Identification of m7G-Related miRNA Signatures Associated with Prognosis, Oxidative Stress, and Immune Landscape in Lung Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Screening of m7G-Related Genes and Prediction of m7G-Related miRNAs
2.3. Screening of Oxidative Stress-Related Genes
2.4. Differential Expression Analysis
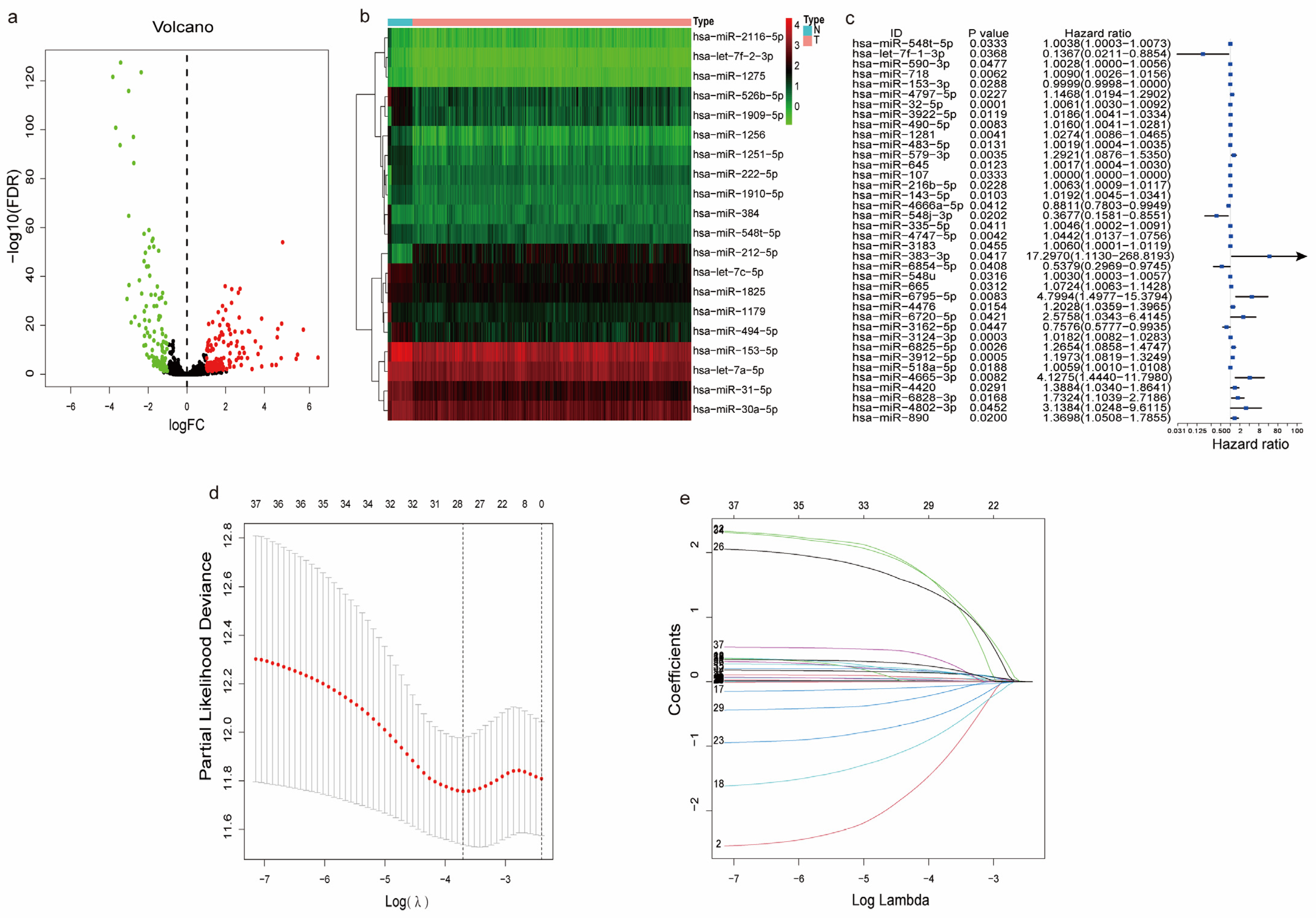
2.5. Construction and Validation of a Prognostic m7G-Related DEMs
2.6. Establishment of a Predictive Nomogram and Calibration
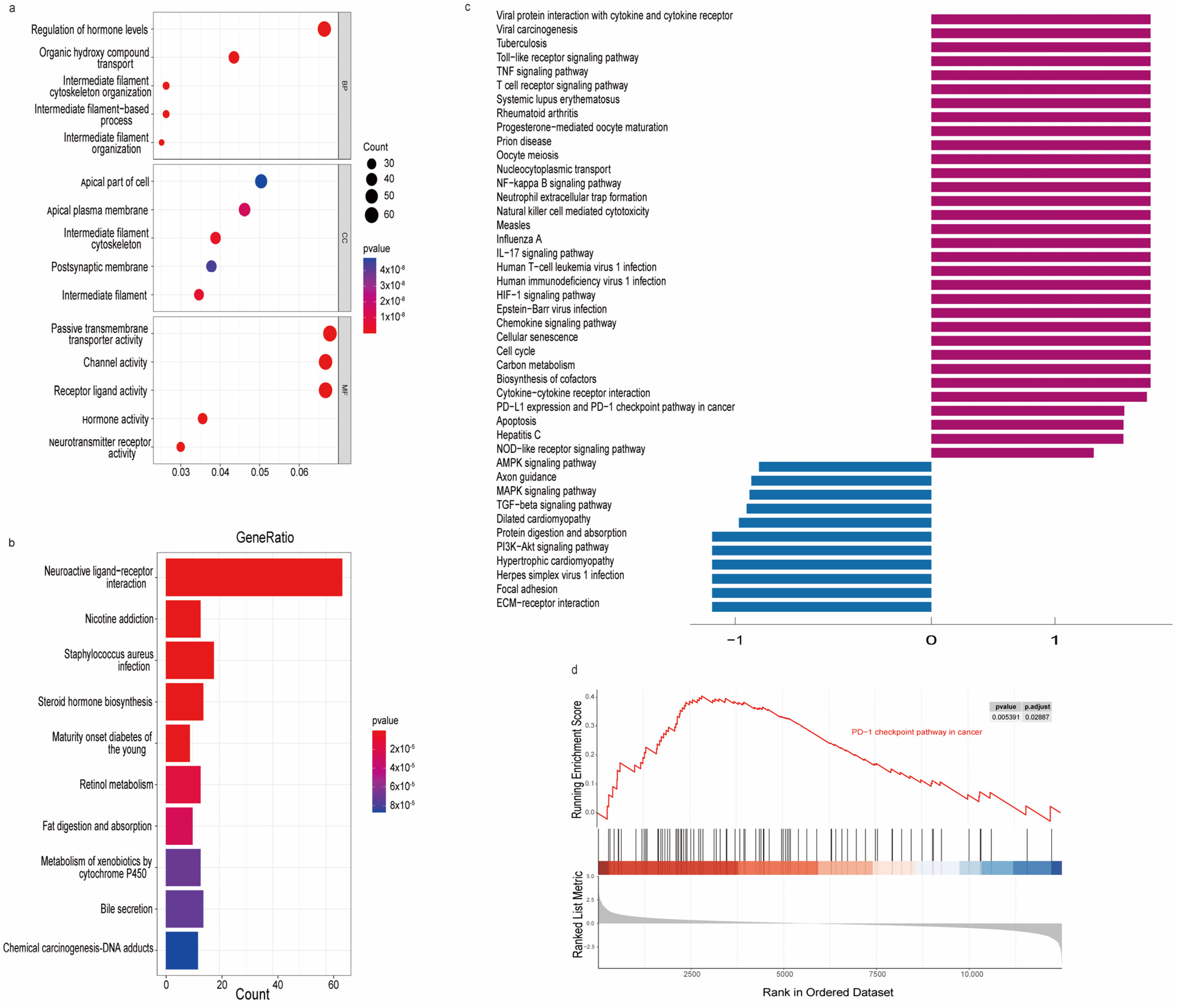
2.7. Functional Enrichment Analysis
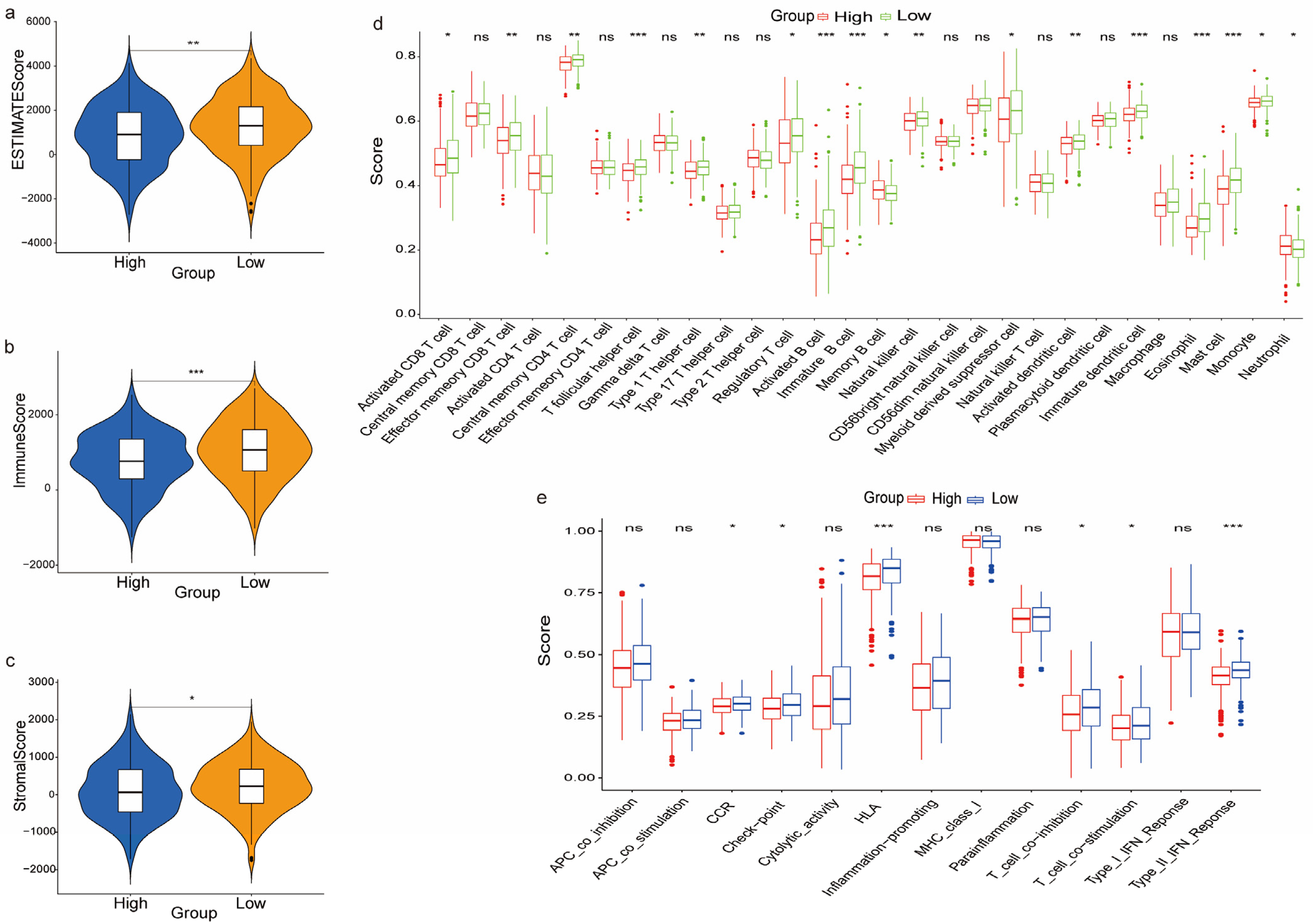
2.8. Tumor Microenvironment and Immune Landscape Analysis
2.9. Protein Expression and Prognostic Power of Oxidative Stress-Related Genes in LUAD
2.10. RNA Isolation
2.11. Quantitative Real-Time PCR
2.12. Subgroup Analyses of Drug Response
2.13. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Identification of m7G-Related miRNAs in LUAD
3.3. Differentially Expressed and Functional Analysis of the m7G-Related miRNAs
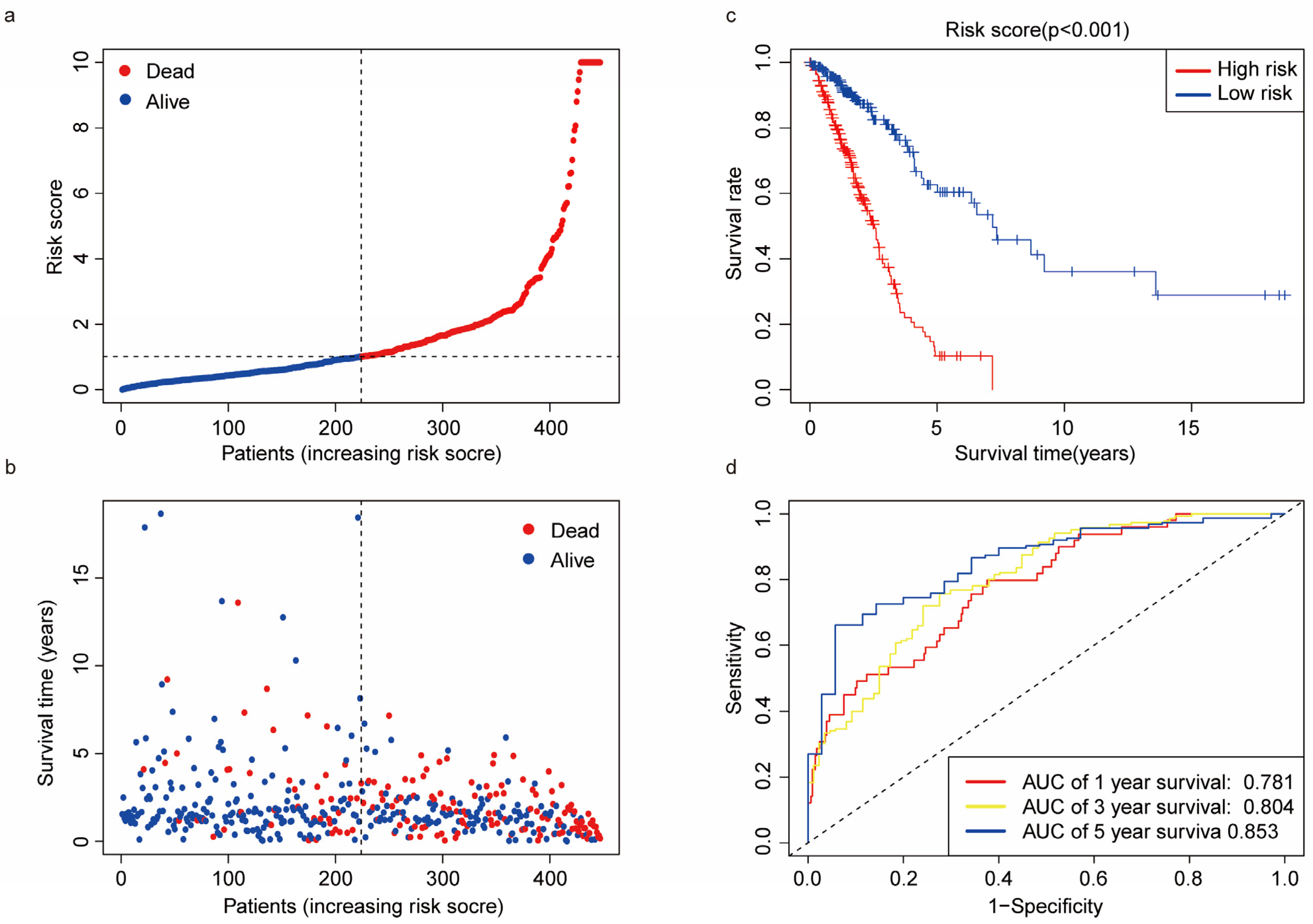
3.4. Predictive Ability of the Risk Score
3.5. Nomogram Construction and Calibration
3.6. Functional Enrichment Analysis
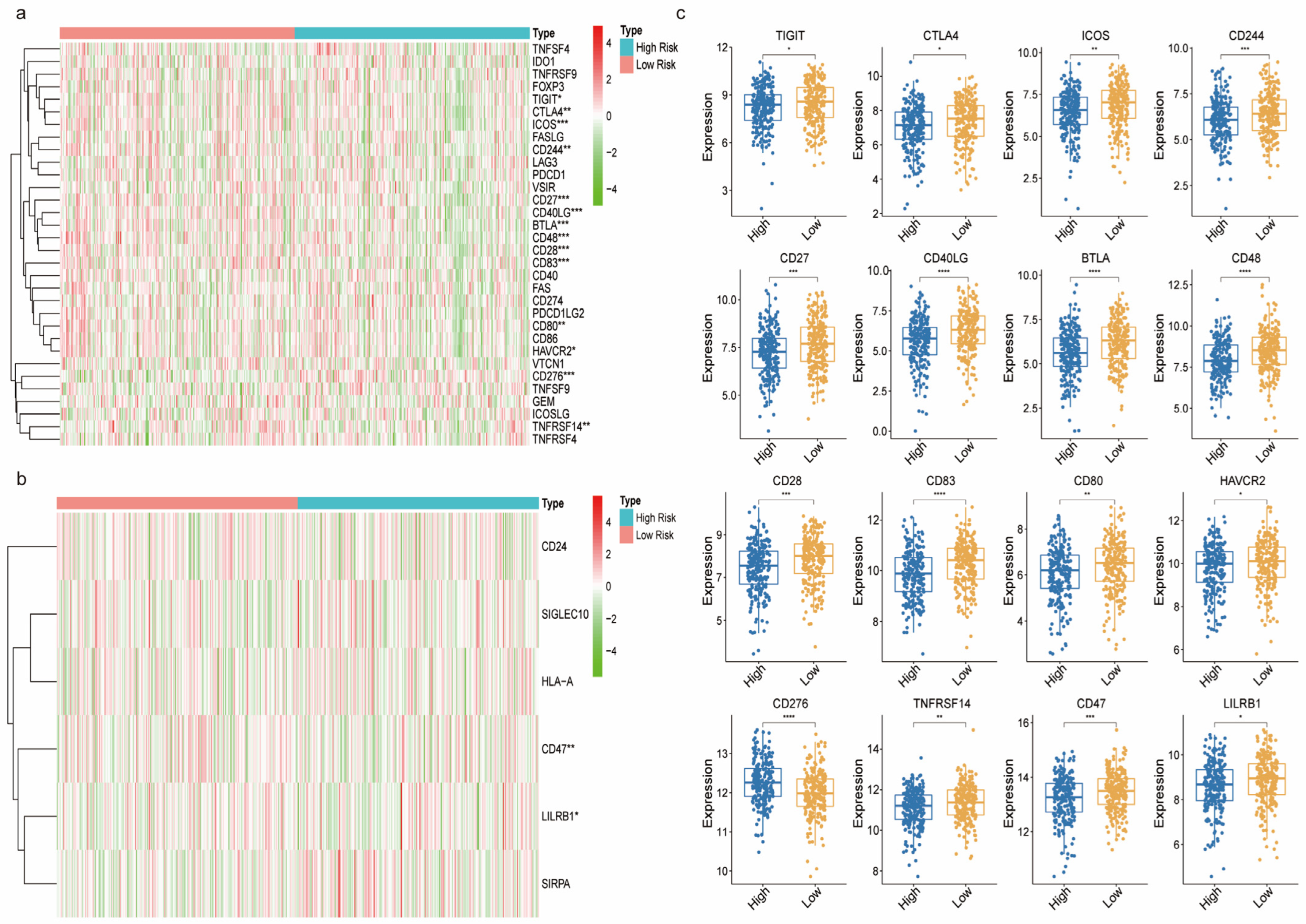
3.7. The Immune Landscape between the High and Low-Risk Groups
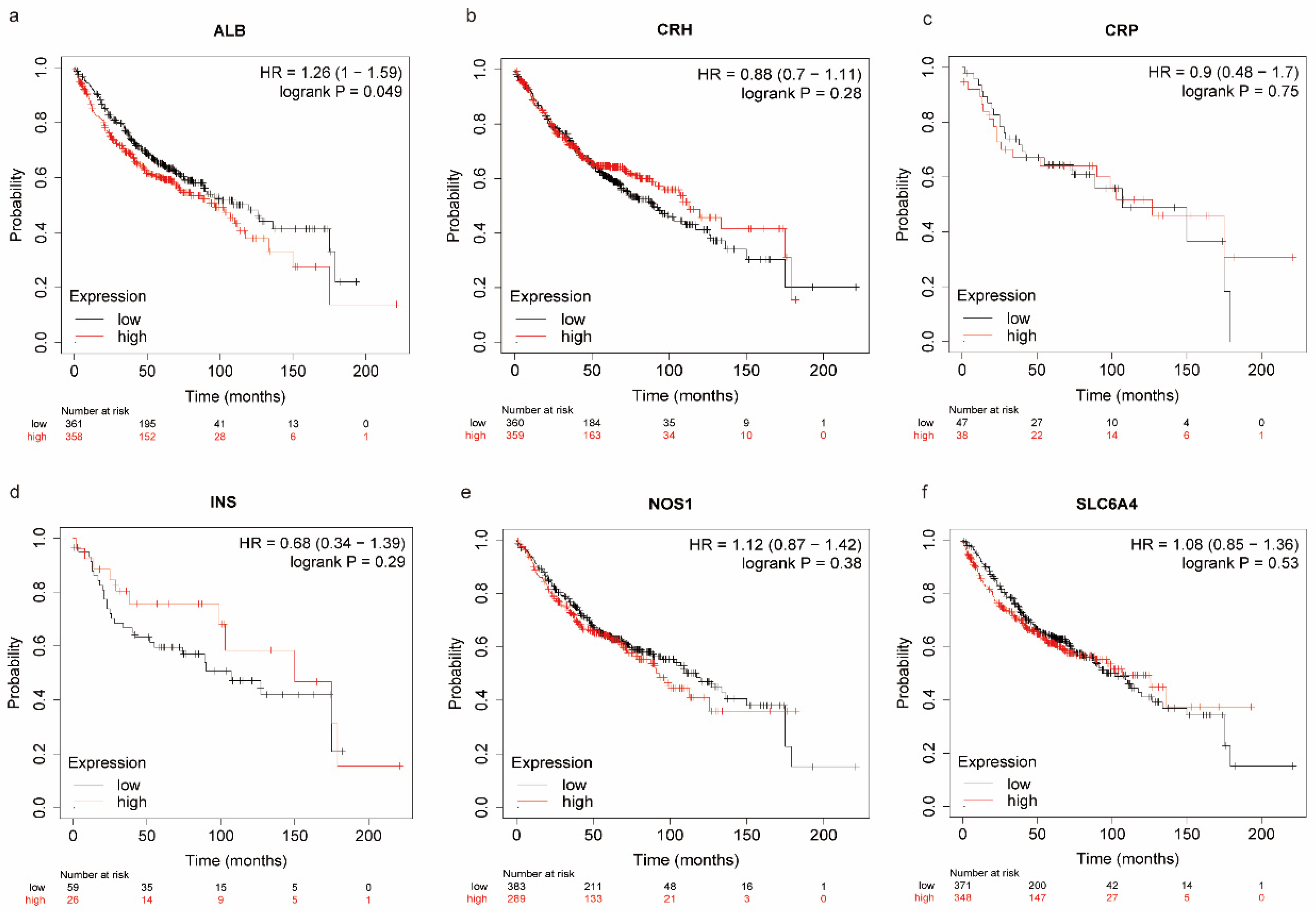
3.8. The Oxidative Stress-Related Genes between the High and Low-Risk Groups
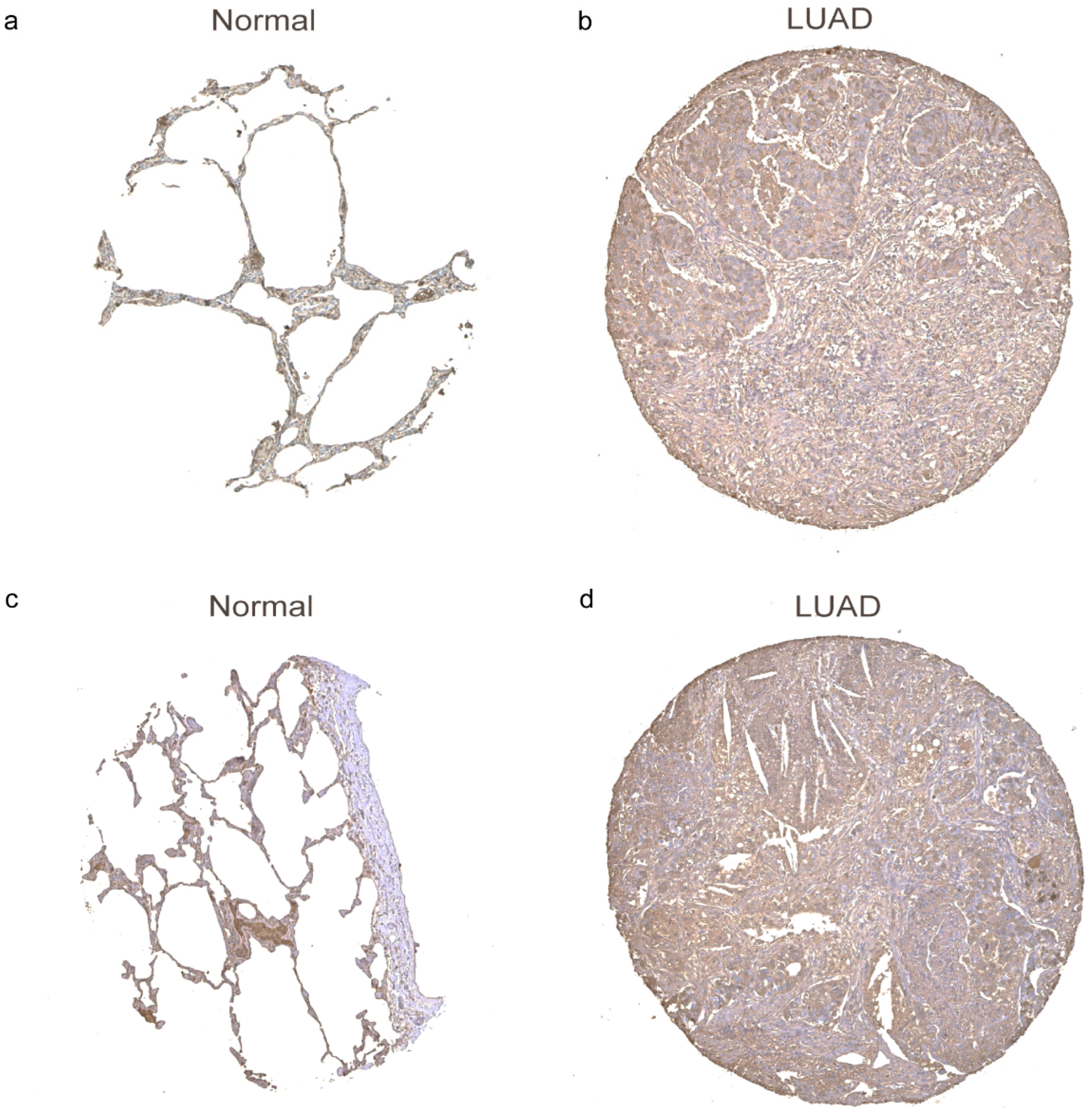
3.9. The Protein Expression Levels of Oxidative Stress-Related Genes in LUAD Tissues
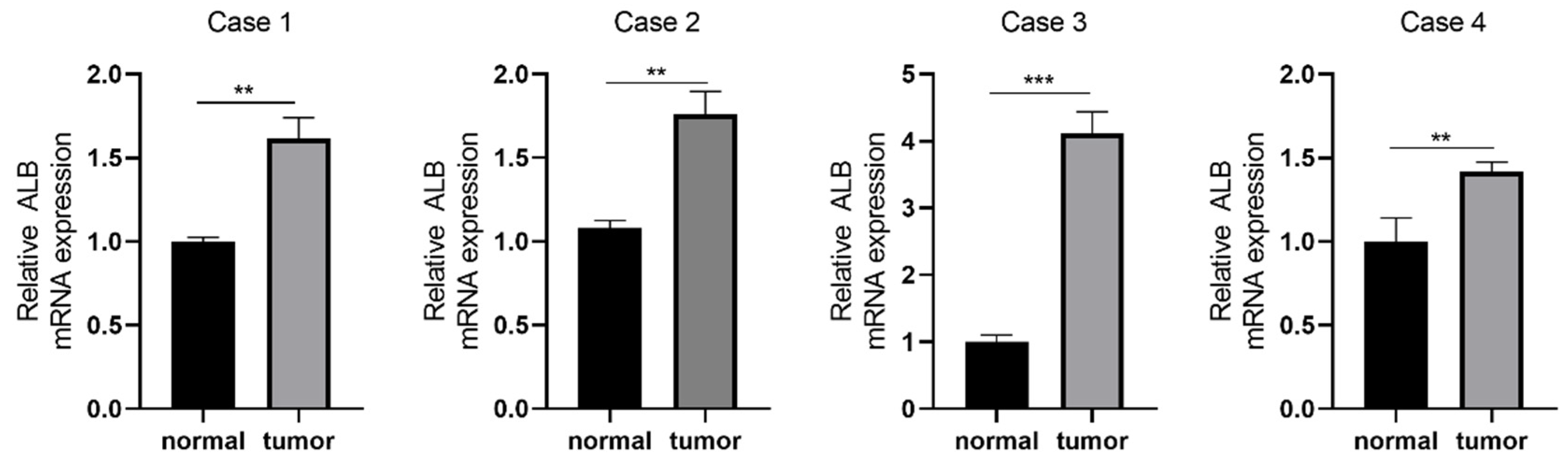
3.10. The Oxidative Stress-Related Genes Validation in LUAD Patients
3.11. Drug Responses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, G.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Brzeziańska, E.; Dutkowska, A.; Antczak, A. The significance of epigenetic alterations in lung carcinogenesis. Mol. Biol. Rep. 2012, 40, 309–325. [Google Scholar] [CrossRef]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Haruehanroengra, P.; Zheng, Y.Y.; Zhou, Y.; Huang, Y.; Sheng, J. RNA modifications and cancer. RNA Biol. 2020, 17, 1560–1575. [Google Scholar] [CrossRef]
- Sprinzl, M.; Hartmann, T.; Meissner, F.; Moll, J.; Vorderwulbecke, T. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1987, 15, r53–r188. [Google Scholar] [CrossRef]
- Edmonds, C.G.; Crain, P.F.; Gupta, R.; Hashizume, T.; Hocart, C.H.; Kowalak, J.A.; Pomerantz, S.C.; Stetter, K.O.; McCloskey, J.A. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 1991, 173, 3138–3148. [Google Scholar] [CrossRef]
