1. Introduction
Breast cancer is a major threat to women worldwide, and early detection and diagnosis are critical to improving cure rates and reducing mortality [
1]. Breast cancer has complex and diverse causes, including environmental factors, hormone secretion, and life stress [
2]. At the molecular level, breast cancer is a highly heterogeneous disease that involves DNA damage, genetic mutations (such as
BRCA gene mutations), activation of hormone receptors (progesterone and estrogen receptors), and expression of human epidermal growth factor receptor 2 (HER2). Early detection plays a crucial role in reducing breast cancer mortality rates in the long term. Identifying early stage breast cancer is essential for achieving optimal prognosis [
3], with imaging examinations being an important means for diagnosis in the early stages.
Breast ultrasound is a non-radiation clinical modality that is well tolerated by patients and is widely used in the diagnosis of breast cancer [
4,
5]. However, its accuracy depends on the expertise of the clinician, which can lead to misdiagnosis [
6]. Other diagnostic modalities, such as magnetic resonance imaging (MRI), mammography, computed tomography, digital breast tomosynthesis and positron emission topography, are also used, but have technical limitations [
7]. To address these challenges, computer-aided diagnosis (CAD) algorithms, based on advanced artificial intelligence (AI) technology, can assist medical staff in accurately interpreting images. This tool has the potential to improve patient outcomes by guiding clinicians to correct diagnoses [
8,
9]. Breast ultrasound classification is crucial for distinguishing between benign and malignant tumours. Segmentation, which divides an image into regions to identify regions of interest (ROI), is a critical notion in image processing. However, segmentation can be problematic and lead to misdiagnosis, for example, when pectoral muscles are present but not part of the breast tissue [
10,
11]. Therefore, it’s important to compare identified cancer ultrasound images with the standard Breast Imaging Reporting and Data System (BI-RADS) from a clinical perspective. BI-RADS provides standardised terms, known as a lexicon, that describe the characteristics of breast masses and effectively differentiate between benign and malignant tumours [
12].
The ultrasound lexicon has several advanced features for inspecting and correlating breast cancer images based on: (1) breast composition (homogeneous fat, homogeneous fibro-glandular or heterogeneous); (2) mass shape (irregular, round, oval), margins (not-circumscribed, circumscribed, indistinct and speculated), and orientation (non-parallel or parallel); (3) calcifications location (outside mass within mass intraductal); and (4) associated features (architectural distortion, edema, skin retraction, etc.). It serves as a guide for radiologists/physicians to compare tumour images with high accuracy [
13,
14]. Recently developed deep learning systems have excellent accuracy in mass segmentation in breast ultrasound images using the Artificial Neural Network (ANN) [
15], making it an advanced tool for the classification and analysis of breast cancer [
16].
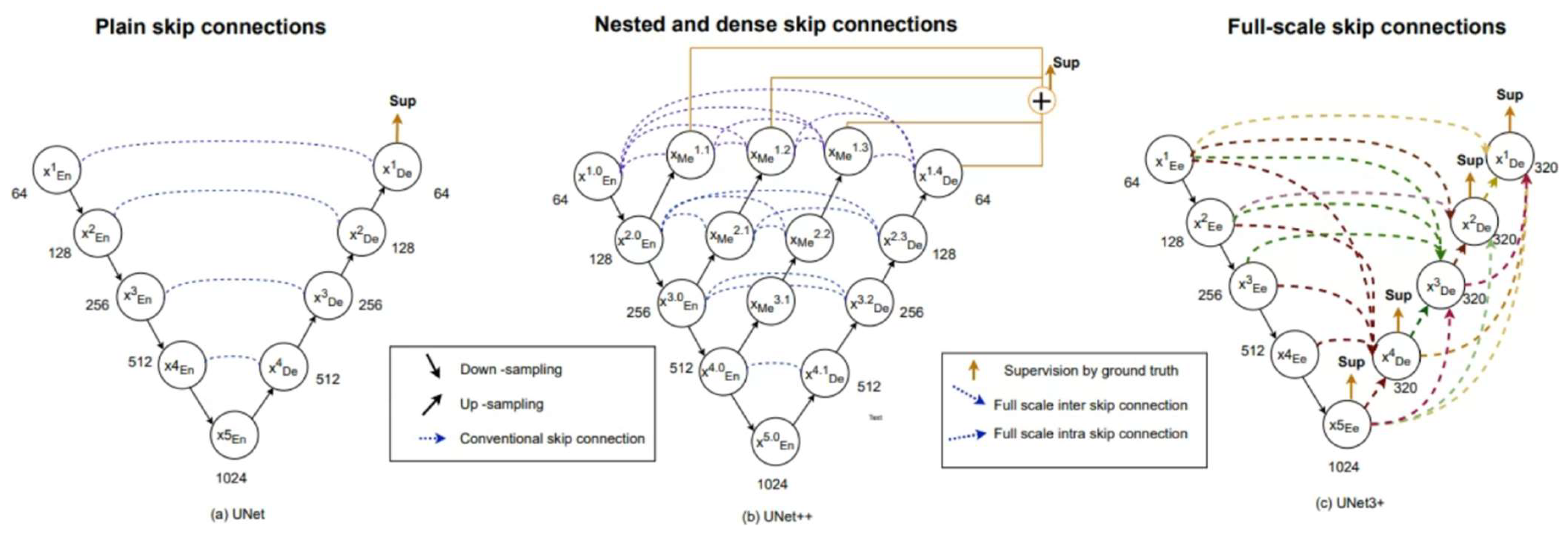
Our study focuses on improving breast cancer detection in ultrasound images using the Unet3+ architecture for semantic segmentation modelling. The Unet3+ architecture is a robust semantic network that shows promising results in medical imaging applications by addressing skip connections to improve the accuracy of Unet segmentation [
