Wnt/β-Catenin Signaling Activation Induces Differentiation in Human Limbal Epithelial Stem Cells Cultured Ex Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Limbal Biopsies and Human Limbal Epithelial Stem Cell (hLESC) Cultures
2.2. Small Molecule LY2090314 Treatment of HLESCs
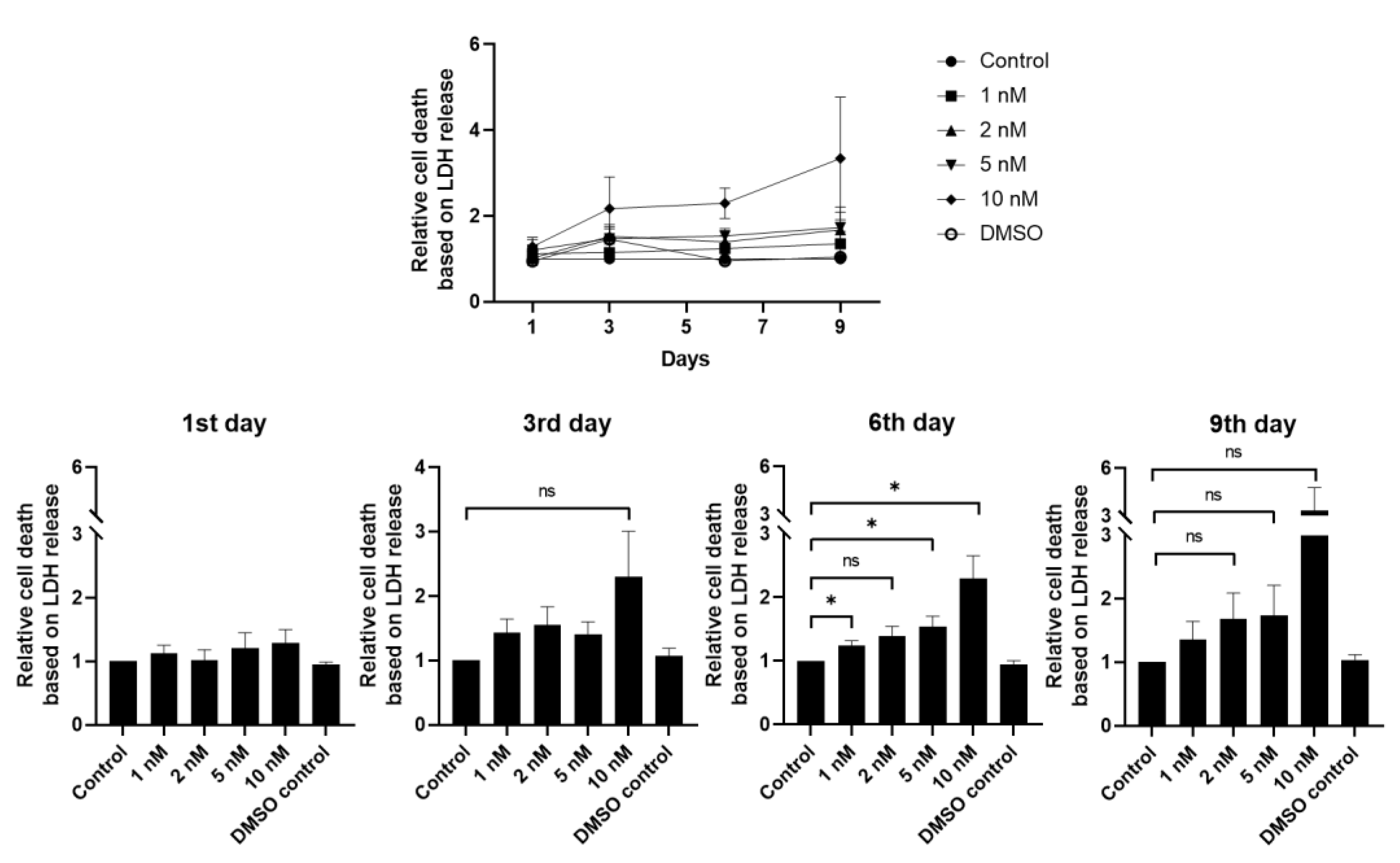
2.3. Cytotoxicity Assay
2.4. RNA Isolation and Real-Time Quantitative RT-PCR (RT-qRT-PCR)
2.5. Microarray and Data Analysis
2.6. Immunocytochemistry (ICC)
2.7. Western Blot (WB)
2.8. Edu Cell Proliferation Assay
2.9. Statistical Analysis
3. Results
3.1. Wnt/β-Catenin Signaling Activation Affects Cell Morphology in hLESC Cultures
3.2. hLESC Toxicity upon Treatment with Increasing Concentrations of Selective Small-Molecule LY2090314
3.3. Real-Time qRT-PCR Identified Wnt/β-Catenin Signaling Activation in hLESC Cultures and Upregulation of Genes for Differentiation and Downregulation of Genes for Limbal Epithelial Stemness Maintenance and Proliferation
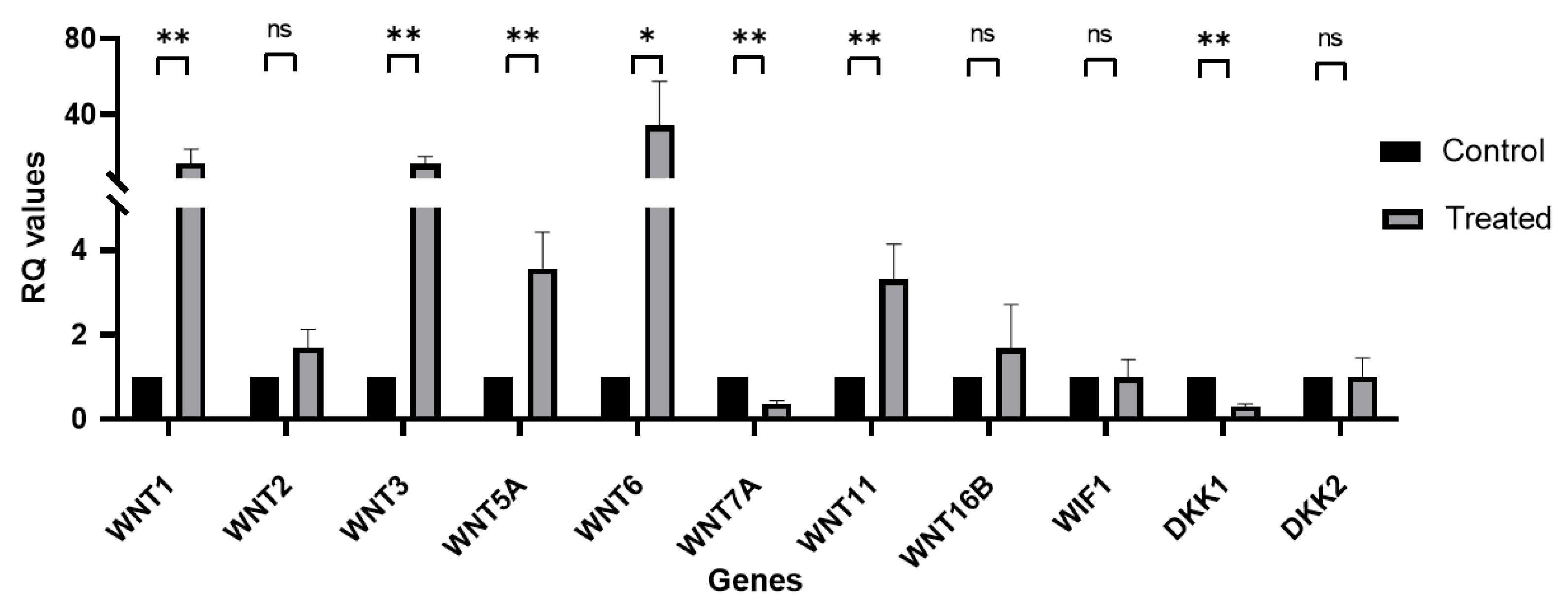
3.4. Wnt Signaling mRNA Expression Profile in Primary hLESC Cultures upon Treatment with LY2090314
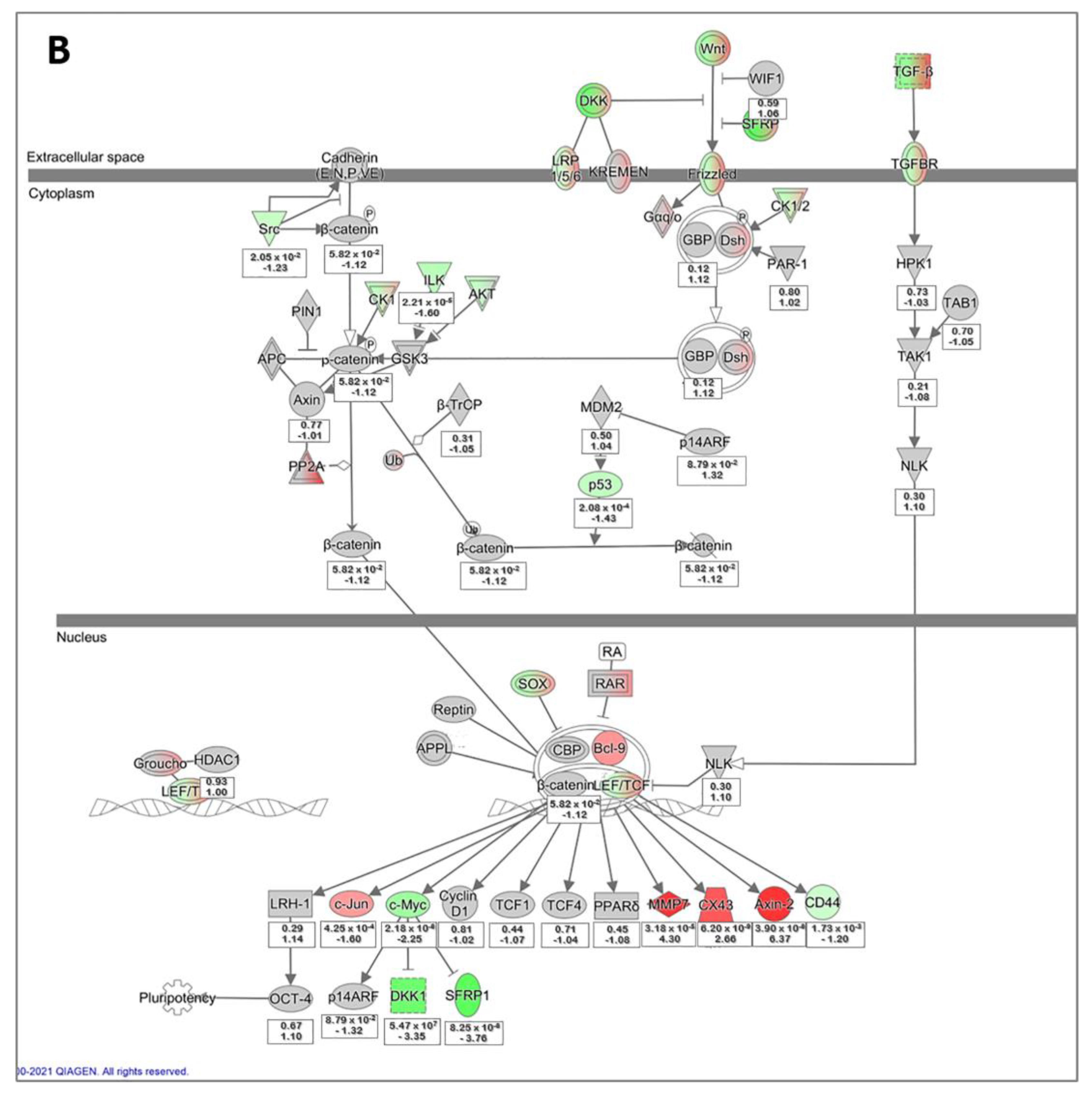
3.5. Microarray Data Analysis of Primary hLESC Cultures upon Treatment with 5 nM Concentration of LY2090314 Compared to Untreated Controls
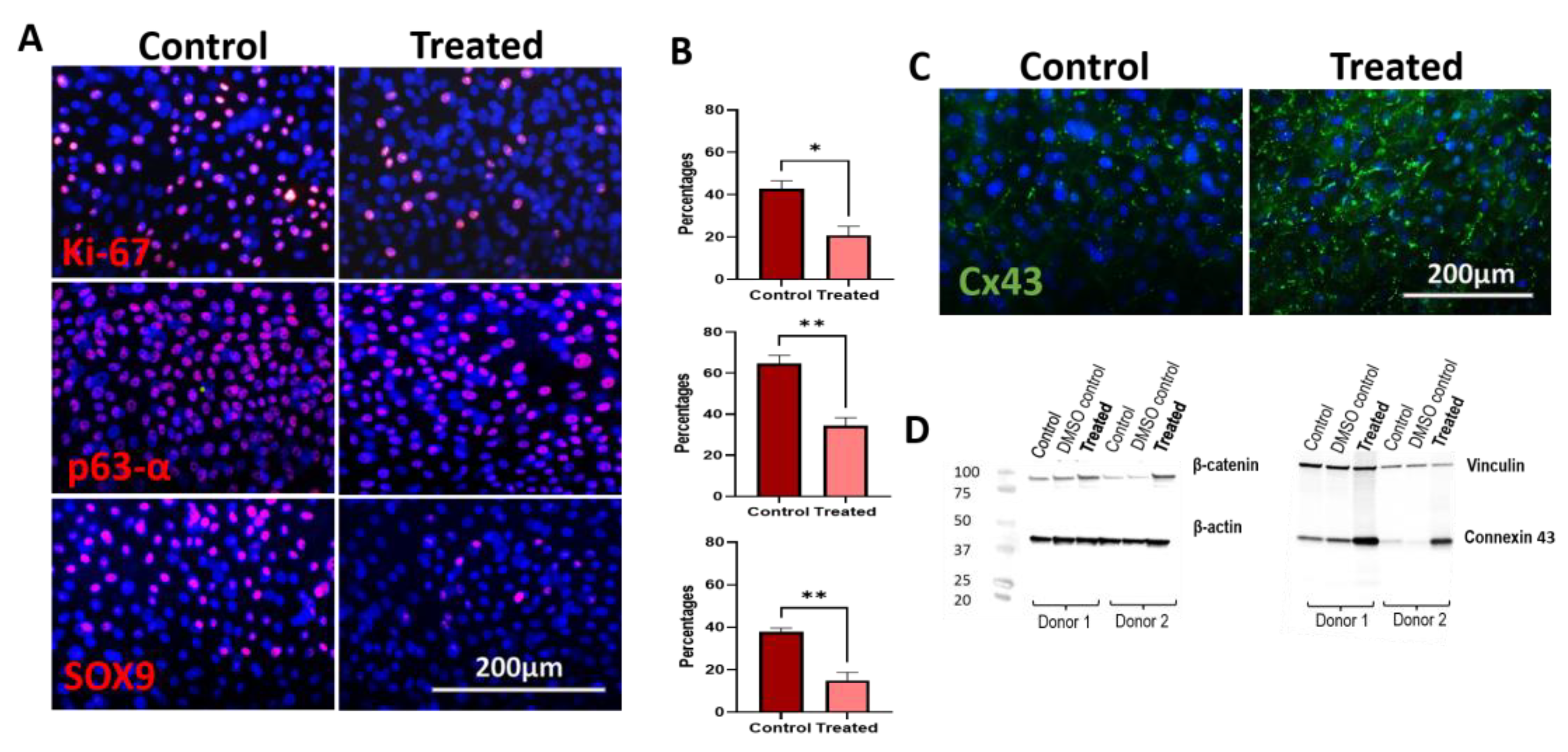
3.6. Immunocytochemistry and Western Blot Analysis of 5 nM LY2090314-Treated hLESC Cultures Revealed Higher Levels of β-Catenin and Cx43 Protein Expression with Low Levels of Ki-67, SOX9, and p63α
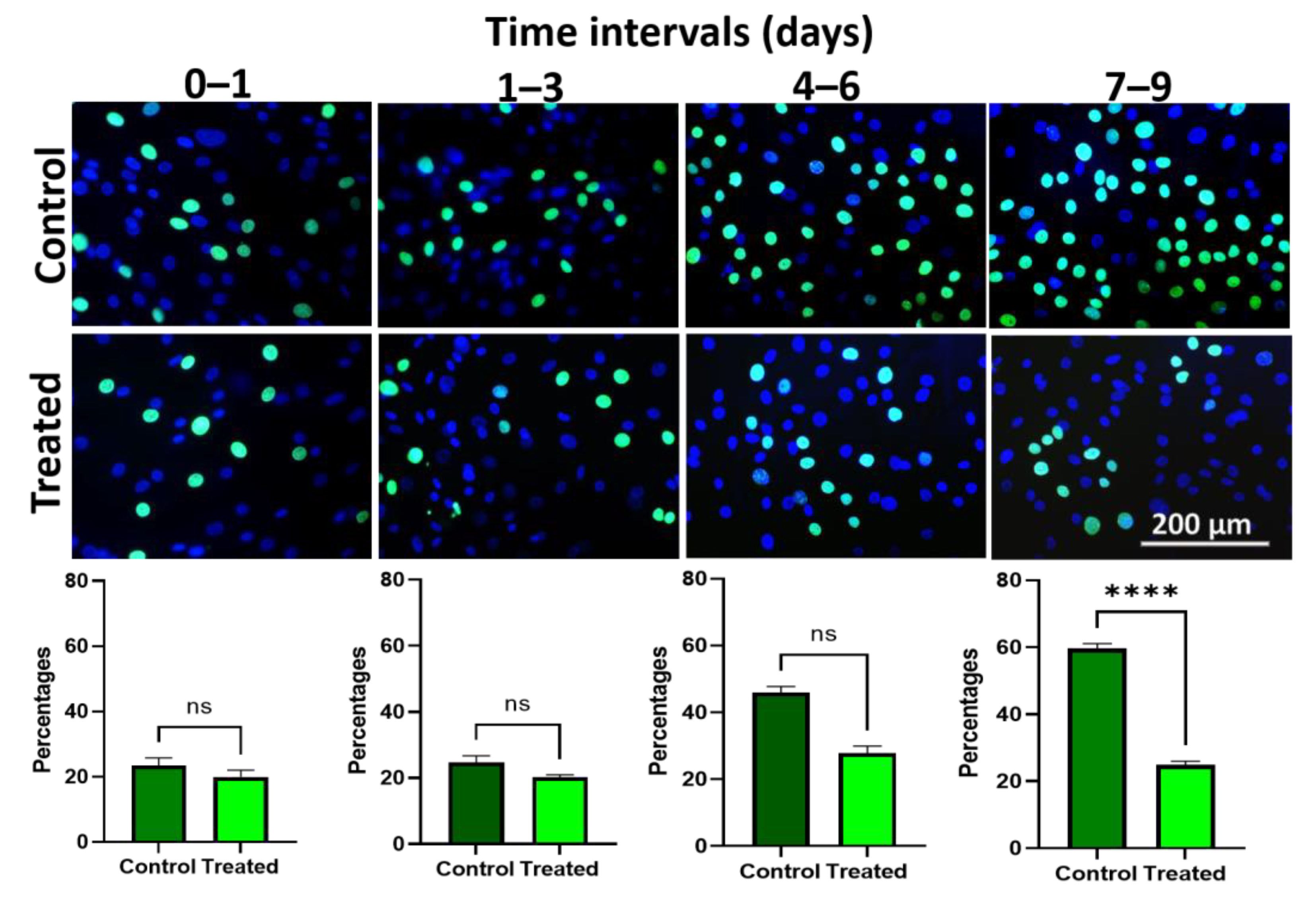
3.7. Edu Proliferation Assay Demonstrates Lower Proliferation upon Wnt/β-Catenin Signaling Activation in Ex Vivo Expanded hLESC Cultures
4. Discussion
4.1. Upon Persistent Activation of Wnt/β-Catenin Signaling the Stemness and Proliferation Decrease, Whereas the Differentiation Increases in Ex Vivo hLESC Cultures Expanded from Limbal Biopsies
4.2. Regulation of Wnt Ligands upon Wnt/β-Catenin Signaling Activation in hLESC Cultures
4.3. Wnt Inhibitors in LY2090314 Treated and Wnt/β-Catenin Signaling Activated hLESCs
4.4. Differentially Expressed Genes Identified by Microarray Comparing hLESC Cultures Untreated or Treated with 5 nM LY2090314
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nosrati, H.; Alizadeh, Z.; Nosrati, A.; Ashrafi-Dehkordi, K.; Banitalebi-Dehkordi, M.; Sanami, S.; Khodaei, M. Stem cell-based therapeutic strategies for corneal epithelium regeneration. Tissue Cell 2021, 68, 101470. [Google Scholar] [CrossRef]