- Pei, Y.; Shuman, S. Interactions between Fission Yeast mRNA Capping Enzymes and Elongation Factor Spt5. J. Biol. Chem. 2002, 277, 19639–19648. [Google Scholar] [CrossRef]
- Qiu, Z.R.; Chico, L.; Chang, J.; Shuman, S.; Schwer, B. Genetic interactions of hypomorphic mutations in the m7G cap-binding pocket of yeast nuclear cap binding complex: An essential role for Cbc2 in meiosis via splicing of MER3 pre-mRNA. RNA 2012, 18, 1996–2011. [Google Scholar] [CrossRef]
- Murthy, K.G.; Park, P.; Manley, J.L. A nuclear micrococcal-sensitive, ATP-dependent exoribonuclease degrades uncapped but not capped RNA substrates. Nucleic Acids Res. 1991, 19, 2685–2692. [Google Scholar] [CrossRef]
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Marchand, V.; Ayadi, L.; Ernst, F.G.M.; Hertler, J.; Bourguignon-Lgel, V.; Galvanin, A.; Kotter, A.; Helm, M.; Lafontaine, D.L.J.; Motorin, Y. AlkAniline-Seq: Profiling of m(7) G and m(3) C RNA Modifications at Single Nucleotide Resolution. Angew. Chem. Int. Ed. Engl. 2018, 57, 16785–16790. [Google Scholar] [CrossRef]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2016, 14, 1138–1152. [Google Scholar] [CrossRef]
- Guy, M.P.; Phizicky, E.M. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014, 11, 1608–1618. [Google Scholar] [CrossRef]
- Tomikawa, C. 7-Methylguanosine Modifications in Transfer RNA (tRNA). Int. J. Mol. Sci. 2018, 19, 4080. [Google Scholar] [CrossRef]
- Haag, S.; Kretschmer, J.; Bohnsack, M.T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 2015, 21, 180–187. [Google Scholar] [CrossRef]
- Figaro, S.; Wacheul, L.; Schillewaert, S.; Graille, M.; Huvelle, E.; Mongeard, R.; Zorbas, C.; Lafontaine, D.L.J.; Heurgué-Hamard, V. Trm112 Is Required for Bud23-Mediated Methylation of the 18S rRNA at Position G1575. Mol. Cell. Biol. 2012, 32, 2254–2267. [Google Scholar] [CrossRef]
- Alexandrov, A.; Martzen, M.R.; Phizicky, E.M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 2002, 8, 1253–1266. [Google Scholar] [CrossRef]
- Zhang, L.S.; Liu, C.; Ma, H.; Dai, Q.; Sun, H.-L.; Luo, G.; Zhang, Z.; Zhang, L.; Hu, L.; Dong, X.; et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell 2019, 74, 1304–1316.e8. [Google Scholar] [CrossRef]
- Bueren-Calabuig, J.A.; Bage, M.G.; Cowling, V.H.; Pisliakov, A.V. Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: Insights from accelerated molecular dynamics simulations. Nucleic Acids Res. 2019, 47, 8675–8692. [Google Scholar] [CrossRef]
- Luo, Y.; Yao, Y.; Wu, P.; Zi, X.; Sun, N.; He, J. The potential role of N7-methylguanosine (m7G) in cancer. J. Hematol. Oncol. 2022, 15, 63. [Google Scholar] [CrossRef]
