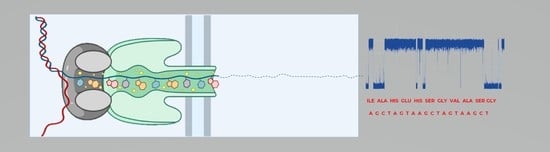
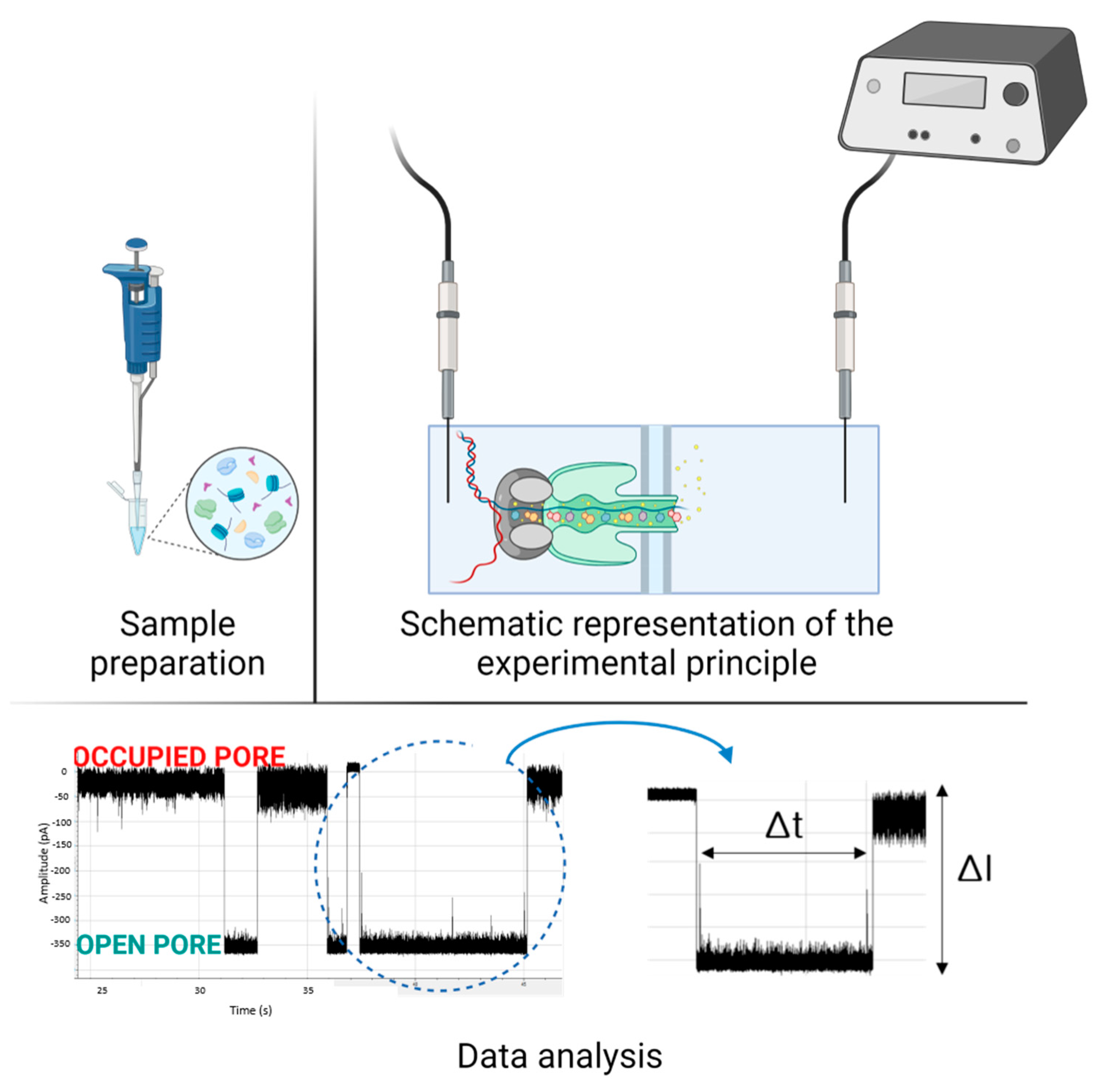
Detection of Biological Molecules Using Nanopore Sensing Techniques
Abstract
:1. Introduction
2. Applications in Sensing and Diagnostic
3. Methodological Approaches and Challenges
3.1. Metabolic Markers
3.1.1. Glucose Detection
3.1.2. Vitamin B1
3.1.3. Uric Acid Detection
3.2. Detection of Nucleic Acids
3.2.1. Bacterial Pathogen Identification
3.2.2. Genetic Markers
3.3. Protein Detection
4. Protein Sequencing
5. Emerging Nanopore Technologies
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kolmogorov, M.; Billingsley, K.J.; Meredith, M.; Monlong, J.; Lorig-Roach, R.; Asri, M.; Jerez, P.A.; Malik, L.; Dewan, R.; Reed, X.; et al. Scalable Nanopore sequencing of human genomes provides a comprehensive view of haplotype-resolved variation and methylation. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Friedman, A.K.; Baker, L.A. Nanopore Sensing. Anal. Chem. 2017, 89, 157–188. [Google Scholar] [CrossRef] [Green Version]
- Smeets, R.M.M.; Keyser, U.F.; Krapf, D.; Wu, M.-Y.; Dekker, N.H.; Dekker, C. Salt Dependence of Ion Transport and DNA Translocation through Solid-State Nanopores. Nano Lett. 2005, 6, 89–95. [Google Scholar] [CrossRef]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; A Benner, S.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Keyser, U.; Koeleman, B.N.; Van Dorp, S.; Krapf, D.; Smeets, R.M.M.; Lemay, S.G.; Dekker, N.; Dekker, C. Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2006, 2, 473–477. [Google Scholar] [CrossRef] [Green Version]
- Crnković, A.; Srnko, M.; Anderluh, G. Biological Nanopores: Engineering on Demand. Life 2021, 11, 27. [Google Scholar] [CrossRef]
- Yuan, W.; Chen, J.; Shi, G. Nanoporous graphene materials. Mater. Today 2014, 17, 77–85. [Google Scholar] [CrossRef]
- Venta, K.; Shemer, G.; Puster, M.; Rodríguez-Manzo, J.A.; Balan, A.; Rosenstein, J.K.; Shepard, K.; Drndić, M. Differentiation of Short, Single-Stranded DNA Homopolymers in Solid-State Nanopores. ACS Nano 2013, 7, 4629–4636. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, T. Progress in single-biomolecule analysis with solid-state nanopores. Curr. Opin. Electrochem. 2017, 4, 159–165. [Google Scholar] [CrossRef]
- Firnkes, M.; Pedone, D.; Knezevic, J.; Döblinger, M.; Rant, U. Electrically Facilitated Translocations of Proteins through Silicon Nitride Nanopores: Conjoint and Competitive Action of Diffusion, Electrophoresis, and Electroosmosis. Nano Lett. 2010, 10, 2162–2167. [Google Scholar] [CrossRef]
- Nehra, A.; Ahlawat, S.; Singh, K.P. A biosensing expedition of nanopore: A review. Sens. Actuators B Chem. 2018, 284, 595–622. [Google Scholar] [CrossRef]
- Li, J.; Golovchenko, J.A. Solid-State Nanopore for Detecting Individual Biopolymers. Micro Nano Technol. Bioanal. Methods Protoc. 2009, 544, 81–93. [Google Scholar] [CrossRef] [Green Version]