- Schermer, A.; Galvin, S.; Sun, T.T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J. Cell Biol. 1986, 103, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; Tseng, S.C.; Lavker, R.M. Location of corneal epithelial stem cells. Nature 2010, 463, E10–E11; discussion E11. [Google Scholar] [CrossRef]
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell 1989, 57, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Dua, H.S.; Shanmuganathan, V.A.; Powell-Richards, A.O.; Tighe, P.J.; Joseph, A. Limbal epithelial crypts: A novel anatomical structure and a putative limbal stem cell niche. Br. J. Ophthalmol. 2005, 89, 529–532. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.F.; Bron, A.J. Limbal palisades of Vogt. Trans. Am. Ophthalmol. Soc. 1982, 80, 155–171. [Google Scholar]
- Townsend, W.M. The limbal palisades of Vogt. Trans. Am. Ophthalmol. Soc. 1991, 89, 721–756. [Google Scholar]
- Schlötzer-Schrehardt, U.; Dietrich, T.; Saito, K.; Sorokin, L.; Sasaki, T.; Paulsson, M.; Kruse, F.E. Characterization of extracellular matrix components in the limbal epithelial stem cell compartment. Exp. Eye Res. 2007, 85, 845–860. [Google Scholar] [CrossRef]
- Tseng, S.C.; He, H.; Zhang, S.; Chen, S.Y. Niche Regulation of Limbal Epithelial Stem Cells: Relationship between Inflammation and Regeneration. Ocul. Surf. 2016, 14, 100–112. [Google Scholar] [CrossRef]
- Haagdorens, M.; Van Acker, S.I.; Van Gerwen, V.; Dhubhghaill, S.N.; Koppen, C.; Tassignon, M.J.; Zakaria, N. Limbal Stem Cell Deficiency: Current Treatment Options and Emerging Therapies. Stem Cells Int. 2016, 2016, 9798374. [Google Scholar] [CrossRef]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [PubMed]
- Schlötzer-Schrehardt, U.; Kruse, F.E. Identification and characterization of limbal stem cells. Exp. Eye Res. 2005, 81, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Matic, M.; Petrov, I.N.; Chen, S.; Wang, C.; Dimitrijevich, S.D.; Wolosin, J.M. Stem cells of the corneal epithelium lack connexins and metabolite transfer capacity. Differentiation 1997, 61, 251–260. [Google Scholar] [CrossRef]
- Bonnet, C.; González, S.; Roberts, J.S.; Robertson, S.Y.T.; Ruiz, M.; Zheng, J.; Deng, S.X. Human limbal epithelial stem cell regulation, bioengineering and function. Prog. Retin. Eye Res. 2021, 85, 100956. [Google Scholar] [CrossRef]
- Clevers, H.; Loh, K.M.; Nusse, R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef]