- Chalbatani, G.M.; Momeni, S.A.; Hadloo, M.H.M.; Karimi, Z.; Hadizadeh, M.; Jalali, S.A.; Miri, S.R.; Memari, F.; Hamblin, M.R. Comprehensive analysis of ceRNA networks to determine genes related to prognosis, overall survival, and immune infiltration in clear cell renal carcinoma. Comput. Biol. Med. 2021, 141, 105043. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef]
- Abbasian, M.H.; Ardekani, A.M.; Sobhani, N.; Roudi, R. The Role of Genomics and Proteomics in Lung Cancer Early Detection and Treatment. Cancers 2022, 14, 5144. [Google Scholar] [CrossRef]
- Mraz, M.; Malinova, K.; Mayer, J.; Pospisilova, S. MicroRNA isolation and stability in stored RNA samples. Biochem. Biophys. Res. Commun. 2009, 390, 1–4. [Google Scholar] [CrossRef]
- Pandolfini, L.; Barbieri, I.; Bannister, A.J.; Hendrick, A.; Andrews, B.; Webster, N.; Murat, P.; Mach, P.; Brandi, R.; Robson, S.C.; et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol. Cell 2019, 74, 1278–1290.e9. [Google Scholar] [CrossRef]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef]
- Diehn, M.; Cho, R.O.; Lobo, N.A.; Kalisky, T.; Dorie, M.J.; Kulp, A.N.; Qian, D.; Lam, J.S.; Ailles, L.E.; Wong, M.; et al. Faculty Opinions recommendation of Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009, 458, 780–783. [Google Scholar] [CrossRef]
- Esme, H.; Cemek, M.; Sezer, M.; Saglam, H.; Demir, A.; Melek, H.; Unlu, M. High levels of oxidative stress in patients with advanced lung cancer. Respirology 2008, 13, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Srivastava, S.; Prasad, R.; Natu, S.M.; Mittal, B.; Negi, M.P.; Srivastava, A.N. Oxidative stress in non-small cell lung cancer patients after chemotherapy: Association with treatment response. Respirology 2010, 15, 349–356. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kern, F.; Fehlmann, T.; Solomon, J.; Schwed, L.; Grammes, N.; Backes, C.; Van Keuren-Jensen, K.; Craig, D.W.; Meese, E.; Keller, A. miEAA 2.0: Integrating multi-species microRNA enrichment analysis and workflow management systems. Nucleic Acids Res. 2020, 48, W521–W528. [Google Scholar] [CrossRef]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R Package for Prediction of Clinical Chemotherapeutic Response from Tumor Gene Expression Levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.-K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef]