- Gorzynski, J.E.; Goenka, S.D.; Shafin, K.; Jensen, T.D.; Fisk, D.G.; Grove, M.E.; Spiteri, E.; Pesout, T.; Monlong, J.; Baid, G.; et al. Ultrarapid Nanopore Genome Sequencing in a Critical Care Setting. N. Engl. J. Med. 2022, 386, 700–702. [Google Scholar] [CrossRef]
- Goenka, S.D.; Gorzynski, J.E.; Shafin, K.; Fisk, D.G.; Pesout, T.; Jensen, T.D.; Monlong, J.; Chang, P.-C.; Baid, G.; Bernstein, J.A.; et al. Accelerated identification of disease-causing variants with ultra-rapid nanopore genome sequencing. Nat. Biotechnol. 2022, 40, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wu, L.; Tu, J.; Lu, Z. Application of Solid-State Nanopore in Protein Detection. Int. J. Mol. Sci. 2020, 21, 2808. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.; Ha, J.-H.; Mayse, L.A.; Presti, M.F.; Wolfe, A.J.; Moody, K.J.; Loh, S.N.; Movileanu, L. A generalizable nanopore sensor for highly specific protein detection at single-molecule precision. Nat. Commun. 2023, 14, 1374. [Google Scholar] [CrossRef]
- Wang, G.; Wang, L.; Han, Y.; Zhou, S.; Guan, X. Nanopore Stochastic Detection: Diversity, Sensitivity, and Beyond. Acc. Chem. Res. 2013, 46, 2867–2877. [Google Scholar] [CrossRef]
- Bayley, H. Nanopore Sequencing: From Imagination to Reality. Clin. Chem. 2015, 61, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Yusko, E.C.; Bruhn, B.R.; Eggenberger, O.; Houghtaling, J.; Rollings, R.C.; Walsh, N.C.; Nandivada, S.; Pindrus, M.; Hall, A.R.; Sept, D.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2016, 12, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Mercado, R.; Moussy, F. In vitro and in vivo mineralization of Nafion membrane used for implantable glucose sensors. Biosens. Bioelectron. 1998, 13, 133–145. [Google Scholar] [CrossRef]
- Li, Q.; Luo, G.; Feng, J.; Zhou, Q.; Zhang, L.; Zhu, Y. Amperometric Detection of Glucose with Glucose Oxidase Absorbed on Porous Nanocrystalline TiO2 Film. Electroanalysis 2001, 13, 413–416. [Google Scholar] [CrossRef]
- Wei, A.; Sun, X.W.; Wang, J.X.; Lei, Y.; Cai, X.P.; Li, C.M.; Dong, Z.L.; Huang, W. Enzymatic glucose biosensor based on ZnO nanorod array grown by hydrothermal decomposition. Appl. Phys. Lett. 2006, 89, 123902. [Google Scholar] [CrossRef]
- Saha, S.; Arya, S.K.; Singh, S.; Sreenivas, K.; Malhotra, B.; Gupta, V. Zinc oxide–potassium ferricyanide composite thin film matrix for biosensing applications. Anal. Chim. Acta 2009, 653, 212–216. [Google Scholar] [CrossRef]
- Lucas, F.L.R.; Piso, T.R.C.; Heide, N.J.; Galenkamp, N.S.; Hermans, J.; Wloka, C.; Maglia, G. Automated Electrical Quantification of Vitamin B1 in a Bodily Fluid using an Engineered Nanopore Sensor. Angew. Chem. Int. Ed. 2021, 60, 22849–22855. [Google Scholar] [CrossRef]
- He, H.; Xu, X.; Wang, P.; Chen, L.; Jin, Y. The facile surface chemical modification of a single glass nanopore and its use in the nonenzymatic detection of uric acid. Chem. Commun. 2014, 51, 1914–1917. [Google Scholar] [CrossRef]
- Cao, S.; Ding, S.; Liu, Y.; Zhu, A.; Shi, G. Biomimetic Mineralization of Gold Nanoclusters as Multifunctional Thin Films for Glass Nanopore Modification, Characterization, and Sensing. Anal. Chem. 2017, 89, 7886–7892. [Google Scholar] [CrossRef]
