Metabolism and Chemical Degradation of New Antidiabetic Drugs (Part II): A Review of Analytical Approaches for Analysis of Gliptins
Abstract
1. Introduction
Drug Metabolism and Drug Degradation Overlapping
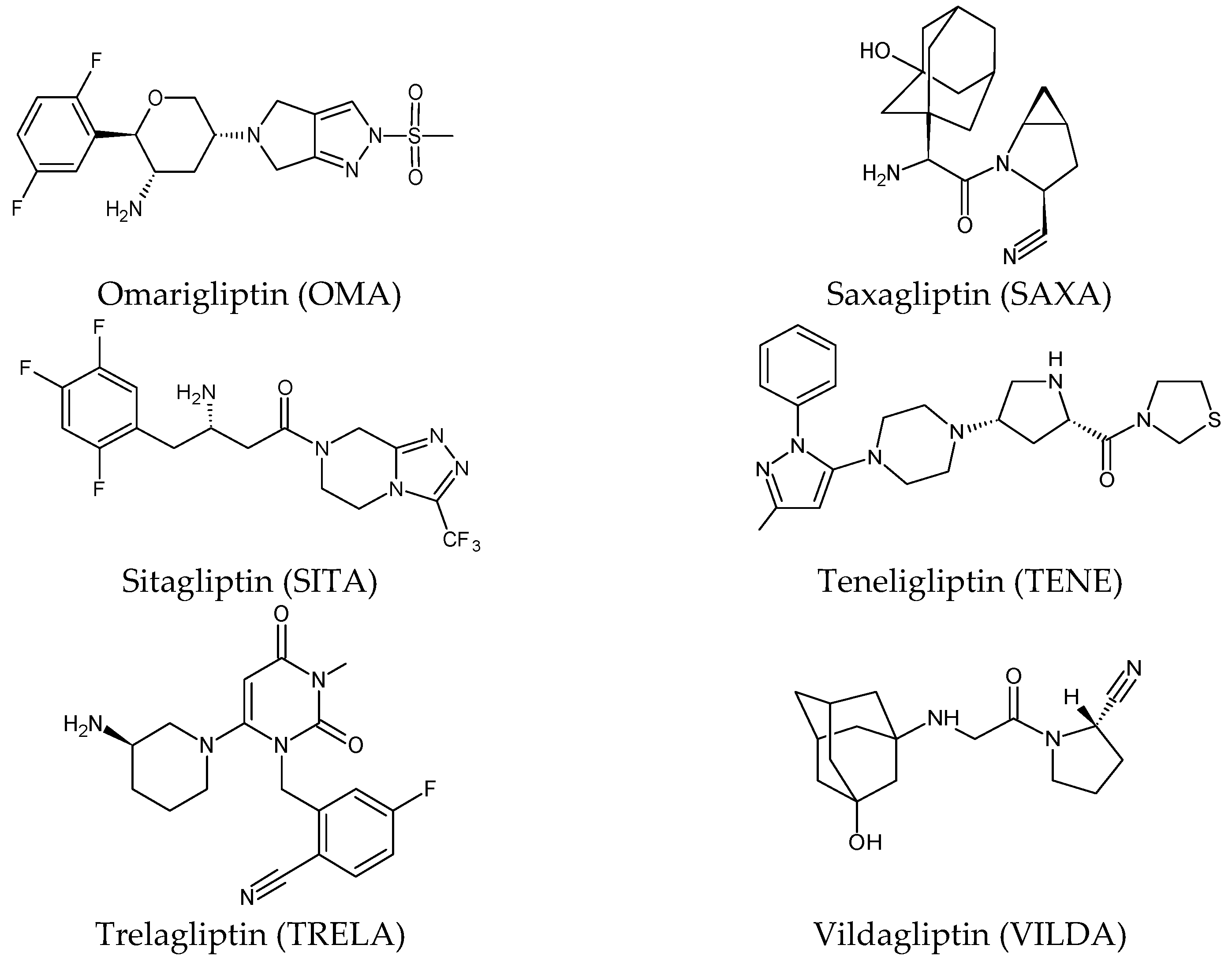
2. Gliptins (DPP-4 Inhibitors)
2.1. Metabolic Transformations of Gliptins (DPP-4 Inhibitors) and the Methods Used for Elucidating Their Metabolic Pathways
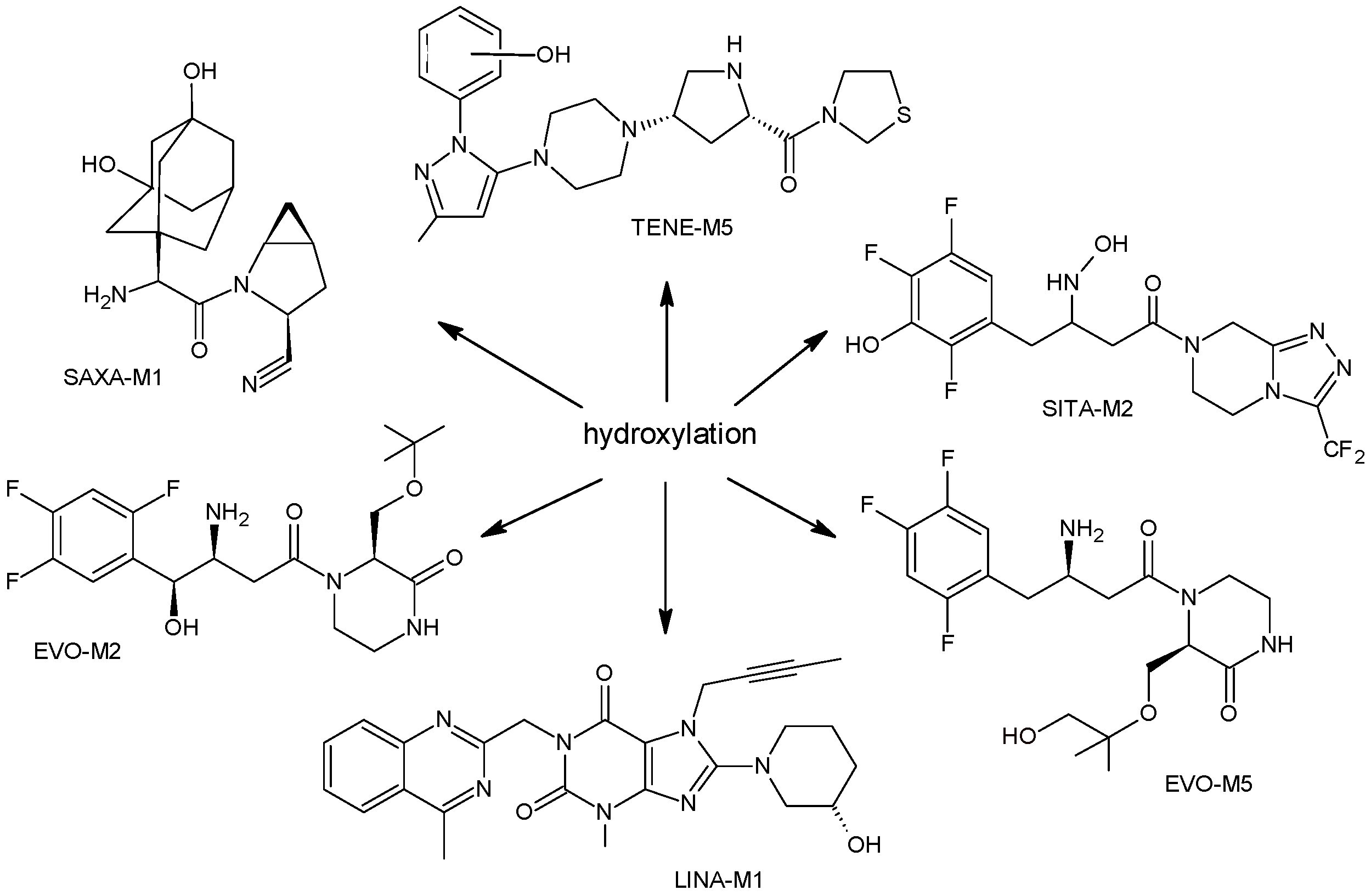
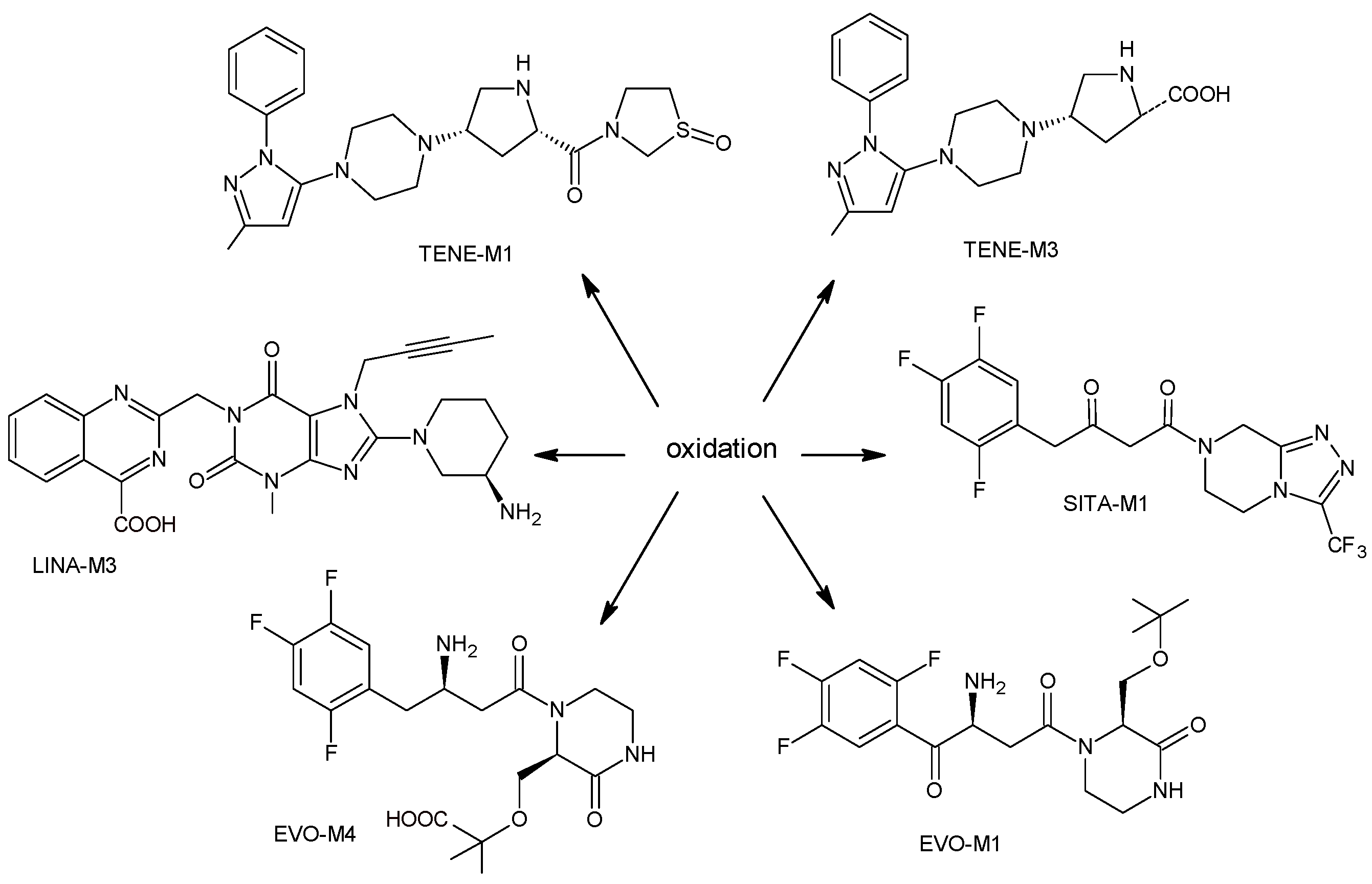
2.1.1. Metabolism of Gliptins
| Metabolite | Structure | m/z [M + H]+ | Ref. |
|---|---|---|---|
| ALO-M1 | 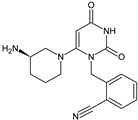 | n.a. | [9] |
| ANA-M1 | 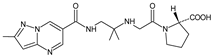 | 366 * | [10] |
| EVO-M1 | 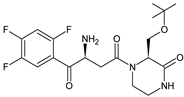 | 416 * | [11] |
| EVO-M2 EVO-M3 (isomers) | 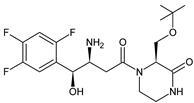 | 418 * | [11,12] |
| EVO-M4 | 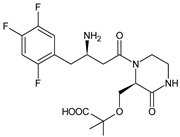 | 418 * | [12] |
| EVO-M5 | 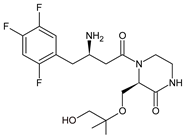 | 432 * | [12] |
| EVO-M6 | 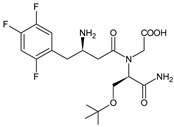 | 434 * | [12] |
| LINA-M1 |  | n.a. | [7] |
| LINA-M2 | 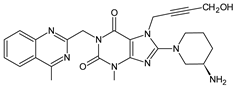 | n.a. | [13] |
| LINA-M3 | 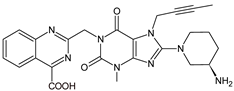 | n.a. | [13] |
| SAXA-M1 | 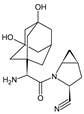 | 332 * | [15,16,17] |
| SITA-M1 | 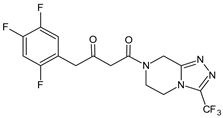 | 408 * | [18] |
| SITA-M2 | 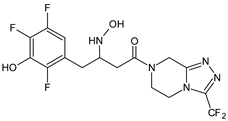 | 422 * | [18] |
| SITA-M3 | 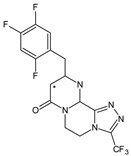 | 406 * | [19] |
| TENE-M1 |  | 443 * | [20,21] |
| TENE-M2 | 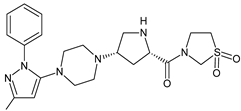 | n.a. | [20] |
| TENE-M3 | 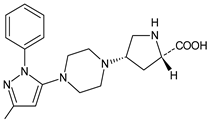 | n.a. | [20] |
| TENE-M4 |  | n.a. | [20] |
| TENE-M5 | 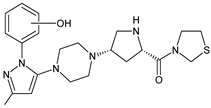 | n.a. | [20] |
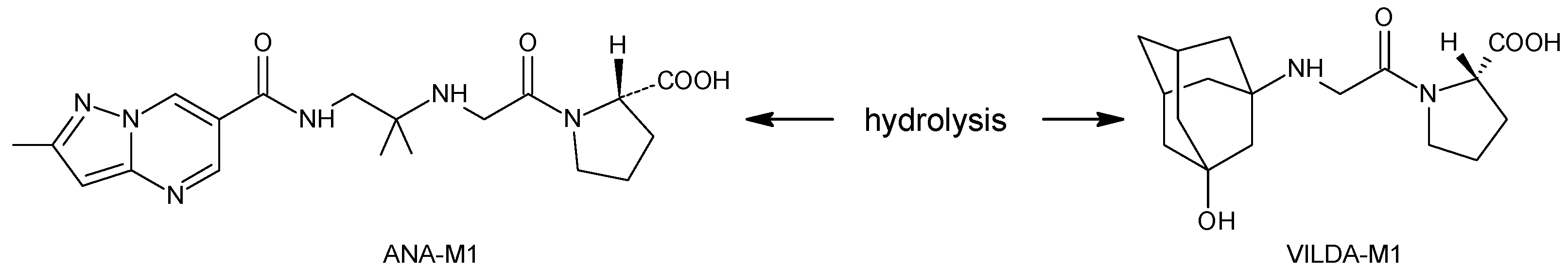
| VILDA-M1 | 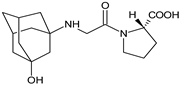 | n.a. | [7] |
2.1.2. Analytical Methods for Elucidating the Metabolism of Gliptins
| Compound | Conditions | Ref. |
|---|---|---|
| ANA | C18 column (2.0 × 150 mm, 3 μm) and isocratic elution using 1% CH3COOH/ACN (80:20, v/v), 1.0 mL/min; MS/MS with positive ESI. C18 column (4.6 × 250 mm, 5 μm) and gradient elution: (A) 50 mM ammonium acetate, (B) ACN, 1.0 mL/min. | [10] |
| EVO | Unison-C8 column (2.0 × 75 mm, 3.0 μm) and gradient elution of A) 5% ACN in 0.1% HCOOH and B) 95% ACN in 0.1% HCOOH, 0.3 mL/min. The column and autosampler temperatures: 40 °C and 6 °C; ESI in positive and negative mode: spray voltage, 4.0 kV in positive mode and −3.0 kV in negative mode; vaporizer temperature, 350 °C; capillary temperature, 330 °C; sheath gas pressure, 35 Arb; and auxiliary gas pressure, 15 Arb; collision energy of 10 to 40 eV. | [11] |
| EVO | Kinetex C18 column (4.6 × 150 mm, 2.6 μm) and gradient elution with (A) 20 mM ammonium acetate (pH 4) and (B) ACN, 1 mL/min. The column temperature 40 °C and UV detection at 268 nm. ESI in the positive mode: spray voltage, 4.5 kV; vaporizer temperature, 350 °C; capillary temperature, 330 °C; sheath gas pressure, 50 Arb; auxiliary gas pressure, 10 Arb; and sweep gas pressure, 5 Arb, the collision gas He, and the normalized collision energy during product ion scanning 35%. | [12] |
| OMA | ACE 5 C8 column (4.6 × 250 mm; 5 µm) and gradient elution with (A) 2 mM ammonium acetate in ACN:H2O (5:95) containing 0.1% HCOOH, and (B) 2 mM ammonium acetate in ACN:H2O (95:5) containing 0.1% HCOOH, 1.0 mL/min; N2 as the nebulizer and auxiliary gas, and Ar as the collision gas. The ESI capillary voltage 1.2 kV. The source and desolvation temperatures 100 and 550 °C; collision energy from 20 to 30 eV. | [14] |
| SAXA | ACE CN column (4.6 × 150 mm, 5 μm) and ACN and 10.0 mM ammonium formate buffer of pH 5.0 (80:20, v/v); triple quadrupole MS detection with positive ESI. | [15] |