17]. Skip connections allow information transfer between encoder and decoder layers of a UNET architecture to improve segmentation performance by more effectively capturing multi-scale contextual information [
18]. We aim to improve the accuracy and efficiency of the Unet3+ model in detecting multiple malignancy-related image features simultaneously.
Table 1 shows the different approaches used for pre-processing, segmentation and classification.
2. Materials and Methods
The paper presents an algorithmic framework, as illustrated in
Figure 1, for analysing medical images of 309 patients. The dataset comprises 151 benign and 158 malignant images that are pre-processed to remove irrelevant information and resized for normalization. Semantic segmentation is then applied to classify each pixel based on its target feature, and precisely outline each object. The study compares the performance of Unet3+ with other models, such as FCN, Unet, Segnet, DeeplabV3 and pspNet, using metrics such as overlap, with ground truth and global accuracy. Finally, the study reports mean accuracy, mean IoU (Intersection over Union), weighted IoU and weighted F1 score for all models including Unet3+.
2.1. Image Data Collection Procedures
This is a retrospective, cross-sectional study approved by the institutional review board (IRB) of Changhua Christian Hospital, Taiwan (No. 181235). Informed consent was waived, and the ethics committee reviewed all experimental methods to ensure compliance with appropriate standards and the Helsinki Declaration. Participants were individuals between 35 and 75 years of age whose tumours had been diagnosed as benign or malignant by fine-needle cytology, core-needle biopsy or open biopsy. Detailed data on patient treatment, such as therapy, histology and radiography, were also collected. Breast ultrasound images were obtained using a GE Voluson 700 (GE Healthcare, Zipf, Austria). For each participant, at least two, different, scan-plane angles were acquired with each image providing the full screen of the scan plane in RGB mode at a resolution of 960 × 720 pixels.
2.2. Image Data Preprocessing
To pre-process the breast ultrasound images, we first removed irrelevant information, such as manufacturer’s labels, directional markings and text fields, which could interfere with image interpretation and lead to incorrect results during noise reduction. This step ensured a clear and focused image [
28]. Next, the images were normalised and transformed to a consistent size and format for efficient processing by machine learning algorithms. As different resolutions and aspect ratios could affect the accuracy of the analysis, it was crucial to resize the images to a standard size and aspect ratio. Finally, we validated the data before feeding it into the algorithm.
2.3. Semantic Segmentation of Images
Image segmentation is the process of dividing an image into memory blocks that are equally related, with no deeper meaning within the divided area. This operation typically relies on low-level representations, such as colour, texture, and boundary smoothness, to cluster and cut output regions or segments. However, this method does not identify regions that belong to the same or related categories. Semantic segmentation, on the other hand, classifies each pixel in an image based on its target feature. The result is a precise outline of each object or feature to understand the overall meaning of the image. Semantic segmentation is crucial to understanding images because it makes them more meaningful and easier to analyse. In essence, semantic segmentation can be viewed as a pixel-by-pixel classification problem, where pixels or super-pixels are classified into different categories for breast cancer detection and segmentation [
29].