- Tang, Y.; Li, Z.; Luo, Q.; Liu, J.; Wu, J. Bacteria detection based on its blockage effect on silicon nanopore array. Biosens. Bioelectron. 2016, 79, 715–720. [Google Scholar] [CrossRef]
- Cheng, M.S.; Lau, S.H.; Chow, V.T.; Toh, C.-S. Membrane-Based Electrochemical Nanobiosensor for Escherichia coli Detection and Analysis of Cells Viability. Environ. Sci. Technol. 2011, 45, 6453–6459. [Google Scholar] [CrossRef]
- Tourancheau, A.; Mead, E.A.; Zhang, X.-S.; Fang, G. Discovering multiple types of DNA methylation from bacteria and microbiome using nanopore sequencing. Nat. Methods 2021, 18, 491–498. [Google Scholar] [CrossRef]
- Schmidt, K.; Mwaigwisya, S.; Crossman, L.C.; Doumith, M.; Munroe, D.; Pires, C.; Khan, A.M.; Woodford, N.; Saunders, N.J.; Wain, J.; et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 2016, 72, 104–114. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.-Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotechnol. 2011, 6, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Xi, D.; Li, Z.; Liu, L.; Ai, S.; Zhang, S. Ultrasensitive Detection of Cancer Cells Combining Enzymatic Signal Amplification with an Aerolysin Nanopore. Anal. Chem. 2017, 90, 1029–1034. [Google Scholar] [CrossRef]
- Liu, L.; Li, T.; Zhang, S.; Song, P.; Guo, B.; Zhao, Y.; Wu, H.-C. Simultaneous Quantification of Multiple Cancer Biomarkers in Blood Samples through DNA-Assisted Nanopore Sensing. Angew. Chem. Int. Ed. 2018, 57, 11882–11887. [Google Scholar] [CrossRef]
- Bhatti, H.; Lu, Z.; Liu, Q. Nanopore Detection of Cancer Biomarkers: A Challenge to Science. Technol. Cancer Res. Treat. 2022, 21, 153303382210766. [Google Scholar] [CrossRef] [PubMed]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Alzghoul, S.; Hailat, M.; Zivanovic, S.; Que, L.; Shah, G.V. Measurement of serum prostate cancer markers using a nanopore thin film based optofluidic chip. Biosens. Bioelectron. 2016, 77, 491–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefureac, R.; Long, Y.T.; Kraatz, H.B.; Howard, P.; Lee, J.S. Transport of α-Helical Peptides through α-Hemolysin and Aerolysin Pores. Biochemistry 2006, 45, 9172–9179. [Google Scholar] [CrossRef] [PubMed]
- Soskine, M.; Biesemans, A.; Moeyaert, B.; Cheley, S.; Bayley, H.; Maglia, G. An Engineered ClyA Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012, 12, 4895–4900. [Google Scholar] [CrossRef] [Green Version]
- Mereuta, L.; Roy, M.; Asandei, A.; Lee, J.K.; Park, Y.; Andricioaei, I.; Luchian, T. Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Sci. Rep. 2014, 4, 3885. [Google Scholar] [CrossRef] [Green Version]
- Freedman, K.J.; Bastian, A.R.; Chaiken, I.; Kim, M.J. Solid-State Nanopore Detection of Protein Complexes: Applications in Healthcare and Protein Kinetics. Small 2012, 9, 750–759. [Google Scholar] [CrossRef]
- Turko, I.V.; Murad, F. Protein Nitration in Cardiovascular Diseases. 2002. Available online: http://pharmrev.aspetjournals.org (accessed on 3 June 2022).