| SAXA | C18 column (2.1 × 50 mm, 5 µm) and gradient elution with (A) 0.1% HCOOH in H2O and (B) 0.1% HCOOH in ACN. TurboIonSpray® source, positive ionization mode, using SRM. N2 as the nebulizer, curtain and collision gas; 450 °C for TIS interface, 5000 V setting for ion spray voltage, 30 setting for the curtain gas and nebulizer gas. | [16] |
| SAXA | HILIC Chrom Matrix HP amide column (3.0 × 100 mm, 5 μm), ACN and 5 mM ammonium formate buffer containing 0.1% HCOOH. Ion spray voltage 5500 V, ion spray temperature 550 °C, ion source gas 1: 50 Arb, ion source gas 2: 55 Arb, curtain gas (N2) 30 Arb, collision gas (N2) 10 Arb, entrance potential 10 V, collision cell exit potential 12 V. | [17] |
| SITA | ZIC-HILIC column (4.6 mm × 150 mm, 5 µm) and gradient elution with (A) HCOOH in H2O (0.1% v/v) and (B) HCOOH (0.1% v/v) in ACN, 0.3 mL/min. The capillary temperature 250 °C, spray voltage +4.5 kV and the sheath and auxiliary gas (N2) flow rates 45 and 15. CID voltage of 40 eV. | [18] |
| TENE | CAPCELL PAK C18 UG120 column (4.6 × 250 mm, 5 µm,) at 40 °C and gradient elution with (A) 20 mM ammonium acetate and (B) ACN, 0.8 mL/min; positive ESI at 3800 V and CID at the collision energy of 35%. | [20] |
| TENE | CAPCELL Pak C18 column (2.0 × 2150 mm, 5 μm) and isocratic elution with ACN, MeOH and H2O, 025 mL/min; temperature of the column and autosampler 50 °C and 10 °C. MS: collision gas 5 psi, curtain gas 10 psi, ion source gas (nebulizer) 30 psi, ion spray voltage 5500 V, and collision energy of 37 eV for TENE; declustering potential, entrance potential, and collision exit potential were 106 V; ESI positive ion mode using MRM. | [21] |
| ANA ALO LINA SITA VILDA | UPLC system and a QQQ mass spectrometer equipped with a switching valve; XBridge C18 column (2.1 × 50 mm, 3.5 μm) and gradient with (A) 0.5 mM ammonium hydrogen carbonate and (B) MeOH for VILDA or (A) 1 mM ammonium acetate and (B) ACN (B) for ANA, ALO, SITA and LINA; 0.55 mL/min. The autosampler 4 °C. ESI positive ion mode using MRM transitions. Orbitrap Fusion MS system coupled with a HPLC system: XBridge C18 column (4.6 × 100 mm, 5 μm) and gradient elution with (A) 20 mM ammonium acetate (A) and (B) ACN (B), 1.0 mL/min; ESI positive and negative ion mode. | [22,23] |
2.2. Chemical Degradation of Gliptins (DPP-4 Inhibitors) and the Methods Used to Elucidate Their Degradation Pathways
2.2.1. Chemical Degradation of Gliptins
| Stress Conditions | Degradant | Structure | m/z [M + H]+ | Ref. |
|---|---|---|---|---|
| [H+] | ALO-D1 |  | 258 ** | [27] |
| [OH−] | ALO-D2 | 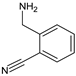 | 133 ** | [27] |
| [OH−] [O] | ALO-D3 | 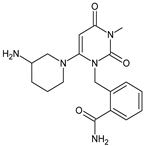 | 358 ** | [27] |
| [T] | ALO-D4 |  | 340 ** | [27] |
| [O] | ALO-D5 | 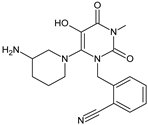 | 356 ** | [27] |
| [H+] | ANA-D1 |  | 225 * | [28] |
| [H+] | ANA-D2 |  | 306 * | [28,29] |
| [H+] [O] | ANA-D3 | 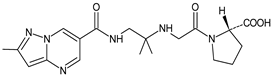 | 403 * | [28,29] |
| [H+] [OH−] [O] | ANA-D4 | 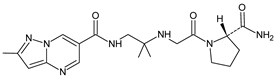 | 402 * | [28,29,31] |
| [H+] | ANA-D5 |  | 384 * | [28] |
| [O] | ANA-D6 |  | 400 * | [28] |
| [O] | ANA-D7 | 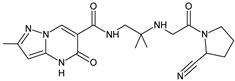 | 400 * | [28] |
| [O] | ANA-D8 | 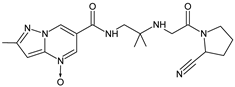 | 398 * | [31] |
| [OH−] | LINA-D1 |  | 672 ** | [32] |
| [H+] | LINA-D2 | 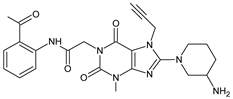 | 492 ** | [32] |
| [H+] | LINA-D3 LINA-D4 (isomers) |  | 509 ** | [32] |
| [O] | LINA-D5 | 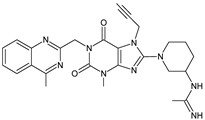 | 514 ** | [32] |
| [O] | LINA-D6 |  | 530 ** | [32] |
| [O] | LINA-D7 LINA-D8 (isomers) | 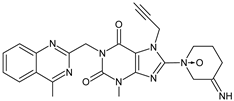 | 487 ** | [32] |
| [O] | LINA-D9 |  | 490 ** | [32] |
| LINA-D10 | 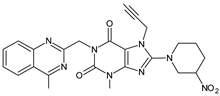 | 503 ** | [32] | |
| LINA-D11 | 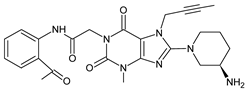 | 492 ** | [32] | |
| LINA-D12 | 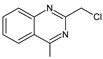 | n.a. | [33] | |
| reducing sugar | LINA-D13 | 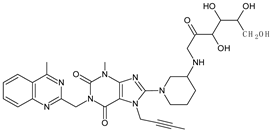 | 635 ** | [34] |