- Tseng, S.C. Concept and application of limbal stem cells. Eye 1989, 3 Pt 2, 141–157. [Google Scholar] [CrossRef]
- Di Girolamo, N. Moving epithelia: Tracking the fate of mammalian limbal epithelial stem cells. Prog. Retin. Eye Res. 2015, 48, 203–225. [Google Scholar] [CrossRef]
- Lavker, R.M.; Tseng, S.C.; Sun, T.T. Corneal epithelial stem cells at the limbus: Looking at some old problems from a new angle. Exp. Eye Res. 2004, 78, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Davanger, M.; Evensen, A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature 1971, 229, 560–561. [Google Scholar] [CrossRef]
- Deng, S.X.; Borderie, V.; Chan, C.C.; Dana, R.; Figueiredo, F.C.; Gomes, J.A.P.; Pellegrini, G.; Shimmura, S.; Kruse, F.E. Global Consensus on Definition, Classification, Diagnosis, and Staging of Limbal Stem Cell Deficiency. Cornea 2019, 38, 364–375. [Google Scholar] [CrossRef]
- Pellegrini, G.; Traverso, C.E.; Franzi, A.T.; Zingirian, M.; Cancedda, R.; De Luca, M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet 1997, 349, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 2007, 13, 4042–4045. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal. Transduct. Target Ther. 2022, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, M.N.; Vartanyan, L.; Vu, D.M.; Ng, M.Y.; Li, X.; Deng, S.X. Preferential biological processes in the human limbus by differential gene profiling. PLoS ONE 2013, 8, e61833. [Google Scholar] [CrossRef]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Nelson, W.J.; Nusse, R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 2004, 303, 1483–1487. [Google Scholar] [CrossRef]
- Nakatsu, M.N.; Ding, Z.; Ng, M.Y.; Truong, T.T.; Yu, F.; Deng, S.X. Wnt/β-catenin signaling regulates proliferation of human cornea epithelial stem/progenitor cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4734–4741. [Google Scholar] [CrossRef]
- Lee, H.J.; Wolosin, J.M.; Chung, S.H. Divergent effects of Wnt/β-catenin signaling modifiers on the preservation of human limbal epithelial progenitors according to culture condition. Sci. Rep. 2017, 7, 15241. [Google Scholar] [CrossRef]
- Zhang, C.; Mei, H.; Robertson, S.Y.T.; Lee, H.J.; Deng, S.X.; Zheng, J.J. A Small-Molecule Wnt Mimic Improves Human Limbal Stem Cell Ex Vivo Expansion. iScience 2020, 23, 101075. [Google Scholar] [CrossRef]