- Chen, P.; Sun, Z.; Wang, J.; Liu, X.; Bai, Y.; Chen, J.; Liu, A.; Qiao, F.; Chen, Y.; Yuan, C.; et al. Portable nanopore-sequencing technology: Trends in development and applications. Front. Microbiol. 2023, 14, 1043967. [Google Scholar] [CrossRef] [PubMed]
- Munro, R.; Holmes, N.; Moore, C.; Carlile, M.; Payne, A.; Tyson, J.R.; Williams, T.; Alder, C.; Snell, L.B.; Nebbia, G.; et al. A framework for real-time monitoring, analysis and adaptive sampling of viral amplicon nanopore sequencing. Front. Genet. 2023, 14, 1138582. [Google Scholar] [CrossRef] [PubMed]
- Fahie, M.A.; Li, F.; Palmer, C.; Yoon, C.; Chen, M. Modifying the pH sensitivity of OmpG nanopore for improved detection at acidic pH. Biophys. J. 2022, 121, 731–741. [Google Scholar] [CrossRef]
- Keyser, U.F. Controlling molecular transport through nanopores. J. R. Soc. Interface 2011, 8, 1369–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moragues-Solanas, L.; Scotti, R.; O’grady, J. Rapid metagenomics for diagnosis of bloodstream and respiratory tract nosocomial infections: Current status and future prospects. Expert Rev. Mol. Diagn. 2021, 21, 371–380. [Google Scholar] [CrossRef]
- Makałowski, W.; Shabardina, V. Bioinformatics of nanopore sequencing. J. Hum. Genet. 2019, 65, 61–67. [Google Scholar] [CrossRef]
- White, L.K.; Hesselberth, J.R. Modification mapping by nanopore sequencing. Front. Genet. 2022, 13, 1037134. [Google Scholar] [CrossRef]
- Wan, Y.K.; Hendra, C.; Pratanwanich, P.N.; Göke, J. Beyond sequencing: Machine learning algorithms extract biology hidden in Nanopore signal data. Trends Genet. 2021, 38, 246–257. [Google Scholar] [CrossRef]
- Wang, D.; Qi, G.; Zhou, Y.; Li, H.; Zhang, Y.; Xu, C.; Hu, P.; Jin, Y. Glucose level determination in single cells in their satiety and starvation states using an enzymatic functional glass nanopore. Chem. Commun. 2020, 56, 5393–5396. [Google Scholar] [CrossRef]
- Ramanavicius, S.; Ramanavicius, A. Charge Transfer and Biocompatibility Aspects in Conducting Polymer-Based Enzymatic Biosensors and Biofuel Cells. Nanomaterials 2021, 11, 371. [Google Scholar] [CrossRef]
- Joo, S.; Park, S.; Chung, T.D.; Kim, H.C. Integration of a Nanoporous Platinum Thin Film into a Microfluidic System for Non-enzymatic Electrochemical Glucose Sensing. Anal. Sci. 2007, 23, 277–281. [Google Scholar] [CrossRef] [Green Version]
- Özel, R.E.; Lohith, A.; Mak, W.H.; Pourmand, N. Single-cell intracellular nano-pH probes. RSC Adv. 2015, 5, 52436–52443. [Google Scholar] [CrossRef] [Green Version]
- Umehara, S.; Pourmand, N.; Webb, C.D.; Davis, R.W.; Yasuda, K.; Karhanek, M. Current Rectification with Poly-l-Lysine-Coated Quartz Nanopipettes. Nano Lett. 2006, 6, 2486–2492. [Google Scholar] [CrossRef] [Green Version]
- Vilozny, B.; Actis, P.; Seger, R.A.; Vallmajo-Martin, Q.; Pourmand, N. Reversible Cation Response with a Protein-Modified Nanopipette. Anal. Chem. 2011, 83, 6121–6126. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, R.A.S.; Özel, R.E.; Mak, W.H.; Mulato, M.; Singaram, B.; Pourmand, N. Single Cell “Glucose Nanosensor” Verifies Elevated Glucose Levels in Individual Cancer Cells. Nano Lett. 2016, 16, 1194–1200. [Google Scholar] [CrossRef] [Green Version]
- Collie, J.T.; Greaves, R.F.; Jones, O.A.; Lam, Q.; Eastwood, G.M.; Bellomo, R. Vitamin B1 in critically ill patients: Needs and challenges. Clin. Chem. Lab. Med. 2017, 55, 1652–1668. [Google Scholar] [CrossRef]
- Hrubša, M.; Siatka, T.; Nejmanová, I.; Vopršalová, M.; Krčmová, L.K.; Matoušová, K.; Javorská, L.; Macáková, K.; Mercolini, L.; Remião, F.; et al. Biological Properties of Vitamins of the B-Complex, Part 1: Vitamins B1, B2, B3, and B5. Nutrients 2022, 14, 484. [Google Scholar] [CrossRef]
- Fattal-Valevski, A. Thiamine (Vitamin B1). J. Evid. Based Complement. Altern. Med. 2011, 16, 12–20. [Google Scholar] [CrossRef]
- Basiri, B.; Sutton, J.M.; Hanberry, B.S.; Zastre, J.A.; Bartlett, M.G. Ion pair liquid chromatography method for the determination of thiamine (vitamin B1) homeostasis. Biomed. Chromatogr. 2015, 30, 35–41. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Z.; Liu, Q.; Luo, H.; Zheng, W. Spectrophotometric determination of vitamin B1 in a pharmaceutical formulation using triphenylmethane acid dyes. J. Pharm. Biomed. Anal. 2002, 30, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Frank, R.A.W.; Leeper, F.J.; Luisi, B.F. Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell. Mol. Life Sci. 2007, 64, 892–905. [Google Scholar] [CrossRef]