| HCOOH | LINA-D14 | 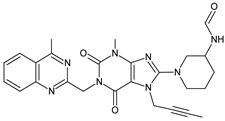 | 501 ** | [34] |
| metformin | LINA-D15 | 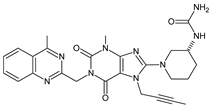 | 516 ** | [34] |
| [O] | OMA-D1 |  | 415 * | [36] |
| [H+] [OH−] | SAXA-D1 | 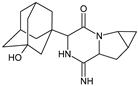 | 316 ** | [37] |
| [H+] [OH−] | SAXA-D2 | 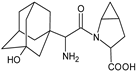 | 335 ** | [37] |
| [H+] [OH−] | SAXA-D3 |  | 335 ** | [37] |
| [H+] [OH−] | SAXA-D4 | 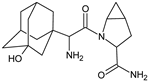 | 334 ** | [37] |
| [O] | SAXA-D5 | 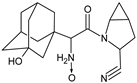 | 332 ** | [37] |
| [O] | SAXA-D6 | 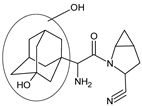 | 332 ** | [37] |
| [O] | SAXA-D7 |  | 348 ** | [37] |
| [H+] [OH−] [T] | SITA-D1 |  | 234 * | [38,39,40] |
| [H+] [OH−] [T] | SITA-D2 |  | 193 * | [38,39,40] |
| [H+] [OH] [O] | SITA-D3 |  | 207 * | [39,40] |
| [O] | SITA-D4 SITA-D5 | 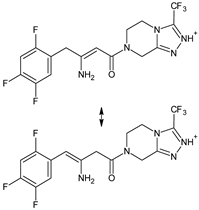 | 406 * | [39] |
| [OH−] [O] | SITA-D6 SITA-D7 | 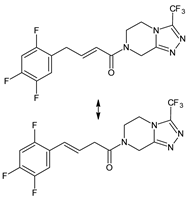 | 391 * | [40] |
| [OH−] [T] | TENE-D1 | 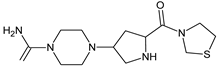 | 310 * | [41] |
| [OH−] [T] | TENE-D2 | 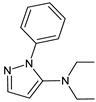 | 214 * | [41] |
| [OH−] [T] | TENE-D3 | 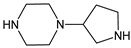 | 156 * | [41] |
| [OH−] | TENE-D4 | 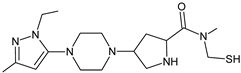 | 355 * | [41] |
| [T] | TENE-D5 | 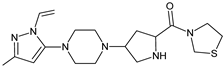 | 376 * | [41] |
| [O] | TENE-D6 | 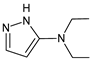 | 139 * | [41] |
| [O] | TENE-D7 | 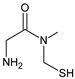 | 137 * | [41] |
| [H+] [O] | TRELA-D1 | 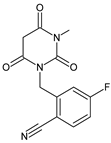 | 276 ** | [43,44,45] |
| [OH−] [O] | TRELA-D2 |  | 376 ** | [44,45] |
| [H+] [O] | TRELA-D3 |  | 358 ** | [44] |
| [O] | TRELA-D4 | 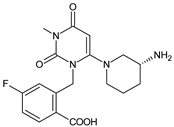 | 378 * | [45] |
| [H+] [OH−] | VILDA-D1 |  | 226 * | [46] |
| [H+] [OH−] [O] | VILDA-D2 | 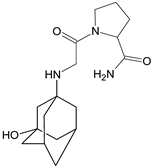 | 322 * | [46] |
| [H+] [OH−] | VILDA-D3 |  | 323 * | [46] |
| [OH−] | VILDA-D4 |  | 115 ** | [47] |
| [OH−] | VILDA-D5 | 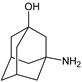 | 168 ** | [47] |
| [H+] | VILDA-D6 | 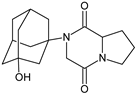 | 304 * | [48] |
| [OH−] | VILDA-D7 | 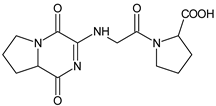 | 323 * | [48] |
| [OH−] | VILDA-D8 | 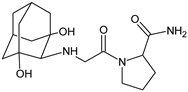 | 337 * | [48] |
| [O] | VILDA-D9 | 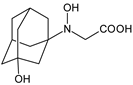 | 241 * | [48] |
2.2.2. Methods for Elucidating Degradation Pathways of Gliptins
| Compound | Conditions | Ref. |
|---|---|---|
| ALO | Gemini-NX C18 column (4.6 mm × 250 mm, 5 µm) and gradient elution with (A) 0.2% HCOOH-0.2% ammonium acetate and (B) ACN and MeOH (60:40, v/v) and PDA detection. ESI source of the Q-TOF unit in positive ionization mode. MS: spray voltage, 3.5 kV; N2 (drying gas) temperature, 350 °C with the nebulizer pressure of 207 kPa; fragmentor voltage, 175 V and collision energies 10–15 eV. | [27] |
| ALO | UPLC-PDA: Kromasil C18 column (4.6 × 250 mm, 5 µm) and gradient with (A) 0.1% HClO4 in H2O (pH adjusted to 3.0 with TEA) and ACN in the ratio of 90:10 and (B) 0.1% HClO4 in H2O (pH adjusted to 3.0 with TEA) and ACN in the ratio of 40:60, 1 mL/min. UV 224 nm and column oven temperature 30 °C. LC-MS: Q-TOF mass spectrometer, (A) 0.1% HCOOH in H2O (pH adjusted to 3.0 with NH4OH) and ACN in the ratio of 90:10 and (B) 0.1% HCOOH in H2O (pH adjusted to 3.0 with NH4OH) and ACN in the ratio of 40:60. MS: positive and negative ESI, capillary voltage 4000 V, gas temperature 400 °C, fragmentor voltage, 125 V; skimmer voltage, 60 V and collision energy, 30 V. | [28] |
| ANA | UPLC-PDA: Aquity BEH C18 column (2.1 × 100 mm, 1.7 µm) and gradient with (A) 10 mM ammonium formate without adjusting pH (measured pH 6.2) and (B) ACN (100% v/v), 0.3 mL/min; UV 248 nm. LC/MS: Q-TOF mass spectrometer, ESI positive mode: capillary voltage, 4.00 KV; fragmentor voltage, 170 V; skimmer voltage, 65 V; desolvation gas flow, 10 L/min; gas temperature, 325 °C, nebulizer gas, 40 psi. | [29] |