- González, S.; Oh, D.; Baclagon, E.R.; Zheng, J.J.; Deng, S.X. Wnt Signaling Is Required for the Maintenance of Human Limbal Stem/Progenitor Cells In Vitro. Investig. Ophthalmol. Vis. Sci. 2019, 60, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.H.; Chen, H.C.; Ma, K.S.; Lai, J.Y.; Yang, U.; Yeh, L.K.; Hsueh, Y.J.; Chu, W.K.; Lai, C.H.; Chen, J.K. Preservation of human limbal epithelial progenitor cells on carbodiimide cross-linked amniotic membrane via integrin-linked kinase-mediated Wnt activation. Acta Biomater. 2016, 31, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Davidson, K.C.; Adams, A.M.; Goodson, J.M.; McDonald, C.E.; Potter, J.C.; Berndt, J.D.; Biechele, T.L.; Taylor, R.J.; Moon, R.T. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA 2012, 109, 4485–4490. [Google Scholar] [CrossRef] [PubMed]
- Szabó, D.J.; Noer, A.; Nagymihály, R.; Josifovska, N.; Andjelic, S.; Veréb, Z.; Facskó, A.; Moe, M.C.; Petrovski, G. Long-Term Cultures of Human Cornea Limbal Explants Form 3D Structures Ex Vivo—Implications for Tissue Engineering and Clinical Applications. PLoS ONE 2015, 10, e0143053. [Google Scholar] [CrossRef] [PubMed]
- Fathke, C.; Wilson, L.; Shah, K.; Kim, B.; Hocking, A.; Moon, R.; Isik, F. Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol. 2006, 7, 4. [Google Scholar] [CrossRef]
- Houschyar, K.S.; Momeni, A.; Pyles, M.N.; Maan, Z.N.; Whittam, A.J.; Siemers, F. Wnt signaling induces epithelial differentiation during cutaneous wound healing. Organogenesis 2015, 11, 95–104. [Google Scholar] [CrossRef]
- Pellegrini, G.; Dellambra, E.; Golisano, O.; Martinelli, E.; Fantozzi, I.; Bondanza, S.; Ponzin, D.; McKeon, F.; De Luca, M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3156–3161. [Google Scholar] [CrossRef]
- Dua, H.S.; Joseph, A.; Shanmuganathan, V.A.; Jones, R.E. Stem cell differentiation and the effects of deficiency. Eye 2003, 17, 877–885. [Google Scholar] [CrossRef]
- Kawasaki, S.; Tanioka, H.; Yamasaki, K.; Connon, C.J.; Kinoshita, S. Expression and tissue distribution of p63 isoforms in human ocular surface epithelia. Exp. Eye Res. 2006, 82, 293–299. [Google Scholar] [CrossRef]
- Gonzalez, G.; Sasamoto, Y.; Ksander, B.R.; Frank, M.H.; Frank, N.Y. Limbal stem cells: Identity, developmental origin, and therapeutic potential. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e303. [Google Scholar] [CrossRef] [PubMed]