- Soriano, E.V.; Rajashankar, K.R.; Hanes, J.W.; Bale, S.; Begley, T.P.; Ealick, S.E. Structural Similarities between Thiamin-Binding Protein and Thiaminase-I Suggest a Common Ancestor. Biochemistry 2008, 47, 1346–1357. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, J.; Chen, Z. Colorimetric determination of uric acid based on the suppression of oxidative etching of silver nanoparticles by chloroauric acid. Microchim. Acta 2019, 187, 18. [Google Scholar] [CrossRef] [PubMed]
- Kayamori, Y.; Katayama, Y.; Matsuyama, T.; Urata, T. Enzymatic Method for Assaying Uric Acid in Serum with a New Tetrazolium Salt Produces Water-Soluble Formazan Dye. Clin. Biochem. 1997, 30, 595–599. [Google Scholar] [CrossRef]
- Mo, Y.; Fei, T. Nanoporous membrane for biosensing applications. Nano Life 2012, 2, 1230003. [Google Scholar] [CrossRef]
- Burillo, A.; Bouza, E. Use of rapid diagnostic techniques in ICU patients with infections. BMC Infect. Dis. 2014, 14, 593. [Google Scholar] [CrossRef] [Green Version]
- Kollef, M.H.; Shorr, A.F.; Bassetti, M.; Timsit, J.-F.; Micek, S.T.; Michelson, A.P.; Garnacho-Montero, J. Timing of antibiotic therapy in the ICU. Crit. Care 2021, 25, 360. [Google Scholar] [CrossRef]
- Xu, Y.; Lewandowski, K.; O Downs, L.; Kavanagh, J.; Hender, T.; Lumley, S.; Jeffery, K.; Foster, D.; Sanderson, N.D.; Vaughan, A.; et al. Nanopore metagenomic sequencing of influenza virus directly from respiratory samples: Diagnosis, drug resistance and nosocomial transmission, United Kingdom, 2018/19 influenza season. Eurosurveillance 2021, 26, 2000004. [Google Scholar] [CrossRef]
- Xu, Y.; Lewandowski, K.; Jeffery, K.; Downs, L.O.; Foster, D.; Sanderson, N.D.; Kavanagh, J.; Vaughan, A.; Salvagno, C.; Vipond, R.; et al. Nanopore metagenomic sequencing to investigate nosocomial transmission of human metapneumovirus from a unique genetic group among haematology patients in the United Kingdom. J. Infect. 2020, 80, 571–577. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Li, A.-D.; Deng, Y.; Jiang, X.-T.; Li, L.-G.; Zhang, T. MinION Nanopore Sequencing Enables Correlation between Resistome Phenotype and Genotype of Coliform Bacteria in Municipal Sewage. Front. Microbiol. 2017, 8, 2105. [Google Scholar] [CrossRef] [PubMed]
- Dever, L.A. Mechanisms of Bacterial Resistance to Antibiotics. Arch. Intern. Med. 1991, 151, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef] [PubMed]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chen, Q.; Xiong, M.; Zhao, J.; Shen, S.; Chen, L.; Pan, Y.; Li, Z.; Li, Y. Clinical Performance of Nanopore Targeted Sequencing for Diagnosing Infectious Diseases. Microbiol. Spectr. 2022, 10, e00270-22. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, M.; Fu, A.; Hu, B.; Shen, G.; Liu, R.; Zhao, W.; Jiang, S.; Cai, X.; Li, C.; et al. Same-day simultaneous diagnosis of bacterial and fungal infections in clinical practice by nanopore targeted sequencing. MedRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://nanoporetech.com/about-us/news/new-nanopore-sequencing-chemistry-developers-hands-set-deliver-q20-99-raw-read (accessed on 21 June 2022).
- Huang, Y.-T.; Liu, P.-Y.; Shih, P.-W. Homopolish: A method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 2021, 22, 95. [Google Scholar] [CrossRef]
- Li, J.-Y.; Shen, G.-G.; Liu, T.-G.; Tang, L.V.; Xia, L.-H.; Hu, Y. Nanopore-targeted sequencing for simultaneous diagnosis of suspected sepsis and early targeted therapy. Ann. Transl. Med. 2021, 9, 1749. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-E.; Kwak, J.-W.; Park, J.W. Nanotechnology for Early Cancer Detection. Sensors 2010, 10, 428–455. [Google Scholar] [CrossRef]
- Freedman, K.J.; Otto, L.M.; Ivanov, A.P.; Barik, A.; Oh, S.-H.; Edel, J.B. Nanopore sensing at ultra-low concentrations using single-molecule dielectrophoretic trapping. Nat. Commun. 2016, 7, 10217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hiratani, M.; Nagaoka, K.; Kawano, R. MicroRNA detection at femtomolar concentrations with isothermal amplification and a biological nanopore. Nanoscale 2017, 9, 16124–16127. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, N.; Kawano, R. MicroRNA detection at femtomolar concentration using a probe-based nanopore technique. Biophys. J. 2022, 121, 539a. [Google Scholar] [CrossRef]
- Tabata, Y.; Matsuo, Y.; Fujii, Y.; Ohta, A.; Hirota, K. Rapid detection of single nucleotide polymorphisms using the MinION nanopore sequencer: A feasibility study for perioperative precision medicine. JA Clin. Rep. 2022, 8, 17. [Google Scholar] [CrossRef]