- Langer, C.J.; Besse, B.; Gualberto, A.; Brambilla, E.; Soria, J.-C. The Evolving Role of Histology in the Management of Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2010, 28, 5311–5320. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, W.; Zhu, S.; Sun, K.; Liao, J.; Liu, H.; Dai, Z.; Han, H.; Ren, X.; Yang, Q.; et al. METTL1 promotes hepatocarcinogenesis via m(7) G tRNA modification-dependent translation control. Clin. Transl. Med. 2021, 11, e661. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2018, 70, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Bi, M.; Zhang, H.; Shi, M. Downregulation of NEAT1 Suppresses Cell Proliferation, Migration, and Invasion in NSCLC Via Sponging miR-153-3p. Cancer Biotherapy Radiopharm. 2020, 35, 362–370. [Google Scholar] [CrossRef]
- Zhang, J.-X.; Yang, W.; Wu, J.-Z.; Zhou, C.; Liu, S.; Shi, H.-B.; Zhou, W.-Z. MicroRNA-32-5p inhibits epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting SMAD family 3. J. Cancer 2021, 12, 2258–2267. [Google Scholar] [CrossRef]
- Kang, B.; Qiu, C.; Zhang, Y. The Effect of lncRNA SNHG3 Overexpression on Lung Adenocarcinoma by Regulating the Expression of miR-890. J. Health Eng. 2021, 2021, 1643788. [Google Scholar] [CrossRef]
- Xie, X.; Ding, F.; Xiao, H. Knockdown of hsa_circ_0000729 Inhibits the Tumorigenesis of Non-Small Cell Lung Cancer Through Mediation of miR-1281/FOXO3 Axis. Cancer Manag. Res. 2021, 13, 8445–8455. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.; Xie, N.; Shao, L.; Sun, H.; Wei, Y.; Sun, Y.; Wang, P.; Yan, Y.; Xie, S.; et al. Anticancer roles of let-7f-1-3p in non-small cell lung cancer via direct targeting of integrin β1. Exp. Ther. Med. 2021, 22, 1305. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qin, F.; Hu, F.; Xu, H.; Sun, G.; Han, G.; Wang, T.; Guo, M. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. 2018, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Lin, M.; He, L.; Qi, H.; Shen, J.; Ying, K. Exosomal lncRNA SCIRT/miR-665 Transferring Promotes Lung Cancer Cell Metastasis through the Inhibition of HEYL. J. Oncol. 2021, 2021, 9813773. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Z.; Li, S.; Han, Z.; Meng, F.; Wei, L. Circ_0089823 reinforces malignant behaviors of non-small cell lung cancer by acting as a sponge for microRNAs targeting SOX4. Neoplasia 2021, 23, 887–897. [Google Scholar] [CrossRef]
