing Landscape of Small Cell Lung Cancer. Cancers 2022, 14, 3772. [Google Scholar] [CrossRef]
- Li, L.; Chen, C.; Liu, C.; Niu, L.; Pan, C. Clinical and molecular impacts of tumor mutational burden in histological and cytological specimens from cancer patients. Ann. Transl. Med. 2022, 10, 214. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Chang, M.; Chang, H.M.; Chang, F. Microsatellite Instability: A Predictive Biomarker for Cancer Immunotherapy. Appl. Immunohistochem. Mol. Morphol. 2018, 26, e15–e21. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves Atezolizumab for Extensive-Stage Small Cell Lung Cancer. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-atezolizumab-extensive-stage-small-cell-lung-cancer (accessed on 27 May 2023).
- FDA Grants Nivolumab Accelerated Approval for Third-Line Treatment of Metastatic Small Cell Lung Cancer. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-nivolumab-accelerated-approval-third-line-treatment-metastatic-small-cell-lung-cancer (accessed on 27 May 2023).
- Facchinetti, F.; Di Maio, M.; Tiseo, M. Adding PD-1/PD-L1 Inhibitors to Chemotherapy for the First-Line Treatment of Extensive Stage Small Cell Lung Cancer (SCLC): A Meta-Analysis of Randomized Trials. Cancers 2020, 12, 2645. [Google Scholar] [CrossRef]
- Wang, B.C.; Xiao, B.Y.; Li, P.C.; Kuang, B.H.; Chen, W.B.; Li, P.D.; Lin, G.H.; Liu, Q. Efficacy and Safety of First-Line Immunotherapy in Combination with Chemotherapy for Patients with Extensive-Stage Small Cell Lung Cancer: A Systematic Review and Network Meta-Analysis. J. Oncol. 2020, 2020, 2368164. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, Z.; Luo, F.; Zhao, Y.; Hou, X.; Liu, T.; Wang, K.; Zhao, H.; Huang, Y.; Zhang, L. Comparison of First-Line Treatments for Patients With Extensive-Stage Small Cell Lung Cancer: A Systematic Review and Network Meta-analysis. JAMA Netw. Open 2020, 3, e2015748. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Xu, H. Comparison of atezolizumab, durvalumab, pembrolizumab, and nivolumab as first-line treatment in patients with extensive-stage small cell lung cancer: A systematic review and network meta-analysis. Medicine 2021, 100, e25180. [Google Scholar] [CrossRef]
- Tempus Sculptor Study: Small Cell Lung Cancer (SCLC) Observational Study. Available online: https://clinicaltrials.gov/ct2/show/NCT05257551 (accessed on 29 May 2023).
- Acheampong, E.; Abed, A.; Morici, M.; Spencer, I.; Beasley, A.B.; Bowyer, S.; Asante, D.B.; Lomma, C.; Lin, W.; Millward, M.; et al. Evaluation of PD-L1 expression on circulating tumour cells in small-cell lung cancer. Transl. Lung Cancer Res. 2022, 11, 440–451. [Google Scholar] [CrossRef]
- Lang, C.; Lantos, A.; Megyesfalvi, Z.; Egger, F.; Hoda, M.A.; Mosleh, B.; Klikovits, T.; Oberndorfer, F.; Timelthaler, G.; Ferencz, B.; et al. Clinical and prognostic implications of CD47 and PD-L1 expression in surgically resected small-cell lung cancer. ESMO Open 2022, 7, 100631. [Google Scholar] [CrossRef]
- Yu, S.; Jia, M.; Li, Y.; Sun, P.L.; Gao, H. Differential Expression of PD-L1 in Central and Peripheral and TTF1-Positive and -Negative Small-Cell Lung Cancer. Front. Med. 2020, 7, 621838. [Google Scholar] [CrossRef]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves Pembrolizumab for Adults and Children with TMB-H Solid Tumors. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors (accessed on 19 March 2023).
- Shao, C.; Li, G.; Huang, L.; Pruitt, S.; Castellanos, E.; Frampton, G.; Carson, K.R.; Snow, T.; Singal, G.; Fabrizio, D.; et al. Prevalence of High Tumor Mutational Burden and Association With Survival in Patients With Less Common Solid Tumors. JAMA Netw. Open 2020, 3, e2025109. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Callahan, M.K.; Awad, M.M.; Calvo, E.; Ascierto, P.A.; Atmaca, A.; Rizvi, N.A.; Hirsch, F.R.; Selvaggi, G.; Szustakowski, J.D.; et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018, 33, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Fan, J.; Li, Z.; Li, P.; Sun, Y.; Yang, Y.; Zhou, X.; Wang, J.; Wang, Y.; Qi, H.; et al. Prognostic impact of tumor mutation burden and the mutation in KIAA1211 in small cell lung cancer. Respir. Res. 2019, 20, 248. [Google Scholar] [CrossRef]
- FDA Grants Accelerated Approval to Pembrolizumab for First Tissue/Site Agnostic Indication. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-pembrolizumab-first-tissuesite-agnostic-indication (accessed on 19 March 2023).