- Dua, H.S.; Azuara-Blanco, A. Limbal stem cells of the corneal epithelium. Surv. Ophthalmol. 2000, 44, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.M.; Rank, K.B.; Zeng, Y.; Capen, A.; Yadav, V.; Manro, J.R.; Engler, T.A.; Chedid, M. Activating the Wnt/β-Catenin Pathway for the Treatment of Melanoma—Application of LY2090314, a Novel Selective Inhibitor of Glycogen Synthase Kinase-3. PLoS ONE 2015, 10, e0125028. [Google Scholar] [CrossRef] [PubMed]
- Kunnimalaiyaan, S.; Schwartz, V.K.; Jackson, I.A.; Clark Gamblin, T.; Kunnimalaiyaan, M. Antiproliferative and apoptotic effect of LY2090314, a GSK-3 inhibitor, in neuroblastoma in vitro. BMC Cancer 2018, 18, 560. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, V.; Testa, A.; Di Iorio, E.; Mavilio, F.; Pellegrini, G.; De Luca, M. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J. Cell Biol. 2007, 177, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.; Denduluri, S.; Zhang, B.; Wang, Z.; Yin, L.; Yan, Z.; Kang, R.; Shi, L.L.; Mok, J.; Lee, M.J.; et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014, 1, 149–161. [Google Scholar] [CrossRef]
- Menzel-Severing, J.; Zenkel, M.; Polisetti, N.; Sock, E.; Wegner, M.; Kruse, F.E.; Schlötzer-Schrehardt, U. Transcription factor profiling identifies Sox9 as regulator of proliferation and differentiation in corneal epithelial stem/progenitor cells. Sci. Rep. 2018, 8, 10268. [Google Scholar] [CrossRef]
- Grueterich, M.; Espana, E.M.; Touhami, A.; Ti, S.E.; Tseng, S.C. Phenotypic study of a case with successful transplantation of ex vivo expanded human limbal epithelium for unilateral total limbal stem cell deficiency. Ophthalmology 2002, 109, 1547–1552. [Google Scholar] [CrossRef]
- Ribeiro-Rodrigues, T.M.; Martins-Marques, T.; Morel, S.; Kwak, B.R.; Girao, H. Role of connexin 43 in different forms of intercellular communication-gap junctions, extracellular vesicles and tunnelling nanotubes. J. Cell Sci. 2017, 130, 3619–3630. [Google Scholar] [CrossRef]
- Wang, I.J.; Tsai, R.J.; Yeh, L.K.; Tsai, R.Y.; Hu, F.R.; Kao, W.W. Changes in corneal basal epithelial phenotypes in an altered basement membrane. PLoS ONE 2011, 6, e14537. [Google Scholar] [CrossRef]
- Shanmuganathan, V.A.; Foster, T.; Kulkarni, B.B.; Hopkinson, A.; Gray, T.; Powe, D.G.; Lowe, J.; Dua, H.S. Morphological characteristics of the limbal epithelial crypt. Br. J. Ophthalmol. 2007, 91, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Matic, M.; Petrov, I.N.; Rosenfeld, T.; Wolosin, J.M. Alterations in connexin expression and cell communication in healing corneal epithelium. Investig. Ophthalmol. Vis. Sci. 1997, 38, 600–609. [Google Scholar]