- Ge, W.L.; Chen, Q.; Meng, L.-D.; Huang, X.-M.; Shi, G.-D.; Zong, Q.-Q.; Sehn, P.; Lu, Y.-C.; Zhang, Y.-H.; Miao, Y.; et al. The YY1/miR-548t-5p/CXCL11 signaling axis regulates cell proliferation and metastasis in human pancreatic cancer. Cell Death Dis. 2020, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.-H.; Jiang, J.; Zhang, W.; Sun, L.; Li, X.-J.; Li, C.; Ge, Q.-D.; Zhuang, Z.-G. A Novel lncRNA HOXC-AS3 Acts as a miR-3922-5p Sponge to Promote Breast Cancer Metastasis. Cancer Investig. 2019, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, M.; Qi, L.; Zu, X.; Li, Y.; Liu, L.; Chen, M.; Li, Y.; He, W.; Hu, X.; et al. ERα-mediated alterations in circ_0023642 and miR-490-5p signaling suppress bladder cancer invasion. Cell Death Dis. 2019, 10, 635. [Google Scholar] [CrossRef]
- Xue, C.; Zhao, Y.; Li, G.; Li, L. Multi-Omic Analyses of the m5C Regulator ALYREF Reveal Its Essential Roles in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 633415. [Google Scholar] [CrossRef]
- Zhang, C.; Li, H.; Wang, J.; Zhang, J.; Hou, X. MicroRNA-338-3p suppresses cell proliferation, migration and invasion in human malignant melanoma by targeting MACC1. Exp. Ther. Med. 2019, 18, 997–1004. [Google Scholar] [CrossRef]
- Wang, W.; Xu, S.; Di, Y.; Zhang, Z.; Li, Q.; Guo, K.; Lv, Y.; Wang, B. Novel role of LINC01013/miR-6795-5p/FMNL3 axis in the regulation of hepatocellular carcinoma stem cell features. Acta Biochim. Biophys. Sin. 2021, 53, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Cao, X.; Jiang, Y.; Xu, J.; Zheng, Y.; Kang, D.; Xu, C. Circulating exosomal microRNAs as diagnostic and prognostic biomarkers in patients with diffuse large B-cell lymphoma. Hematol. Oncol. 2021, 40, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Ni, Z.; He, J.; Jiang, S.; Li, X.; Gong, W.; Zheng, L.; Chen, S.; Li, B.; Zhang, N.; et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36, 3528–3540. [Google Scholar] [CrossRef] [PubMed]
- ZZhang, X.; Tan, Z.; Kang, T.; Zhu, C.; Chen, S. Arsenic sulfide induces miR-4665-3p to inhibit gastric cancer cell invasion and migration. Drug Des. Dev. Ther. 2019, 13, 3037–3049. [Google Scholar] [CrossRef]
- Doroshow, D.B.; Sanmamed, M.F.; Hastings, K.; Politi, K.; Rimm, D.L.; Chen, L.; Melero, I.; Schalper, K.A.; Herbst, R.S. Immunotherapy in Non–Small Cell Lung Cancer: Facts and Hopes. Clin. Cancer Res. 2019, 25, 4592–4602. [Google Scholar] [CrossRef]
- Bonavida, B.; Chouaib, S. Resistance to anticancer immunity in cancer patients: Potential strategies to reverse resistance. Ann. Oncol. 2017, 28, 457–467. [Google Scholar] [CrossRef]
- Spella, M.; Stathopoulos, G.T. Immune Resistance in Lung Adenocarcinoma. Cancers 2021, 13, 384. [Google Scholar] [CrossRef]
- Cui, C.; Wang, J.; Fagerberg, E.; Chen, P.-M.; Connolly, K.A.; Damo, M.; Cheung, J.F.; Mao, T.; Askari, A.S.; Chen, S.; et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell 2021, 184, 6101–6118.e13. [Google Scholar] [CrossRef]