- Watson, C.M.; Holliday, D.L.; Crinnion, L.A.; Bonthron, D.T. Long-read nanopore DNA sequencing can resolve complex intragenic duplication/deletion variants, providing information to enable preimplantation genetic diagnosis. Prenat. Diagn. 2022, 42, 226–232. [Google Scholar] [CrossRef]
- Norris, A.L.; Workman, R.E.; Fan, Y.; Eshleman, J.R.; Timp, W. Nanopore sequencing detects structural variants in cancer. Cancer Biol. Ther. 2016, 17, 246–253. [Google Scholar] [CrossRef]
- Norris, A.L.; Kamiyama, H.; Makohon-Moore, A.; Pallavajjala, A.; Morsberger, L.A.; Lee, K.; Batista, D.; Iacobuzio-Donahue, C.A.; Lin, M.-T.; Klein, A.P.; et al. Transflip mutations produce deletions in pancreatic cancer. Genes Chromosom. Cancer 2015, 54, 472–481. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Zaha, S.; Nagasawa, S.; Miyake, S.; Kojima, Y.; Suzuki, A.; Suzuki, Y.; Seki, M. Long-read whole-genome methylation patterning using enzymatic base conversion and nanopore sequencing. Nucleic Acids Res. 2021, 49, e81. [Google Scholar] [CrossRef]
- Li, Y.; Tollefsbol, T.O. DNA Methylation Detection: Bisulfite Genomic Sequencing Analysis. Epigenet. Protoc. 2011, 791, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Staunstrup, N.H.; Starnawska, A.; Nyegaard, M.; Christiansen, L.; Nielsen, A.L.; Børglum, A.; Mors, O. Genome-wide DNA methylation profiling with MeDIP-seq using archived dried blood spots. Clin. Epigenet. 2016, 8, 81. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://nanoporetech.com/applications/investigation/epigenetics (accessed on 21 April 2023).
- Liu, Y.; Rosikiewicz, W.; Pan, Z.; Jillette, N.; Wang, P.; Taghbalout, A.; Foox, J.; Mason, C.; Carroll, M.; Cheng, A.; et al. DNA methylation-calling tools for Oxford Nanopore sequencing: A survey and human epigenome-wide evaluation. Genome Biol. 2021, 22, 295. [Google Scholar] [CrossRef]
- Simpson, J.T.; E Workman, R.; Zuzarte, P.C.; David, M.; Dursi, L.J.; Timp, W. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 2017, 14, 407–410. [Google Scholar] [CrossRef]
- McIntyre, A.B.R.; Alexander, N.; Grigorev, K.; Bezdan, D.; Sichtig, H.; Chiu, C.Y.; Mason, C.E. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 2019, 10, 579. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Huang, N.; Zhang, Z.; Wang, D.-P.; Liang, F.; Miao, Y.; Xiao, C.-L.; Luo, F.; Wang, J. DeepSignal: Detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 2019, 35, 4586–4595. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Hannon, E.; Schendel, D.; Ladd-Acosta, C.; Grove, J.; Hansen, C.S.; Hougaard, D.M.; Bresnahan, M.; Mors, O.; Hollegaard, M.V.; Bækvad-Hansen, M.; et al. Variable DNA methylation in neonates mediates the association between prenatal smoking and birth weight. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180120. [Google Scholar] [CrossRef] [Green Version]
- Flynn, R.; Washer, S.; Jeffries, A.R.; Andrayas, A.; Shireby, G.; Kumari, M.; Schalkwyk, L.C.; Mill, J.; Hannon, E. Evaluation of nanopore sequencing for epigenetic epidemiology: A comparison with DNA methylation microarrays. Hum. Mol. Genet. 2022, 31, 3181–3190. [Google Scholar] [CrossRef]
- Dronina, J.; Bubniene, U.S.; Ramanavicius, A. The application of DNA polymerases and Cas9 as representative of DNA-modifying enzymes group in DNA sensor design (review). Biosens. Bioelectron. 2021, 175, 112867. [Google Scholar] [CrossRef]
- Dronina, J.; Samukaite-Bubniene, U.; Ramanavicius, A. Towards application of CRISPR-Cas12a in the design of modern viral DNA detection tools (Review). J. Nanobiotechnol. 2022, 20, 41. [Google Scholar] [CrossRef]
- Liu, P. Direct and high-throughput detection of DNA methylation and demethylation intermediates using biological nanopore. Biophys. J. 2023, 122, 288a. [Google Scholar] [CrossRef]
- Powers, A.D.; Palecek, S.P. Protein Analytical Assays for Diagnosing, Monitoring, and Choosing Treatment for Cancer Patients. J. Healthc. Eng. 2012, 3, 503–534. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhao, Z.; Haque, F.; Guo, P. Engineering of protein nanopores for sequencing, chemical or protein sensing and disease diagnosis. Curr. Opin. Biotechnol. 2018, 51, 80–89. [Google Scholar] [CrossRef]
- Plesa, C.; Kowalczyk, S.W.; Zinsmeester, R.; Grosberg, A.Y.; Rabin, Y.; Dekker, C. Fast Translocation of Proteins through Solid State Nanopores. Nano Lett. 2013, 13, 658–663. [Google Scholar] [CrossRef] [Green Version]