| ANA | HPLC-PDA: a Daisogel-SP-100–10-ODS-P column (20 × 250 mm, 10 μm), 5 mL/min, UV 247 nm. LC-MS system with ion trap in positive ESI with ion source voltage of 5000 V and a source temperature of 450 °C; the nebulizer 20 psi using N2. | [31] |
| LINA in the presence of metformin | HPLC-PDA: InertSustain® C8 column (4.6 × 150 mm, 5 μm) and gradient elution with (A) 10 mM ammonium acetate (pH adjusted to 5.5 with CH3COOH) and (B) mixture of ACN and MeOH in the ratio of 80:20 (v/v), 0.9 mL/min; UV 254 nm. LC/Q-TOF: Acquity CSH C8 column (2.1 × 100 mm, 1.7 μm), 0.3 mL/min. Positive ESI: fragmentor voltage, 70 V; capillary voltage, 3500 V; skimmer voltage, 60 V; drying gas flow rate, 10 L/min; drying gas heater temperature, 320 °C; nebulizer gas pressure, 45 psi. N2 as drying, nebulizing and collision gas for CID. | [32] |
| LINA | LC-MS: Zorbax SB-Aq column (4.6 × 250 mm, 5 µm) and gradient elution with (A) phosphate buffer (0.02 M) of pH 3.0 and (B) ACN:MeOH (90:10, v/v), UV 225 nm. The column temperature 45 °C. UPLC-TOF/MS: Acquity BEH shield RP18 column (2.1 × 100 mm, 1.7 µm) and gradient elution with (A) ammonium acetate buffer (pH 3.8) and (B) ACN:MeOH (90:10, v/v), 0.21 mL/min. Positive ESI: capillary voltage at 3.0 KV, source and desolvation temperature 120 °C and 500 °C. The cone and desolvation gas flows 60 and 800 L/h. | [34] |
| SITA | LC-PDA: Zorbax Eclipse Plus C 18 column (4.6 × 100 mm, 5 µm) and gradient elution with (A) 10 mM ammonium formate and (B) MeOH, 0.5 mL/min. ESI-Q-TOF-MS/MS: positive ESI: spray voltage, 4.8 kV; CV, 20 V; capillary temperature, 300 °C and tube lens offset voltage, 10 V. N2 as a sheath gas (40 psi), and He as a damping and collision gas. CID experiments: collision energies between 15 and 25 eV. | [37] |
| SITA | LC-MS: RP18 column and gradient elution with A) ACN-H2O (1:99, v/v) with 10 mM ammonium formate (0.1%) and B) ACN-H2O (95:5, v/v) with 10 mM ammonium formate (0.1%), 0.4 mL/min. Positive ESI: N2, flow rate 12 L/min, nebulizer pressure, 30 psi; gas temperature, 200 °C; sheath gas temperature, 350 °C; sheath gas flow, 12 mL/min; VCap, skimmer, 65 V; fragmentor voltage, 150 V; octopole RF Peak, 750 V and CID, 20 eV. | [38] |
| SITA | UPLC-UV: Acquity BEH C-18 column (2.1 × 50 mm, 1.7 µm) and gradient program with (A) 10 mM ammonium formate, pH 6.4 and (B) CAN, 0.15 mL/min, UV 267 nm. Positive ESI: cone voltage, 20 V; CV, 3 KV; desolvation gas N2, 1000 L/h, cone gas N2, 50 L/h, source temperature, 180 °C; desolvation temperature, 500 °C. For fragmentation of selected ion collision energy 30 V. | [39] |
| SITA | LC-PDA: Gemini C18 110 A column (2 × 100 mm, 3 µm) and gradient elution with (A) ACN-H2O (1:99, v/v) with 10 mM ammonium formate (0.1%) and (B) ACN-H2O (95:5, v/v) with 10 mM ammonium formate (0.1%), 0.4 mL/min. LC-Q-TOF: positive ESI; N2 flow rate 12 L/min; nebulizer pressure, 30 psig; gas temperature, 200 °C; sheath gas temperature, 350 °C, sheath gas flow, 12 mL/min; VCap, 4000 V; skimmer, 65 V; fragmentor voltage, 150 V and octopole RF Peak, 750 V and CID, 20 eV. | [40] |
| TENE | HPLC-UV: Kromasil® 100-5 C18 (4.6 × 250 mm, 5 μm) and isocratic elution with pH 6.0 phosphate buffer and ACN (60:40 v/v), 1 mL/min. UPLC-PDA: Acquity BEH C18 column (2.1 × 50 mm, 1.7 μm) and gradient elution with A (10% ACN in H2O with 0.1% HCOOH) and B (90% ACN with 0.1% HCOOH), 0.3 mL/min. | [41] |
| TRELA | UPLC-UV: BDS HYPERSIL C18 column (3 mm × 50 mm, 1.9 μm) and HYPERSIL Gold C18 column (3 mm × 50 mm, 1.9 μm), UHPLC-UV on SYMMETRY C18 column (2.1 mm × 100 mm, 2.2 μm) and isocratic elution with ACN-potassium dihydrogen pH 3.5 phosphate buffer 0.05 M (50:50, v/v). UPLC-MS/MS: Agilent SB-C18 column (2.1 × 50 mm 1.8 μm) and a mixture of ACN-HCOOH 0.1% (80:20, v/v); 120 °C, capillary voltage of 3 KV, dwell at 0.161 s, desolvation gas flow rate at 500 L/h, desolvation temperature at 400 °C. | [43] |