- Rodrigues, M.; Ben-Zvi, A.; Krachmer, J.; Schermer, A.; Sun, T.T. Suprabasal expression of a 64-kilodalton keratin (no. 3) in developing human corneal epithelium. Differentiation 1987, 34, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Bian, F.; Zhang, X.; Qi, H.; Chuang, E.Y.; Pflugfelder, S.C.; Li, D.Q. The β-catenin/Tcf4/survivin signaling maintains a less differentiated phenotype and high proliferative capacity of human corneal epithelial progenitor cells. Int. J. Biochem. Cell Biol. 2011, 43, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Clémence, B.; Denise, O.; Hua, M.; Sarah, R.; Derek, C.; Jean-Louis, B.; Francine, B.-C.; Sophie, X.D.; Jie, J.Z. Wnt6 Plays a Complex Role in Maintaining Human Limbal Stem/Progenitor Cells. Sci. Rep. 2021; under review. [Google Scholar] [CrossRef]
- Zhao, S.; Wan, X.; Dai, Y.; Gong, L.; Le, Q. WNT16B enhances the proliferation and self-renewal of limbal epithelial cells via CXCR4/MEK/ERK signaling. Stem Cell Rep. 2022, 17, 864–878. [Google Scholar] [CrossRef]
- Mei, H.; Nakatsu, M.N.; Baclagon, E.R.; Deng, S.X. Frizzled 7 maintains the undifferentiated state of human limbal stem/progenitor cells. Stem Cells 2014, 32, 938–945. [Google Scholar] [CrossRef]
- Peng, H.; Park, J.K.; Katsnelson, J.; Kaplan, N.; Yang, W.; Getsios, S.; Lavker, R.M. microRNA-103/107 Family Regulates Multiple Epithelial Stem Cell Characteristics. Stem Cells 2015, 33, 1642–1656. [Google Scholar] [CrossRef]
- Ouyang, H.; Xue, Y.; Lin, Y.; Zhang, X.; Xi, L.; Patel, S.; Cai, H.; Luo, J.; Zhang, M.; Zhang, M.; et al. WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis. Nature 2014, 511, 358–361. [Google Scholar] [CrossRef]
- Lyu, J.; Joo, C.K. Wnt-7a up-regulates matrix metalloproteinase-12 expression and promotes cell proliferation in corneal epithelial cells during wound healing. J. Biol. Chem. 2005, 280, 21653–21660. [Google Scholar] [CrossRef]
- Shah, R.; Spektor, T.M.; Punj, V.; Turjman, S.; Ghiam, S.; Kim, J.; Tolstoff, S.; Amador, C.; Chun, S.T.; Weisenberger, D.J.; et al. Wnt5a promotes diabetic corneal epithelial wound healing and limbal stem cell expression. Investig. Ophthalmol. Vis. Sci. 2021, 62, 847. [Google Scholar]
- Han, B.; Chen, S.Y.; Zhu, Y.T.; Tseng, S.C. Integration of BMP/Wnt signaling to control clonal growth of limbal epithelial progenitor cells by niche cells. Stem Cell Res. 2014, 12, 562–573. [Google Scholar] [CrossRef]