- Merlo, A.; Gabrielson, E.; Mabry, M.; Vollmer, R.; Baylin, S.B.; Sidransky, D. Homozygous deletion on chromosome 9p and loss of heterozygosity on 9q, 6p, and 6q in primary human small cell lung cancer. Cancer Res. 1994, 54, 2322–2326. [Google Scholar]
- Lin, W.; Chen, R.; Yi, Y.; Lin, R.; Zhao, J.; Yuan, M.; Guan, Y.; Xia, X. P2.04-54 Characteristic of MSI-H Lung Cancer Patients Identified with Targeted Next-Generation Sequencing. J. Thorac. Oncol. 2019, 14, S729. [Google Scholar] [CrossRef]
- Hansen, L.T.; Thykjaer, T.; Orntoft, T.F.; Rasmussen, L.J.; Keller, P.; Spang-Thomsen, M.; Edmonston, T.B.; Schmutte, C.; Fishel, R.; Petersen, L.N. The role of mismatch repair in small-cell lung cancer cells. Eur. J. Cancer 2003, 39, 1456–1467. [Google Scholar] [CrossRef]
- Yanagawa, N.; Yamada, N.; Sugimoto, R.; Osakabe, M.; Uesugi, N.; Shiono, S.; Endoh, M.; Ogata, S.Y.; Saito, H.; Maemondo, M.; et al. The Frequency of DNA Mismatch Repair Deficiency Is Very Low in Surgically Resected Lung Carcinoma. Front. Oncol. 2021, 11, 752005. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Zhang, Y.; Yu, Y.; Chen, H.; Liu, K.; Yao, M.; Wang, K.; Gu, W.; Shou, T. Comprehensive genomic profiling of small cell lung cancer in Chinese patients and the implications for therapeutic potential. Cancer Med. 2019, 8, 4338–4347. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Bergsland, E.; Aggarwal, R.; Aparicio, A.; Beltran, H.; Crabtree, J.S.; Hann, C.L.; Ibrahim, T.; Byers, L.A.; Sasano, H.; et al. DLL3 as an Emerging Target for the Treatment of Neuroendocrine Neoplasms. Oncologist 2022, 27, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.H.; Giffin, M.J.; Bailis, J.M.; Smit, M.D.; Carbone, D.P.; He, K. DLL3: An emerging target in small cell lung cancer. J. Hematol. Oncol. 2019, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Augustyn, A.; Borromeo, M.; Wang, T.; Fujimoto, J.; Shao, C.; Dospoy, P.D.; Lee, V.; Tan, C.; Sullivan, J.P.; Larsen, J.E.; et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 14788–14793. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.; Di Stefano, I.; Poma, A.M.; Ricci, S.; Proietti, A.; Davini, F.; Lucchi, M.; Melfi, F.; Fontanini, G. Prevalence of Delta-Like Protein 3 in a Consecutive Series of Surgically Resected Lung Neuroendocrine Neoplasms. Front. Oncol. 2021, 11, 729765. [Google Scholar] [CrossRef]
- Cimic, A.; Vranic, S.; Arguello, D.; Contreras, E.; Gatalica, Z.; Swensen, J. Molecular Profiling Reveals Limited Targetable Biomarkers in Neuroendocrine Carcinoma of the Cervix. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 299–304. [Google Scholar] [CrossRef]
- Hwang, J.; McGrath, J.; Lozada, J.R.; Brodskiy, P.; Xiu, J.; Wei, S.; Heath, E.I.; Carneiro, B.A.; Lou, E.; Soares, H.P.; et al. Molecular correlates of Delta-like-ligand 3 (DLL3) expression in neuroendocrine neoplasms (NENs). J. Clin. Oncol. 2022, 40, 4127. [Google Scholar] [CrossRef]
- Vranic, S.; Palazzo, J.; Sanati, S.; Florento, E.; Contreras, E.; Xiu, J.; Swensen, J.; Gatalica, Z. Potential Novel Therapy Targets in Neuroendocrine Carcinomas of the Breast. Clin. Breast Cancer 2019, 19, 131–136. [Google Scholar] [CrossRef]
- Morgensztern, D.; Besse, B.; Greillier, L.; Santana-Davila, R.; Ready, N.; Hann, C.L.; Glisson, B.S.; Farago, A.F.; Dowlati, A.; Rudin, C.M.; et al. Efficacy and Safety of Rovalpituzumab Tesirine in Third-Line and Beyond Patients with DLL3-Expressing, Relapsed/Refractory Small-Cell Lung Cancer: Results From the Phase II TRINITY Study. Clin. Cancer Res. 2019, 25, 6958–6966. [Google Scholar] [CrossRef]