| TRELA | LC-UV: XSelect CSH C18 column (4.6 mm × 250 mm, 5 μm) and 0.05% TFA in H2O or ACN containing 0.05% TFA; UV 224 nm and 275 nm. UPLC-MS: BDS Hypersil C18 column (2.1 × 150 mm, 2.4 μm) and gradient elution with (A) 0.1% HCOOH in H2O and (B) ACN, 0.3 mL/min, column temperature 35 °C. UHPLC-LTQ-Orbitrap: ESI, the capillary temperature 250 °C, source voltage and spray voltage were 5 kV, sheath gas (N2) flow 35 psi. | [44] |
| VILDA | UHPLC-PDA-MS: Kinetex XB-C18 column (2.1 × 150 mm, 1.7 µm) and gradient elution with (A) 0.1% HCOOH in H2O and (B) 0.1% HCOOH in ACN, 0.3 mL/min, column temperature 25 °C. MS: CP, 4500 V; endplate, 500 V, nebulizer pressure, 40 psi; drying gas temperature, 145 °C and gas flow rate, 9 L/min. | [46] |
| VILDA | UPLC/Q-TOF: BEH C8 column (2.1 × 50 mm, 1.7 μm) and gradient with (A) H2O with 0.1% HCOOH and (B) MeOH containing 0.1% HCOOH, 0.3 mL/min, column temperature 35 °C. MS: positive ESI, CV 2.50 kV, cone voltage 30 V, extractor cone voltage 3 V, desolvation gas 400 L/h and cone gas 10 L/h; desolvation temperature 400 °C, source temperature 120 °C. | [47] |
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yao, M.; Chen, B.; Zhao, W.; Mehl, J.T.; Li, L.; Zhu, M. LC-MS differential analysis for fast and sensitive determination of biotransformation of therapeutic proteins. Drug Metab. Dispos. 2018, 46, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Rakusanova, S.; Cajka, T. Current analytical methods to monitor type 2 diabetes medication in biological samples. TrAC Trends Anal. Chem. 2023, 158, 116831. [Google Scholar] [CrossRef]
- Neagu, A.-N.; Jayathirtha, M.; Baxter, E.; Donnelly, M.; Petre, B.A.; Darie, C.C. Applications of Tandem Mass Spectrometry (MS/MS) in Protein Analysis for Biomedical Research. Molecules 2022, 27, 2411. [Google Scholar] [CrossRef] [PubMed]
- ICH Topic Q 3 A (R2) “Impurities in New Drug Substances”. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/ich-q-3-r2-impurities-new-drug-substances-step-5_en.pdf (accessed on 10 May 2023).
- Charoo, N.A.; Untoo, S.; Rahman, Z. Using metabolite data to develop patient centric specification for amide impurity in vildagliptin tablets. Sci. Pharm. 2022, 90, 1. [Google Scholar] [CrossRef]
- Rameshrad, M.; Razavi, B.M.; Ferns, G.A.A.; Hosseinzadeh, H. Pharmacology of dipeptidyl peptidase-4 inhibitors and its use in the management of metabolic syndrome: A comprehensive review on drug repositioning. J. Pharm. Sci. 2019, 27, 341–360. [Google Scholar] [CrossRef]
- Chen, X.-W.; He, Z.-X.; Zhou, Z.-W.; Yang, T.; Zhang, X.; Yang, Y.-X.; Duan, W.; Zhou, S.-F. Clinical pharmacology of dipeptidyl peptidase 4 inhibitors indicated for the treatment of type 2 diabetes mellitus. Clin. Exp. Pharmacol. Physiol. 2015, 42, 999–1024. [Google Scholar] [CrossRef]
- Ahrén, B. DPP-4 Inhibition and the Path to Clinical Proof. Front. Endocrinol. 2019, 10, 376. [Google Scholar] [CrossRef]
- Kaku, K.; Kisanuki, K.; Shibata, M.; Oohira, T. Benefit-risk assessment of alogliptin for the treatment of type 2 diabetes mellitus. Drug Saf. 2019, 42, 1311–1327. [Google Scholar] [CrossRef]
- Furuta, S.; Smart, C.; Hackett, A.; Benning, R.; Warrington, S. Pharmacokinetics and metabolism of [14C]anagliptin, a novel dipeptidyl peptidase-4 inhibitor, in humans. Xenobiotica 2013, 43, 432–442. [Google Scholar] [CrossRef]
- Jeong, H.U.; Kim, J.H.; Lee, D.Y.; Shim, H.J.; Lee, H.S.; McPhee, D.J. In vitro metabolic pathways of the new anti-diabetic drug evogliptin in human liver preparations. Molecules 2015, 20, 21802–21815. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Kim, J.H.; Shim, H.J.; Jeong, H.U.; Lee, H.S. Absorption, metabolism, and excretion of [14C]evogliptin tartrate in male rats and dogs. J. Toxicol. Environ. Health—A: Curr. Issues 2018, 81, 453–464. [Google Scholar] [CrossRef] [PubMed]