- Di Muccio, G.; Rossini, A.E.; Di Marino, D.; Zollo, G.; Chinappi, M. Insights into protein sequencing with an α-Hemolysin nanopore by atomistic simulations. Sci. Rep. 2019, 9, 6440. [Google Scholar] [CrossRef] [Green Version]
- Kroll, F.; Dimitriadis, A.; Campbell, T.; Darwent, L.; Collinge, J.; Mead, S.; Vire, E. Prion protein gene mutation detection using long-read Nanopore sequencing. Sci. Rep. 2022, 12, 8284. [Google Scholar] [CrossRef]
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020, 38, 433–438. [Google Scholar] [CrossRef]
- Stevanovski, I.; Chintalaphani, S.R.; Gamaarachchi, H.; Ferguson, J.M.; Pineda, S.S.; Scriba, C.K.; Tchan, M.; Fung, V.; Ng, K.; Cortese, A.; et al. Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci. Adv. 2022, 8, eabm5386. [Google Scholar] [CrossRef]
- Alzheimer’s Association. 2021 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2021, 17, 327–406. [Google Scholar] [CrossRef]
- Lenhart, B.; Wei, X.; Watson, B.; Wang, X.; Zhang, Z.; Li, C.-Z.; Moss, M.; Liu, C. In vitro biosensing of β-Amyloid peptide aggregation dynamics using a biological nanopore. Sens. Actuators B Chem. 2021, 338, 129863. [Google Scholar] [CrossRef]
- Sharma, N. Exploring Biomarkers for Alzheimer’s Disease. J. Clin. Diagn. Res. 2016, 10, KE01. [Google Scholar] [CrossRef]
- Giamblanco, N.; Coglitore, D.; Janot, J.-M.; Coulon, P.E.; Charlot, B.; Balme, S. Detection of protein aggregate morphology through single antifouling nanopore. Sens. Actuators B Chem. 2018, 260, 736–745. [Google Scholar] [CrossRef]
- Meyer, N.; Abrao-Nemeir, I.; Janot, J.-M.; Torrent, J.; Lepoitevin, M.; Balme, S. Solid-state and polymer nanopores for protein sensing: A review. Adv. Colloid Interface Sci. 2021, 298, 102561. [Google Scholar] [CrossRef]
- INir, I.; Huttner, D.; Meller, A. Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores. Biophys. J. 2015, 108, 2340–2349. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhao, J.; Si, W.; Kan, Y.; Xu, Z.; Sha, J.; Chen, Y. Electroosmotic Facilitated Protein Capture and Transport through Solid-State Nanopores with Diameter Larger than Length. Small Methods 2020, 4, 1900893. [Google Scholar] [CrossRef]
- Wei, R.; Gatterdam, V.; Wieneke, R.; Tampé, R.; Rant, U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 2012, 7, 257–263. [Google Scholar] [CrossRef]
- Das, N.; Ray, R.; Ray, S.; Roychaudhuri, C. Intelligent Quantification of Picomolar Protein Concentration in Serum by Functionalized Nanopores. IEEE Sens. J. 2018, 18, 10183–10191. [Google Scholar] [CrossRef]
- Meyer, N.; Janot, J.-M.; Torrent, J.; Balme, S. Real-Time Fast Amyloid Seeding and Translocation of α-Synuclein with a Nanopipette. ACS Cent. Sci. 2022, 8, 441–448. [Google Scholar] [CrossRef]
- Euskirchen, P.; Bielle, F.; Labreche, K.; Kloosterman, W.P.; Rosenberg, S.; Daniau, M.; Schmitt, C.; Masliah-Planchon, J.; Bourdeaut, F.; Dehais, C.; et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017, 134, 691–703. [Google Scholar] [CrossRef] [Green Version]
- Shah, G. Human Cancer Cell Specific Gene Transcript. Google Patents 7,713,693, 11 May 2010. [Google Scholar]
- Ouellette, R.J.; Rawn, J.D. Amino Acids, Peptides, and Proteins. In Organic Chemistry; Elsevier: Amsterdam, The Netherlands, 2018; pp. 929–971. [Google Scholar] [CrossRef]
- Huang, G.; Corstiaan, R.; Versloot, A.; Maglia, G. Detection of single amino acid differences in haemoglobin from blood samples using a nanopore. ChemRxiv 2021. [Google Scholar] [CrossRef]
- Farrokhnia, M.; Amoabediny, G.; Ebrahimi, M.; Ganjali, M.; Arjmand, M. Ultrasensitive early detection of insulin antibody employing novel electrochemical nano-biosensor based on controllable electro-fabrication process. Talanta 2021, 238, 122947. [Google Scholar] [CrossRef]
- Rutkowska, A.; Freedman, K.; Skalkowska, J.; Kim, M.J.; Edel, J.B.; Albrecht, T. Electrodeposition and Bipolar Effects in Metallized Nanopores and Their Use in the Detection of Insulin. Anal. Chem. 2015, 87, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Lastra, L.S.; Sharma, V.; Farajpour, N.; Nguyen, M.; Freedman, K.J. Nanodiagnostics: A review of the medical capabilities of nanopores. Nanomed. Nanotechnol. Biol. Med. 2021, 37, 102425. [Google Scholar] [CrossRef] [PubMed]