- Blackhall, F.; Jao, K.; Greillier, L.; Cho, B.C.; Penkov, K.; Reguart, N.; Majem, M.; Nackaerts, K.; Syrigos, K.; Hansen, K.; et al. Efficacy and Safety of Rovalpituzumab Tesirine Compared With Topotecan as Second-Line Therapy in DLL3-High SCLC: Results From the Phase 3 TAHOE Study. J. Thorac. Oncol. 2021, 16, 1547–1558. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Champiat, S.; Lai, W.V.; Izumi, H.; Govindan, R.; Boyer, M.; Hummel, H.D.; Borghaei, H.; Johnson, M.L.; Steeghs, N.; et al. Tarlatamab, a First-in-Class DLL3-Targeted Bispecific T-Cell Engager, in Recurrent Small-Cell Lung Cancer: An Open-Label, Phase I Study. J. Clin. Oncol. 2023, 41, 2893–2903. [Google Scholar] [CrossRef] [PubMed]
- Polley, E.; Kunkel, M.; Evans, D.; Silvers, T.; Delosh, R.; Laudeman, J.; Ogle, C.; Reinhart, R.; Selby, M.; Connelly, J.; et al. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J. Natl. Cancer Inst. 2016, 108, djw122. [Google Scholar] [CrossRef] [PubMed]
- Miri, M.R.; Zare, A.; Saberzadeh, J.; Baghban, N.; Nabipour, I.; Tamadon, A. Anti-lung Cancer Marine Compounds: A Review. Ther. Innov. Regul. Sci. 2022, 56, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.P.; Kumar, V.B.; Ponnusamy, V.K.; Mai, T.T.T.; Nhat, P.T.; Brindhadevi, K.; Pugazhendhi, A. Phytochemicals intended for anticancer effects at preclinical levels to clinical practice: Assessment of formulations at nanoscale for non-small cell lung cancer (NSCLC) therapy. Process Biochem. 2021, 104, 55–75. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; DeCamp, M.; et al. National Comprehensive Cancer Network. Small Cell Lung Cancer. version 3.2023. J. Natl. Compr. Canc. Netw. 2023, 21, 340–350. [Google Scholar] [CrossRef]
- Faivre-Finn, C.; Snee, M.; Ashcroft, L.; Appel, W.; Barlesi, F.; Bhatnagar, A.; Bezjak, A.; Cardenal, F.; Fournel, P.; Harden, S.; et al. Concurrent once-daily versus twice-daily chemoradiotherapy in patients with limited-stage small-cell lung cancer (CONVERT): An open-label, phase 3, randomised, superiority trial. Lancet Oncol. 2017, 18, 1116–1125. [Google Scholar] [CrossRef]
- Turrisi, A.T., 3rd; Kim, K.; Blum, R.; Sause, W.T.; Livingston, R.B.; Komaki, R.; Wagner, H.; Aisner, S.; Johnson, D.H. Twice-daily compared with once-daily thoracic radiotherapy in limited small-cell lung cancer treated concurrently with cisplatin and etoposide. N. Engl. J. Med. 1999, 340, 265–271. [Google Scholar] [CrossRef]
- Auperin, A.; Arriagada, R.; Pignon, J.P.; Le Pechoux, C.; Gregor, A.; Stephens, R.J.; Kristjansen, P.E.; Johnson, B.E.; Ueoka, H.; Wagner, H.; et al. Prophylactic cranial irradiation for patients with small-cell lung cancer in complete remission. Prophylactic Cranial Irradiation Overview Collaborative Group. N. Engl. J. Med. 1999, 341, 476–484. [Google Scholar] [CrossRef]
- Horn, L.; Mansfield, A.S.; Szczesna, A.; Havel, L.; Krzakowski, M.; Hochmair, M.J.; Huemer, F.; Losonczy, G.; Johnson, M.L.; Nishio, M.; et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2220–2229. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Dvorkin, M.; Chen, Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Ozguroglu, M.; Ji, J.H.; et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): A randomised, controlled, open-label, phase 3 trial. Lancet 2019, 394, 1929–1939. [Google Scholar] [CrossRef]