- ICH Topic Q 3 A (R2) “Impurities in New Drug Substances Blech, S.; Ludwig-Schwellinger, E.; Gräfe-Mody, E.U.; Withopf, B.; Wagner, K. The metabolism and disposition of the oral dipeptidyl peptidase-4 inhibitor, linagliptin, in humans. Drug Metab. Dispos. 2010, 38, 667–678. [Google Scholar]
- Xu, S.; Tatosian, D.; Mcintosh, I.; Caceres, M.; Matthews, C.; Samuel, K.; Selverian, D.; Kumar, S.; Kauh, E. Absorption, metabolism and excretion of [14C]omarigliptin, a once-weekly DPP-4 inhibitor, in humans. Xenobiotica 2018, 48, 584–591. [Google Scholar] [CrossRef]
- Shah, P.A.; Shah, J.V.; Sanyal, M.; Shrivastav, P.S. LC-MS/MS analysis of metformin, saxagliptin and 5-hydroxy saxagliptin in human plasma and its pharmacokinetic study with a fixed-dose formulation in healthy Indian subjects. Biomed. Chromatogr. 2017, 31, e3809. [Google Scholar] [CrossRef]
- Xu, X.S.; Demers, R.; Gu, H.; Christopher, L.J.; Su, H.; Cojocaru, L.; Boulton, D.W.; Kirby, M.; Stouffer, B.; Humphreys, W.G.; et al. Liquid chromatography and tandem mass spectrometry method for the quantitative determination of saxagliptin and its major pharmacologically active 5-monohydroxy metabolite in human plasma: Method validation and overcoming specific and non-specific binding at low concentrations. J. Chromatogr. B 2012, 889–890, 77–86. [Google Scholar]
- Peng, Y.; Chang, Q.; Yang, N.; Gu, S.; Zhou, Y.; Yin, L.; Aa, J.; Wang, G.; Sun, J. Quantitative determination of metformin, saxagliptin and 5-hydroxy saxagliptin simultaneously by hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry and its application to a bioequivalence study with a single-pill combination in human. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1081–1082, 109–117. [Google Scholar]
- Khreit, O.I.G.; Grant, H.M.; Henderson, C.; Watson, D.G.; Sutcliffec, O.B. Identification of Novel Metabolic Pathways of Sitagliptin (STG) by LC/MS and LC/MS2 after incubations with rat Hepatocytes. J. Drug Metab. Toxicol. 2016, 7, 220. [Google Scholar] [CrossRef]
- Vincent, S.H.; Reed, J.R.; Bergman, A.J.; Elmore, C.S.; Zhu, B.; Xu, S.; Ebel, D.; Larson, P.; Zeng, W.; Chen, L.; et al. Metabolism and excretion of the dipeptidyl peptidase 4 inhibitor [ 14C]sitagliptin in humans. Drug Metab. Dispos. 2007, 35, 533–538. [Google Scholar] [CrossRef]
- Nakamaru, Y.; Hayashi, Y.; Ikegawa, R.; Kinoshita, S.; Madera, B.P.; Gunput, D.; Kawaguchi, A.; Davies, M.; Mair, S.; Yamazaki, H.; et al. Metabolism and disposition of the dipeptidyl peptidase IV inhibitor teneligliptin in humans. Xenobiotica 2014, 44, 242–253. [Google Scholar] [CrossRef]
- Park, J.-W.; Kim, K.-A.; Park, J.-Y. Development of a liquid chromatography/tandem-mass spectrometry assay for the simultaneous determination of teneligliptin and its active metabolite teneligliptin sulfoxide in human plasma. Biomed. Chromatogr. 2020, 34, e4721. [Google Scholar] [CrossRef]
- Mizuno, K.; Takeuchi, K.; Umehara, K.; Nakajima, M. Identification of a novel metabolite of vildagliptin in humans: Cysteine targets the nitrile moiety to form a thiazoline ring. Biochem. Pharmacol. 2018, 156, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Keck, B.D.; Ognibene, T.; Vogel, J.S. Analytical validation of accelerator mass spectrometry for pharmaceutical development. Bioanalysis 2010, 2, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Takeuchi, K.; Umehara, K.; Nakajima, M. Identification of novel metabolites of vildagliptin in rats: Thiazoline-containing thiol adducts formed via cysteine or glutathione conjugation. Drug Metab. Dispos. 2019, 47, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Makarov, A.; Denisov, E.; Kholomeev, A.; Balschun, W.; Lange, O.; Strupat, K.; Horning, S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006, 78, 2113–2120. [Google Scholar] [CrossRef]