- Bonnet, C.; Ruiz, M.; Gonzalez, S.; Tseng, C.H.; Bourges, J.L.; Behar-Cohen, F.; Deng, S.X. Single mRNA detection of Wnt signaling pathway in the human limbus. Exp. Eye Res. 2023, 229, 109337. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, M.; Gorivodsky, M.; Shtrom, S.; Grinberg, A.; Niehrs, C.; Morasso, M.I.; Westphal, H. Dkk2 plays an essential role in the corneal fate of the ocular surface epithelium. Development 2006, 133, 2149–2154. [Google Scholar] [CrossRef] [PubMed]
- Bondestam, J.; Huotari, M.A.; Morén, A.; Ustinov, J.; Kaivo-Oja, N.; Kallio, J.; Horelli-Kuitunen, N.; Aaltonen, J.; Fujii, M.; Moustakas, A.; et al. cDNA cloning, expression studies and chromosome mapping of human type I serine/threonine kinase receptor ALK7 (ACVR1C). Cytogenet. Cell Genet. 2001, 95, 157–162. [Google Scholar] [CrossRef]
- Ibáñez, C.F. Regulation of metabolic homeostasis by the TGF-β superfamily receptor ALK7. Febs J. 2022, 289, 5776–5797. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, D.J.; Guilherme, A.; Danai, L.V.; Heyda, L.; Matevossian, A.; Cohen, J.; Nicoloro, S.M.; Straubhaar, J.; Noh, H.L.; Jung, D.; et al. A major role of insulin in promoting obesity-associated adipose tissue inflammation. Mol. Metab. 2015, 4, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.Y.T.; Roberts, J.S.; Deng, S.X. Regulation of Limbal Epithelial Stem Cells: Importance of the Niche. Int. J. Mol. Sci. 2021, 22, 11975. [Google Scholar] [CrossRef] [PubMed]
- Portal, C.; Wang, Z.; Scott, D.K.; Wolosin, J.M.; Iomini, C. The c-Myc Oncogene Maintains Corneal Epithelial Architecture at Homeostasis, Modulates p63 Expression, and Enhances Proliferation During Tissue Repair. Investig. Ophthalmol. Vis. Sci. 2022, 63, 3. [Google Scholar] [CrossRef] [PubMed]
- Grumolato, L.; Liu, G.; Mong, P.; Mudbhary, R.; Biswas, R.; Arroyave, R.; Vijayakumar, S.; Economides, A.N.; Aaronson, S.A. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010, 24, 2517–2530. [Google Scholar] [CrossRef]
- Kulkarni, B.B.; Tighe, P.J.; Mohammed, I.; Yeung, A.M.; Powe, D.G.; Hopkinson, A.; Shanmuganathan, V.A.; Dua, H.S. Comparative transcriptional profiling of the limbal epithelial crypt demonstrates its putative stem cell niche characteristics. BMC Genom. 2010, 11, 526. [Google Scholar] [CrossRef]