- Liu, S.V.; Reck, M.; Mansfield, A.S.; Mok, T.; Scherpereel, A.; Reinmuth, N.; Garassino, M.C.; De Castro Carpeno, J.; Califano, R.; Nishio, M.; et al. Updated Overall Survival and PD-L1 Subgroup Analysis of Patients With Extensive-Stage Small-Cell Lung Cancer Treated With Atezolizumab, Carboplatin, and Etoposide (IMpower133). J. Clin. Oncol. 2021, 39, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, L.; Chen, Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Ozguroglu, M.; Ji, J.H.; Garassino, M.C.; et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open 2022, 7, 100408. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Han, L.; Wu, L.; Chen, J.; Sun, H.; Wen, G.; Ji, Y.; Dvorkin, M.; Shi, J.; Pan, Z.; et al. Effect of First-Line Serplulimab vs Placebo Added to Chemotherapy on Survival in Patients With Extensive-Stage Small Cell Lung Cancer: The ASTRUM-005 Randomized Clinical Trial. JAMA 2022, 328, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, C.; Yao, W.; Wang, Q.; Min, X.; Chen, G.; Xu, X.; Li, X.; Xu, F.; Fang, Y.; et al. Adebrelimab or placebo plus carboplatin and etoposide as first-line treatment for extensive-stage small-cell lung cancer (CAPSTONE-1): A multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 739–747. [Google Scholar] [CrossRef]
- Rudin, C.M.; Awad, M.M.; Navarro, A.; Gottfried, M.; Peters, S.; Csoszi, T.; Cheema, P.K.; Rodriguez-Abreu, D.; Wollner, M.; Yang, J.C.; et al. Pembrolizumab or Placebo Plus Etoposide and Platinum as First-Line Therapy for Extensive-Stage Small-Cell Lung Cancer: Randomized, Double-Blind, Phase III KEYNOTE-604 Study. J. Clin. Oncol. 2020, 38, 2369–2379. [Google Scholar] [CrossRef]
- Reck, M.; Luft, A.; Szczesna, A.; Havel, L.; Kim, S.W.; Akerley, W.; Pietanza, M.C.; Wu, Y.L.; Zielinski, C.; Thomas, M.; et al. Phase III Randomized Trial of Ipilimumab Plus Etoposide and Platinum Versus Placebo Plus Etoposide and Platinum in Extensive-Stage Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3740–3748. [Google Scholar] [CrossRef]
- Trigo, J.; Subbiah, V.; Besse, B.; Moreno, V.; Lopez, R.; Sala, M.A.; Peters, S.; Ponce, S.; Fernandez, C.; Alfaro, V.; et al. Lurbinectedin as second-line treatment for patients with small-cell lung cancer: A single-arm, open-label, phase 2 basket trial. Lancet Oncol. 2020, 21, 645–654. [Google Scholar] [CrossRef]
- Petty, W.J.; Paz-Ares, L. Emerging Strategies for the Treatment of Small Cell Lung Cancer: A Review. JAMA Oncol. 2023, 9, 419–429. [Google Scholar] [CrossRef]
- Rudin, C.M.; Pietanza, M.C.; Bauer, T.M.; Ready, N.; Morgensztern, D.; Glisson, B.S.; Byers, L.A.; Johnson, M.L.; Burris, H.A., 3rd; Robert, F.; et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: A first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017, 18, 42–51. [Google Scholar] [CrossRef]
- Farago, A.F.; Yeap, B.Y.; Stanzione, M.; Hung, Y.P.; Heist, R.S.; Marcoux, J.P.; Zhong, J.; Rangachari, D.; Barbie, D.A.; Phat, S.; et al. Combination Olaparib and Temozolomide in Relapsed Small-Cell Lung Cancer. Cancer Discov. 2019, 9, 1372–1387. [Google Scholar] [CrossRef]
- Owonikoko, T.K.; Niu, H.; Nackaerts, K.; Csoszi, T.; Ostoros, G.; Mark, Z.; Baik, C.; Joy, A.A.; Chouaid, C.; Jaime, J.C.; et al. Randomized Phase II Study of Paclitaxel plus Alisertib versus Paclitaxel plus Placebo as Second-Line Therapy for SCLC: Primary and Correlative Biomarker Analyses. J. Thorac. Oncol. 2020, 15, 274–287. [Google Scholar] [CrossRef] [PubMed]
- Stefani, A.; Piro, G.; Schietroma, F.; Strusi, A.; Vita, E.; Fiorani, S.; Barone, D.; Monaca, F.; Sparagna, I.; Valente, G.; et al. Unweaving the mitotic spindle: A focus on Aurora kinase inhibitors in lung cancer. Front. Oncol. 2022, 12, 1026020. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).