- Bertol, C.D.; Friedrich, M.T.; Carlos, G.; Froehlich, P.E. Analytical stability-indicating methods for alogliptin in tablets by LC-CAD and LC-UV. J. AOAC Int. 2017, 100, 400–405. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, D.; Li, Z.; Hang, T.; Song, M. Isolation and characterization of related substances in alogliptin benzoate by LC-QTOF mass spectrometric techniques. J. Pharm. Biomed. Anal. 2016, 128, 253–263. [Google Scholar] [CrossRef]
- Baira, S.M.; Sigalapalli, D.K.; Bathini, N.B.; Srinivas, R.; Talluri, M.V.N.K. LC/QTOF/MS/MS characterization, molecular docking and in silico toxicity prediction studies on degradation products of anagliptin. J. Pharm. Biomed. Anal. 2018, 159, 92–99. [Google Scholar] [CrossRef]
- Furuta, S.; Tamura, M.; Hirooka, H.; Mizuno, Y.; Miyoshi, M.; Furuta, Y. Pharmacokinetic disposition of anagliptin, a novel dipeptidyl peptidase-4 inhibitor, in rats and dogs. Eur. J. Drug Metab. Pharmacokinet. 2013, 38, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.E.; Piizzi, G. gem-Disubstituent effect: Theoretical basis and synthetic applications. Chem. Rev. 2005, 105, 1735–1766. [Google Scholar] [CrossRef]
- Pandya, C.P.; Rajput, S.J. Stress degradation studies of anagliptin, development of validated stability indicating method, degradation kinetics study, identification and isolation of degradation products. Chromatographia 2018, 81, 1533–1550. [Google Scholar] [CrossRef]
- Yadav, A.S.; Dornala, D.; Swain, D.; Prabha, A.; Samanthula, G. Application of online liquid chromatography/quadrupole time-of-flight electrospray ionization tandem mass spectrometry for structural characterization of linagliptin degradation products and related impurities. Rapid Commun. Mass Spectrom. 2020, 34, e8874. [Google Scholar] [CrossRef]
- Huang, Y.; Lu, H.; Zhang, F.; Min, C. Identification, isolation, characterization, and UHPLC quantification of potential genotoxic impurities in linagliptin. J. Sep. Sci. 2018, 41, 3985–3994. [Google Scholar] [CrossRef]
- Jadhav, S.B.; Reddy, P.S.; Narayanan, K.L.; Bhosale, P.N. Development of RP-HPLC, stability indicating method for degradation products of linagliptin in presence of metformin HCl by applying 2 level factorial design; and identification of impurity-VII, VIII and IX and synthesis of impurity-VII. Sci. Pharm. 2017, 85, 25. [Google Scholar] [CrossRef] [PubMed]
- Heleno Ferreira, R.B.; Duarte, J.A.; Ferreira, F.D.; Oliveira, L.F.S.D.; Machado, M.M.; Malesuik, M.D.; Paula, F.R.; Steppe, M.; Schapoval, E.E.S.; Paim, C.S. Biological safety studies and simultaneous determination of linagliptin and synthetic impurities by LC-PDA. J. Anal. Methods Chem. 2019, 2019, e7534609. [Google Scholar] [CrossRef] [PubMed]
- Tantawy, M.A.; Hassan, A.M.; Hegazy, M.A.; Kelani, K.M. Quality and stability profile assessment of the recent Antidiabetic Omarigliptin by using different chromatographic methods. J. Chromatogr. Sci. 2021, 59, 762–769. [Google Scholar] [CrossRef]
- Sridhar, L.; Goutami, P.; Darshan, D.V.; Ramakrishna, K.; Rao, R.N.; Prabhakar, S. LC-ESI-MS/MS studies on saxagliptin and its forced degradation products. Anal. Methods. 2014, 6, 8212–8221. [Google Scholar] [CrossRef]
- Mowaka, S.; Mohamed, D. Novel contribution to the simultaneous analysis of certain hypoglycemic drugs in the presence of their impurities and degradation products utilizing UPLC-MS/MS. RSC Adv. 2015, 5, 60467–60481. [Google Scholar] [CrossRef]
- Vishnuvardhan, C.; Radhakrishnanand, P.; Navalgund, S.G.; Satheeshkumar, N. Liquid chromatography/electrospray ionisation tandem mass spectrometric study of sitagliptin and its stressed degradation products. Drug Res. 2014, 64, 668–674. [Google Scholar] [CrossRef]
- Gumieniczek, A.; Berecka, A.; Mroczek, T.; Wojtanowski, K.; Dąbrowska, K.; Stępień, K. Determination of chemical stability of sitagliptin by LC-UV, LC-MS and FT-IR methods. J. Pharm. Biomed. Anal. 2019, 164, 789–807. [Google Scholar] [CrossRef]