- Den Haan, J.M.; Lehar, S.M.; Bevan, M.J. Cd8+ but Not Cd8− Dendritic Cells Cross-Prime Cytotoxic T Cells in Vivo. J. Exp. Med. 2000, 192, 1685–1696. [Google Scholar] [CrossRef]
- Grisaru-Tal, S.; Dulberg, S.; Beck, L.; Zhang, C.; Itan, M.; Hediyeh-Zadeh, S.; Caldwell, J.; Rozenberg, P.; Dolitzky, A.; Avlas, S.; et al. Metastasis-Entrained Eosinophils Enhance Lymphocyte-Mediated Antitumor Immunity. Cancer Res. 2021, 81, 5555–5571. [Google Scholar] [CrossRef]
- Munder, M.; Schneider, H.; Luckner, C.; Giese, T.; Langhans, C.D.; Fuentes, J.M.; Kropf, P.; Mueller, I.; Kolb, A.; Modolell, M.; et al. Suppression of T-cell functions by human granulocyte arginase. Blood 2006, 108, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Schmielau, J.; Finn, O.J. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001, 61, 4756–4760. [Google Scholar] [PubMed]
- Radsak, M.; Iking-Konert, C.; Stegmaier, S.; Andrassy, K.; Hänsch, G.M. Polymorphonuclear neutrophils as accessory cells for T-cell activation: Major histocompatibility complex class II restricted antigen-dependent induction of T-cell proliferation. Immunology 2000, 101, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Reali, E.; Guerrini, R.; Moretti, S.; Spisani, S.; Lanza, F.; Tomatis, R.; Traniello, S.; Gavioli, R. Polymorphonuclear neutrophils pulsed with synthetic peptides efficiently activate memory cytotoxic T lymphocytes. J. Leukoc. Biol. 1996, 60, 207–213. [Google Scholar] [CrossRef]
- Eruslanov, E.B.; Bhojnagarwala, P.S.; Quatromoni, J.G.; Stephen, T.L.; Ranganathan, A.; Deshpande, C.; Akimova, T.; Vachani, A.; Litzky, L.; Hancock, W.W.; et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J. Clin. Investig. 2014, 124, 5466–5480. [Google Scholar] [CrossRef]
- Leveque, E.; Rouch, A.; Syrykh, C.; Mazières, J.; Brouchet, L.; Valitutti, S.; Espinosa, E.; Lafouresse, F. Phenotypic and Histological Distribution Analysis Identify Mast Cell Heterogeneity in Non-Small Cell Lung Cancer. Cancers 2022, 14, 1394. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Ganesan, A.P.; Johansson, M.; Ruffell, B.; Beltran, A.; Lau, J.; Jablons, D.M.; Coussens, L.M. Tumor-infiltrating regulatory T cells inhibit endogenous cytotoxic T cell responses to lung adenocarcinoma. J. Immunol. 2013, 191, 2009–2017. [Google Scholar] [CrossRef]
- Gettinger, S.; Choi, J.; Hastings, K.; Truini, A.; Datar, I.; Sowell, R.; Wurtz, A.; Dong, W.; Cai, G.; Melnick, M.A.; et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov. 2017, 7, 1420–1435. [Google Scholar] [CrossRef]