- Cardozo, N.; Zhang, K.; Doroschak, K.; Nguyen, A.; Siddiqui, Z.; Bogard, N.; Strauss, K.; Ceze, L.; Nivala, J. Multiplexed direct detection of barcoded protein reporters on a nanopore array. Nat. Biotechnol. 2021, 40, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.L.; Huo, M.Z.; Ying, Y.L.; Long, Y.T. Biological Nanopore Approach for Single-Molecule Protein Sequencing. Angew. Chem. Int. Ed. Engl. 2020, 60, 14738–14749. [Google Scholar] [CrossRef] [PubMed]
- Ouldali, H.; Sarthak, K.; Ensslen, T.; Piguet, F.; Manivet, P.; Pelta, J.; Behrends, J.C.; Aksimentiev, A.; Oukhaled, A. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 2019, 38, 176–181. [Google Scholar] [CrossRef]
- Piguet, F.; Ouldali, H.; Pastoriza-Gallego, M.; Manivet, P.; Pelta, J.; Oukhaled, A. Identification of single amino acid differences in uniformly charged homopolymeric peptides with aerolysin nanopore. Nat. Commun. 2018, 9, 996. [Google Scholar] [CrossRef]
- Chavis, A.E.; Brady, K.T.; Hatmaker, G.A.; Angevine, C.E.; Kothalawala, N.; Dass, A.; Robertson, J.W.F.; Reiner, J.E. Single Molecule Nanopore Spectrometry for Peptide Detection. ACS Sens. 2017, 2, 1319–1328. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wu, X.; Li, M.; Ying, Y.; Long, Y. Diversified exploitation of aerolysin nanopore in single-molecule sensing and protein sequencing. View 2020, 1, 20200006. [Google Scholar] [CrossRef]
- Restrepo-Pérez, L.; Joo, C.; Dekker, C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 2018, 13, 786–796. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhou, J.; Maccaferri, N.; Krahne, R.; Wang, K.; Garoli, D. Enhanced Optical Spectroscopy for Multiplexed DNA and Protein-Sequencing with Plasmonic Nanopores: Challenges and Prospects. Anal. Chem. 2022, 94, 503–514. [Google Scholar] [CrossRef]
- Zhao, Y.; Iarossi, M.; De Fazio, A.F.; Huang, J.-A.; De Angelis, F. Label-Free Optical Analysis of Biomolecules in Solid-State Nanopores: Toward Single-Molecule Protein Sequencing. ACS Photonics 2022, 9, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Garoli, D.; Yamazaki, H.; Maccaferri, N.; Wanunu, M. Plasmonic Nanopores for Single-Molecule Detection and Manipulation: Toward Sequencing Applications. Nano Lett. 2019, 19, 7553–7562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Gu, L.-Q.; Tian, K. The aerolysin nanopore: From peptidomic to genomic applications. Nanoscale 2018, 10, 13857–13866. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Krapp, L.F.; Al Ouahabi, A.; König, N.F.; Cirauqui, N.; Radenovic, A.; Lutz, J.-F.; Peraro, M.D. Aerolysin nanopores decode digital information stored in tailored macromolecular analytes. Sci. Adv. 2020, 6, eabc2661. [Google Scholar] [CrossRef]
- Patil, S.; Augustine, D.; Sowmya, S.; Haragannavar, V.C.; Gujjar, N.; Yousef, A.; Kashyap, S. Nanopore Sequencing Technology in Oral Oncology: A Comprehensive Insight. J. Contemp. Dent. Pr. 2022, 23, 268–275. [Google Scholar] [CrossRef]

| Molecule of Interest | Disease/Physiological Status | Experimental Setup | Reference | |
|---|---|---|---|---|
| Metabolic markers | Glucose | Diabetes, cancer | Glass nanopore based on nano pipettes | [22,23,24,25] |
| Vitamin B1 | Normal functioning of the human body | ClyA | [26] | |
| Uric acid | Gout, arthritis, kidney disease | Glass capillary nano-pipette | [27,28] | |
| Detection of nucleic acids | Bacteria | Infectious diseases | α-HL, silicon nanopore, MinION™ | [29,30,31,32] |
| Genetic markers: microRNA, pancreatic cancer marker, breast cancer marker | Cancer | Biological and solid-state nanopores | [33,34,35,36,37,38] | |
| Protein detection | Prion gene, ᏸ-amyloid, α-synuclein, PSA, insulin, hemoglobin | Neurologic diseases, cancer, autoimmune diseases, etc. | α-HL, ClyA, aerolysin, SiN, graphene Al2O3, SiO2, PlyAB | [17,39,40,41,42,43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Șoldănescu, I.; Lobiuc, A.; Covașă, M.; Dimian, M. Detection of Biological Molecules Using Nanopore Sensing Techniques. Biomedicines 2023, 11, 1625. https://doi.org/10.3390/biomedicines11061625
Șoldănescu I, Lobiuc A, Covașă M, Dimian M. Detection of Biological Molecules Using Nanopore Sensing Techniques. Biomedicines. 2023; 11(6):1625. https://doi.org/10.3390/biomedicines11061625
Chicago/Turabian StyleȘoldănescu, Iuliana, Andrei Lobiuc, Mihai Covașă, and Mihai Dimian. 2023. "Detection of Biological Molecules Using Nanopore Sensing Techniques" Biomedicines 11, no. 6: 1625. https://doi.org/10.3390/biomedicines11061625
APA StyleȘoldănescu, I., Lobiuc, A., Covașă, M., & Dimian, M. (2023). Detection of Biological Molecules Using Nanopore Sensing Techniques. Biomedicines, 11(6), 1625. https://doi.org/10.3390/biomedicines11061625