2.4. Training Protocol and Infrastructure
The learning rate was set to 0.01 with a maximum epoch of 100 and a batch size of two. Gaussian blur was applied to the images to reduce noise and smooth edges, while random rotation up to 90° and elastic transformation were used for distortion correction. The image size was fixed at 360 × 360 pixels with pixel normalisation using mean and standard deviation.
The computational platform used in this study consisted of an Intel Core i5-11400F (2.6 GHz hexa-core with up to 4.4 GHz turbo boost and 12 MB cache) (Intel Corp., Santa Clara, CA, USA), NVIDIA RTX3060 graphics card with 12 GB video RAM (Nvidia Corp., San Jose, CA, USA) and Compute Unified Device Architecture (CUDA) version 11.2 (Nvidia Corp., San Jose, CA, USA). This setup provided an accelerated computational environment through the use of the NVIDIA processing unit. All programmes related to this study were implemented using the Pytorch framework version 3.7 (PyTorch, San Francisco, CA, USA).
2.5. Semantic Segmentation Network Model
The Unet3+ [
17] architecture is an advanced version of the original Unet architecture originally designed for biomedical image segmentation. It incorporates several features that improve the accuracy of the semantic segmentation results. The architecture consists of two networks: an encoder network and a decoder network. The encoder network reduces the spatial dimensions of an input image using convolutional and pooling layers, while the decoder network upsamples the reduced feature map to its original size using convolutional and upsampling layers.
Figure 2 is the structural diagram of the Unet model and its related variants (such as Unet++, Unet3+). When compared to Unet, a notable aspect of Unet3+ is the dense connections between the encoder and decoder layers within a block, which connect all subsequent blocks in the network. This facilitates the flow of information between layers, thereby improving the accuracy of semantic segmentation. In addition, residual links are used to connect distant layers in the network to allow information propagation across multiple layers, further improving performance [
30,
31].
Both Unet and Unet++ do not explore enough information from full scale, resulting in the inability to explicitly determine the position and boundary of an organ. However, each decoder layer in Unet3+ contains smaller and equal scale feature maps from the encoder, as well as larger scale feature maps from the decoder. This allows fine-grained detail and coarse-grained semantics to be captured at full scale. Unet 3+ produces a side output from each decoder stage that is supervised by the ground truth. To achieve deep supervision, the last layer of each decoder stage undergoes a simple 3 × 3 convolution followed by bilinear upsampling and a sigmoid function.
2.6. Compared to Related Semantic Segmentation Network Model
In our study, we investigated six related segmentation models. We compared the performance of breast cancer segmentation on medical images, including SegNet [
11], DeepLabv3+ [
32], Fully Convolutional Neural Networks (FCNs) [
33], pspNet (Pyramid Scene Parsing Network) [
34] and EfficientNet. SegNet is a deep learning model that excels at semantic segmentation tasks due to its encoder–decoder architecture that extracts high-level features while preserving spatial information. DeepLabv3+ extends DeepLabv3 by adding an encoder–decoder structure. The encoder module processes multiscale contextual information through extended convolution at multiple scales, while the decoder module refines segmentation results along object boundaries. The FCN model is a well-known classification network model based on the VGG16 architecture and has shown potential in identifying and localising breast tumours [
35]. The Pyramid Scene Parsing Network (PspNet) [
34] and EfficientNet are utilised for precise breast cancer detection in medical images. PspNet incorporates a pyramid pooling module to gather contextual information at various scales, while EfficientNet employs scaling and compound scaling techniques to attain superior accuracy with fewer parameters than other models.
3. An Analysis and Evaluation
The performance of the semantic segmentation was evaluated by measuring the overlap with the ground-truth image dataset. This evaluation included global accuracy, mean accuracy, mean/frequency weighted intersection over union (IU) and mean F1 score. Indicators, such as accuracy, precision, recall and comprehensive evaluation index (F-measure), were used to evaluate the proposed method. The number of predicted true positives (TP), false positives (FP), true negatives (TN) and false negatives (FN) were also considered.