- Rowshanravan, B.; Halliday, N.; Sansom, D.M. CTLA-4: A moving target in immunotherapy. Blood 2018, 131, 58–67. [Google Scholar] [CrossRef]
- Contardi, E.; Palmisano, G.L.; Tazzari, P.L.; Martelli, A.M.; Falà, F.; Fabbi, M.; Kato, T.; Lucarelli, E.; Donati, D.; Polito, L.; et al. CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int. J. Cancer 2005, 117, 538–550. [Google Scholar] [PubMed]
- Zhang, H.; Dutta, P.; Liu, J.; Sabri, N.; Song, Y.; Li, W.; Li, J. Tumour cell-intrinsic CTLA4 regulates PD-L1 expression in non-small cell lung cancer. J. Cell. Mol. Med. 2018, 23, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Fan, J.; Chen, W.; Luan, J.; Mei, X.; Wang, S.; Li, Y.; Ye, L.; Li, S.; et al. Blocking CD47 efficiently potentiated therapeutic effects of anti-angiogenic therapy in non-small cell lung cancer. J. Immunother. Cancer 2019, 7, 346. [Google Scholar] [CrossRef]
- Kang, X.; Kim, J.; Deng, M.; John, S.; Chen, H.; Wu, G.; Phan, H.; Zhang, C.C. Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle 2016, 15, 25–40. [Google Scholar] [CrossRef]
- Barkal, A.A.; Weiskopf, K.; Kao, K.S.; Gordon, S.R.; Rosental, B.; Yiu, Y.Y.; George, B.M.; Markovic, M.; Ring, N.G.; Tsai, J.M.; et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 2018, 19, 76–84. [Google Scholar] [CrossRef]
- Nygren, M.K.; Tekle, C.; Ingebrigtsen, V.A.; Makela, R.; Krohn, M.; Aure, M.R.; Nunes-Xavier, C.E.; Perala, M.; Tramm, T.; Alsner, J.; et al. Identifying microRNAs regulating B7-H3 in breast cancer: The clinical impact of microRNA-29c. Br. J. Cancer 2014, 110, 2072–2080. [Google Scholar] [CrossRef]
- Yang, J.C.-H.; Sequist, L.V.; Geater, S.L.; Tsai, C.-M.; Mok, T.S.K.; Schuler, M.; Yamamoto, N.; Yu, C.-J.; Ou, S.-H.I.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef]
- Belani, C.P.; Yamamoto, N.; Bondarenko, I.M.; Poltoratskiy, A.; Novello, S.; Tang, J.; Bycott, P.; Niethammer, A.G.; Ingrosso, A.; Kim, S.; et al. Randomized phase II study of pemetrexed/cisplatin with or without axitinib for non-squamous non-small-cell lung cancer. BMC Cancer 2014, 14, 290. [Google Scholar] [CrossRef]
- Solomon, B.; Callejo, A.; Bar, J.; Berchem, G.; Bazhenova, L.; Saintigny, P.; Raymond, E.; Girard, N.; Sulaiman, R.; Bresson, C.; et al. Survival prolongation by rationale innovative genomics (SPRING): An international WIN consortium phase I study exploring safety and efficacy of avelumab, palbociclib, and axitinib in advanced non-small cell lung cancer (NSCLC) with integrated genomic and transcriptomic correlates. Ann. Oncol. 2019, 30, v648. [Google Scholar]