- Kumar, T.N.V.G.; Vidyadhara, S.; Narkhede, N.A.; Silpa, Y.S.; Lakshmi, M.R. Method development, validation, and stability studies of teneligliptin by RP-HPLC and identification of degradation products by UPLC tandem mass spectroscopy. J. Anal. Sci. Tech. 2016, 7, 27. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, W.; Sun, L.; Zou, Q.; Wei, P.; Ouyang, P. Characterization of process-related impurities including forced degradation products of alogliptin benzoate and the development of the corresponding reversed-phase high-performance liquid chromatography method. J. Sep. Sci. 2014, 37, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Zaghary, W.A.; Mowaka, S.; Hassan, M.A.; Ayoub, B.M. Suitability of various chromatographic and spectroscopic techniques for analysis and kinetic degradation study of trelagliptin. Sci. Rep. 2017, 7, 17255. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Chen, X.; Wang, G.; Du, Z.; Ma, X.; Wang, H.; Yu, G.; Liu, A.; Li, M.; Peng, W.; et al. Development of a validated HPLC method for the quantitative determination of trelagliptin succinate and its related substances in pharmaceutical dosage forms. Eur. J. Pharm. Sci. 2018, 111, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, L.; Zou, L.; Hui, W.; Liu, L.; Zou, Q.; Ouyang, P. Identification, characterization and HPLC quantification of process-related impurities in Trelagliptin succinate bulk drug: Six identified as new compounds. J. Pharm. Biomed. Anal. 2016, 128, 18–27. [Google Scholar] [CrossRef]
- Gumieniczek, A.; Berecka-Rycerz, A.; Fornal, E.; Żyżyńska-Granica, B.; Granica, S. Comprehensive insight into chemical stability of important antidiabetic drug vildagliptin using chromatography (LC-UV and UHPLC-DAD-MS) and spectroscopy (mid-ir and nir with pca). Molecules 2021, 26, 5632. [Google Scholar] [CrossRef]
- Giordani, C.F.A.; Campanharo, S.; Wingert, N.R.; Bueno, L.M.; Manoel, J.W.; Garcia, C.V.; Volpato, N.M.; Iop, G.D.; de Azevedo Mello, P.; de Moraes Flores, E.M.; et al. UPLC-ESI/Q-TOF MS/MS method for determination of vildagliptin and its organic impurities. J. Chromatogr. Sci. 2020, 58, 718–725. [Google Scholar] [CrossRef]
- Arar, S.; Al-Qudah, E.; Alzweiri, M.; Sweidan, K. New forced degradation products of vildagliptin: Identification and structural elucidation using LC-MS, with proposed formation mechanisms. J. Liq. Chromatogr. Relat. Technol. 2020, 43, 633–644. [Google Scholar] [CrossRef]
- Vehovec, T.; Obreza, A. Review of operating principle and applications of the charged aerosol detector. J. Chromatogr. A 2010, 1217, 1549–1556. [Google Scholar] [CrossRef]
- Novakova, L.; Vlckova, H.; Solich, P. Evaluation of new mixed-mode UHPLC stationary phase and the importance of stationary phase choice when using low ionic-strength mobile phase additives. Talanta 2012, 93, 99–105. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gumieniczek, A.; Berecka-Rycerz, A. Metabolism and Chemical Degradation of New Antidiabetic Drugs (Part II): A Review of Analytical Approaches for Analysis of Gliptins. Biomedicines 2023, 11, 1956. https://doi.org/10.3390/biomedicines11071956
Gumieniczek A, Berecka-Rycerz A. Metabolism and Chemical Degradation of New Antidiabetic Drugs (Part II): A Review of Analytical Approaches for Analysis of Gliptins. Biomedicines. 2023; 11(7):1956. https://doi.org/10.3390/biomedicines11071956
Chicago/Turabian StyleGumieniczek, Anna, and Anna Berecka-Rycerz. 2023. "Metabolism and Chemical Degradation of New Antidiabetic Drugs (Part II): A Review of Analytical Approaches for Analysis of Gliptins" Biomedicines 11, no. 7: 1956. https://doi.org/10.3390/biomedicines11071956
APA StyleGumieniczek, A., & Berecka-Rycerz, A. (2023). Metabolism and Chemical Degradation of New Antidiabetic Drugs (Part II): A Review of Analytical Approaches for Analysis of Gliptins. Biomedicines, 11(7), 1956. https://doi.org/10.3390/biomedicines11071956