- van Amerongen, R.; Fuerer, C.; Mizutani, M.; Nusse, R. Wnt5a can both activate and repress Wnt/β-catenin signaling during mouse embryonic development. Dev. Biol. 2012, 369, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Mikels, A.J.; Nusse, R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006, 4, e115. [Google Scholar] [CrossRef] [PubMed]



| Symbol | Entrez Gene Name | p-Value | Fold Change |
|---|---|---|---|
| Differentially expressed genes that increase in the hLESCs treated with 5 nM LY2090314 | |||
| ACVR1C | Activin A receptory type 1C | 3.1 × 10−3 | 2.0 |
| AXIN 2 | Axin 2 | 3.9 × 10−6 | 6.36 |
| FZD7 | Frizzled class receptor 7 | 1.3 × 10−4 | 2.24 |
| GJA1 | Gap junction protein alpha 1 | 6.2 × 10−9 | 3.66 |
| MMP7 | Matrix metallopeptidase 7 | 3.2 × 10−5 | 4.3 |
| PPP2R2B | Protein phosphatase 2 regulatory subunit B beta | 6.4 × 10−5 | 4.07 |
| TCF7 | Transcription factor 7 | 2.3 × 10−4 | 2.16 |
| TGFB3 | Transforming growth factor beta 3 | 3.7 × 10−5 | 2.69 |
| WNT5A | Wnt family member 5A | 7.4 × 10−5 | 2.28 |
| Differentially expressed genes that decrease in the hLESCs treated with 5 nM LY2090314 | |||
| DKK1 | Dickkopf Wnt signaling pathway inhibitor | 5.5 × 10−7 | −3.34 |
| MYC | MYC proto-oncogene, bHLH transcription factor | 2.2 × 10−6 | −2.25 |
| SRFP1 | Secreted frizzled related protein 1 | 2.3 × 10−4 | −3.76 |
| TGFB2 | Transforming growth factor beta 2 | 1.1 × 10−3 | −2.42 |
| WNT7A | Wnt family member 7A | 8.3 × 10−4 | −2.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bisevac, J.; Katta, K.; Petrovski, G.; Moe, M.C.; Noer, A. Wnt/β-Catenin Signaling Activation Induces Differentiation in Human Limbal Epithelial Stem Cells Cultured Ex Vivo. Biomedicines 2023, 11, 1829. https://doi.org/10.3390/biomedicines11071829
Bisevac J, Katta K, Petrovski G, Moe MC, Noer A. Wnt/β-Catenin Signaling Activation Induces Differentiation in Human Limbal Epithelial Stem Cells Cultured Ex Vivo. Biomedicines. 2023; 11(7):1829. https://doi.org/10.3390/biomedicines11071829
Chicago/Turabian StyleBisevac, Jovana, Kirankumar Katta, Goran Petrovski, Morten Carstens Moe, and Agate Noer. 2023. "Wnt/β-Catenin Signaling Activation Induces Differentiation in Human Limbal Epithelial Stem Cells Cultured Ex Vivo" Biomedicines 11, no. 7: 1829. https://doi.org/10.3390/biomedicines11071829
APA StyleBisevac, J., Katta, K., Petrovski, G., Moe, M. C., & Noer, A. (2023). Wnt/β-Catenin Signaling Activation Induces Differentiation in Human Limbal Epithelial Stem Cells Cultured Ex Vivo. Biomedicines, 11(7), 1829. https://doi.org/10.3390/biomedicines11071829