- Gao, W.; Wang, M.; Wang, L.; Lu, H.; Wu, S.; Dai, B.; Ou, Z.; Zhang, L.; Heymach, J.V.; Gold, K.A.; et al. Selective Antitumor Activity of Ibrutinib in EGFR-Mutant Non–Small Cell Lung Cancer Cells. Gynecol. Oncol. 2014, 106, dju204. [Google Scholar] [CrossRef]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Schrump, D.S. Cytotoxicity Mediated by Histone Deacetylase Inhibitors in Cancer Cells: Mechanisms and Potential Clinical Implications. Clin. Cancer Res. 2009, 15, 3947–3957. [Google Scholar] [CrossRef] [PubMed]
- Greve, G.; Schiffmann, I.; Pfeifer, D.; Pantic, M.; Schüler, J.; Lübbert, M. The pan-HDAC inhibitor panobinostat acts as a sensitizer for erlotinib activity in EGFR-mutated and -wildtype non-small cell lung cancer cells. BMC Cancer 2015, 15, 947. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Lu, W.; Chen, G.; Cheng, F.; Su, H.; Chen, Y.; Liu, M.; Pang, X. Inhibition of histone deacetylases sensitizes EGF receptor-TK inhibitor-resistant non-small-cell lung cancer cells to erlotinib in vitro and in vivo. Br. J. Pharmacol. 2017, 174, 3608–3622. [Google Scholar] [CrossRef]
- Zheng, H.; Zhao, W.; Yan, C.; Watson, C.C.; Massengill, M.; Xie, M.; Massengill, C.; Noyes, D.R.; Maetinez, G.V.; Afzal, R.; et al. HDAC Inhibitors Enhance T-Cell Chemokine Expression and Augment Response to PD-1 Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2016, 22, 4119–4132. [Google Scholar] [CrossRef]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Xiao, M.; Shi, Y.; Wang, Y.; Xu, Z.; Wang, K. Identification of m7G-Related miRNA Signatures Associated with Prognosis, Oxidative Stress, and Immune Landscape in Lung Adenocarcinoma. Biomedicines 2023, 11, 1569. https://doi.org/10.3390/biomedicines11061569
Jiang S, Xiao M, Shi Y, Wang Y, Xu Z, Wang K. Identification of m7G-Related miRNA Signatures Associated with Prognosis, Oxidative Stress, and Immune Landscape in Lung Adenocarcinoma. Biomedicines. 2023; 11(6):1569. https://doi.org/10.3390/biomedicines11061569
Chicago/Turabian StyleJiang, Sujing, Mingshu Xiao, Yueli Shi, Yongfang Wang, Zhiyong Xu, and Kai Wang. 2023. "Identification of m7G-Related miRNA Signatures Associated with Prognosis, Oxidative Stress, and Immune Landscape in Lung Adenocarcinoma" Biomedicines 11, no. 6: 1569. https://doi.org/10.3390/biomedicines11061569
APA StyleJiang, S., Xiao, M., Shi, Y., Wang, Y., Xu, Z., & Wang, K. (2023). Identification of m7G-Related miRNA Signatures Associated with Prognosis, Oxidative Stress, and Immune Landscape in Lung Adenocarcinoma. Biomedicines, 11(6), 1569. https://doi.org/10.3390/biomedicines11061569