Accuracy is a measure of the probability of correctly classifying samples. The formula for accuracy is (TP + TN)/(TP + FP + TN + FN). Precision measures how well a sample predicts a particular category. The precision formula is TP/(TP + FP). Recall measures the proportion of true positives that were correctly identified in the sample. The recall formula is TP/(TP + FN). F-measure, also known as F-score, considers both precision and recall to evaluate the accuracy of a model in binary classification problems. The formula for the F-measure is 2 × ((Precision × Recall)/(Precision + Recall)). Global Accuracy refers to the percentage of pixels that are correctly classified regardless of category, and can be calculated quickly and easily using scoring criteria. Mean Accuracy or Average Class Accuracy calculates the ratio of correctly classified pixels to the total number of pixels in each class. This metric can be misleading if one category has significantly more labelled pixels than others. Intersection over union (IoU), also known as the Jaccard similarity coefficient, evaluates semantic segmentation by calculating the correlation between correctly classified pixels and the ratio of ground truth to predicted pixels. IoU values range from 0 to 1 with higher coefficients indicating greater sample similarity.
4. Results
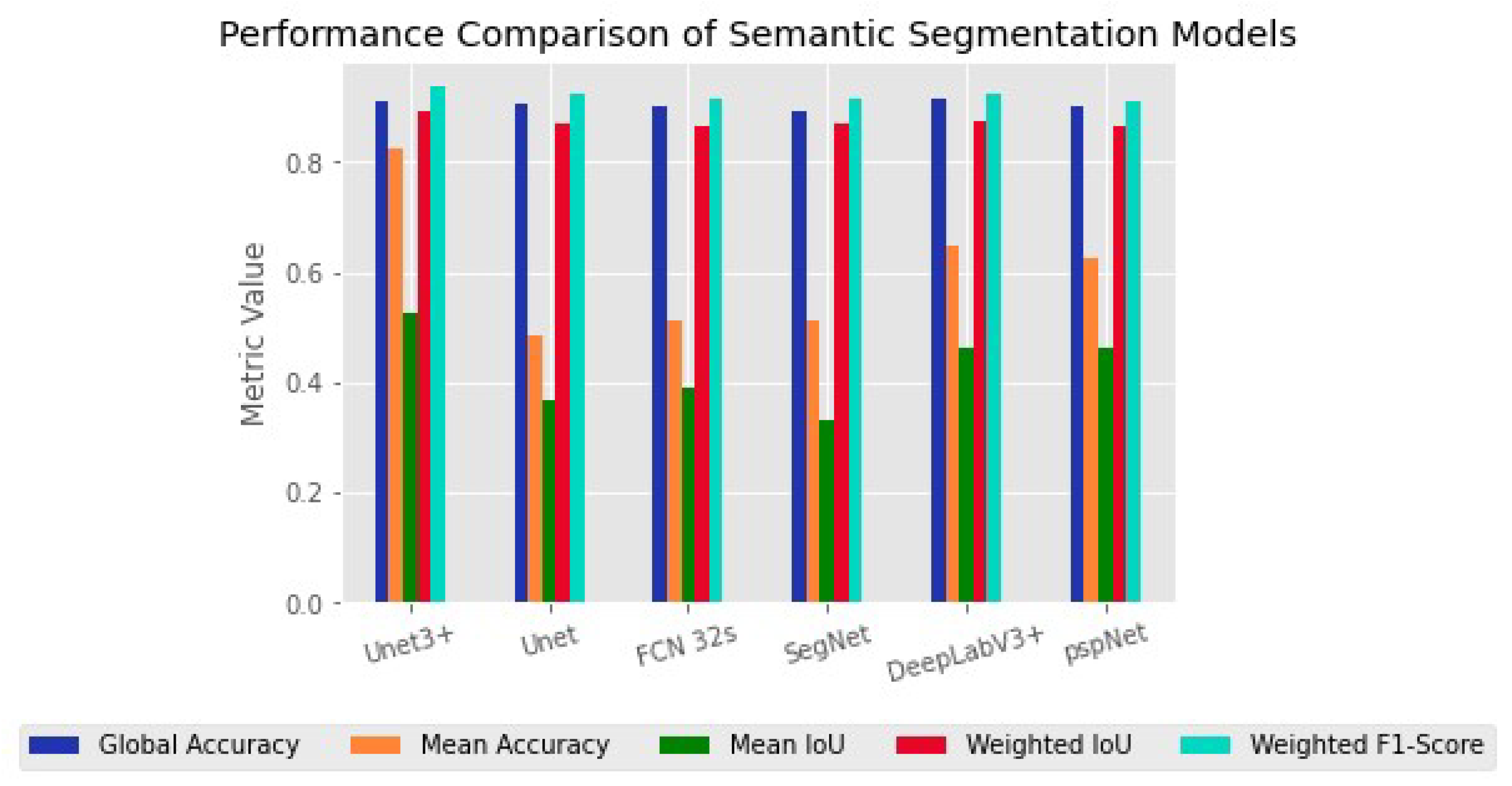
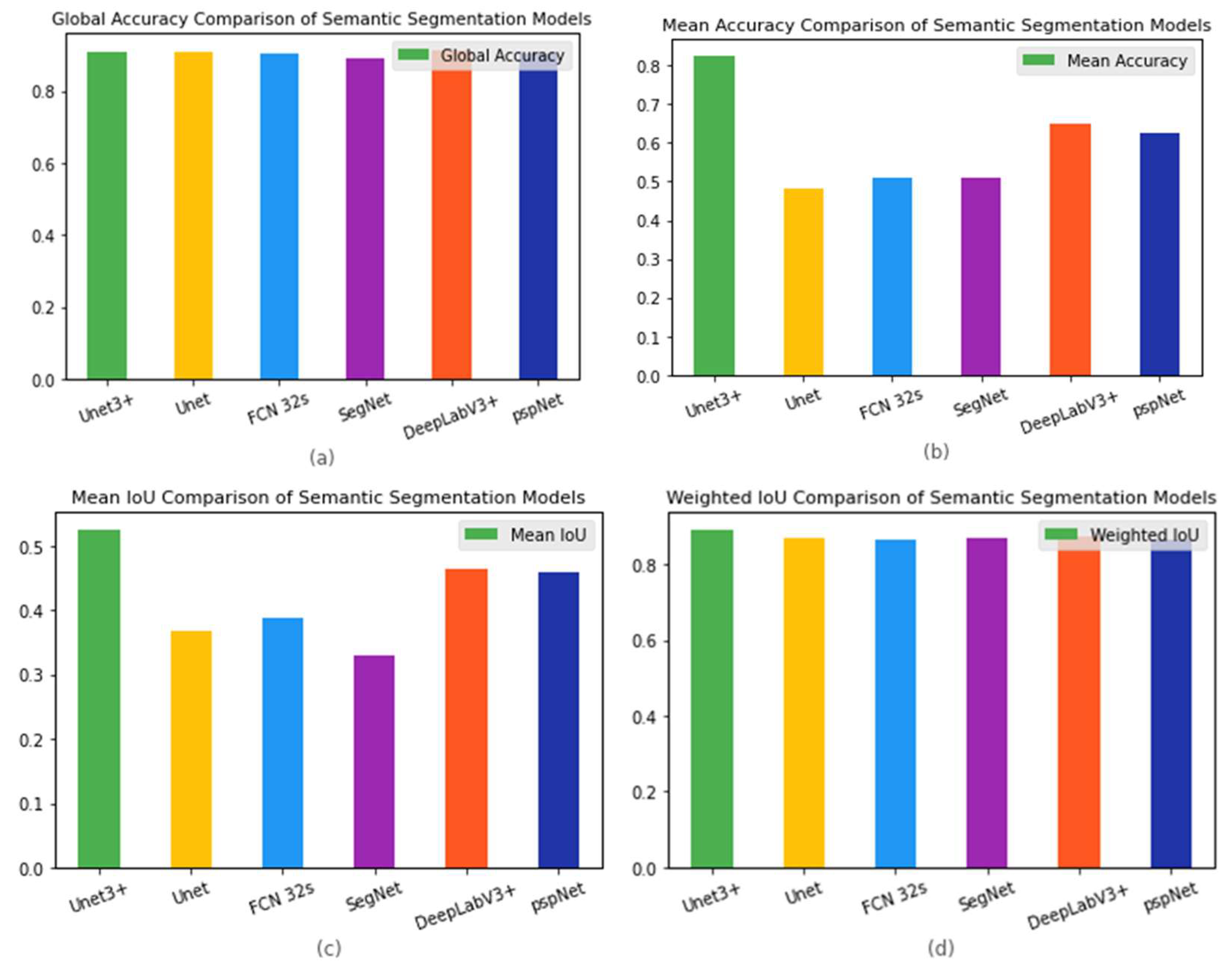
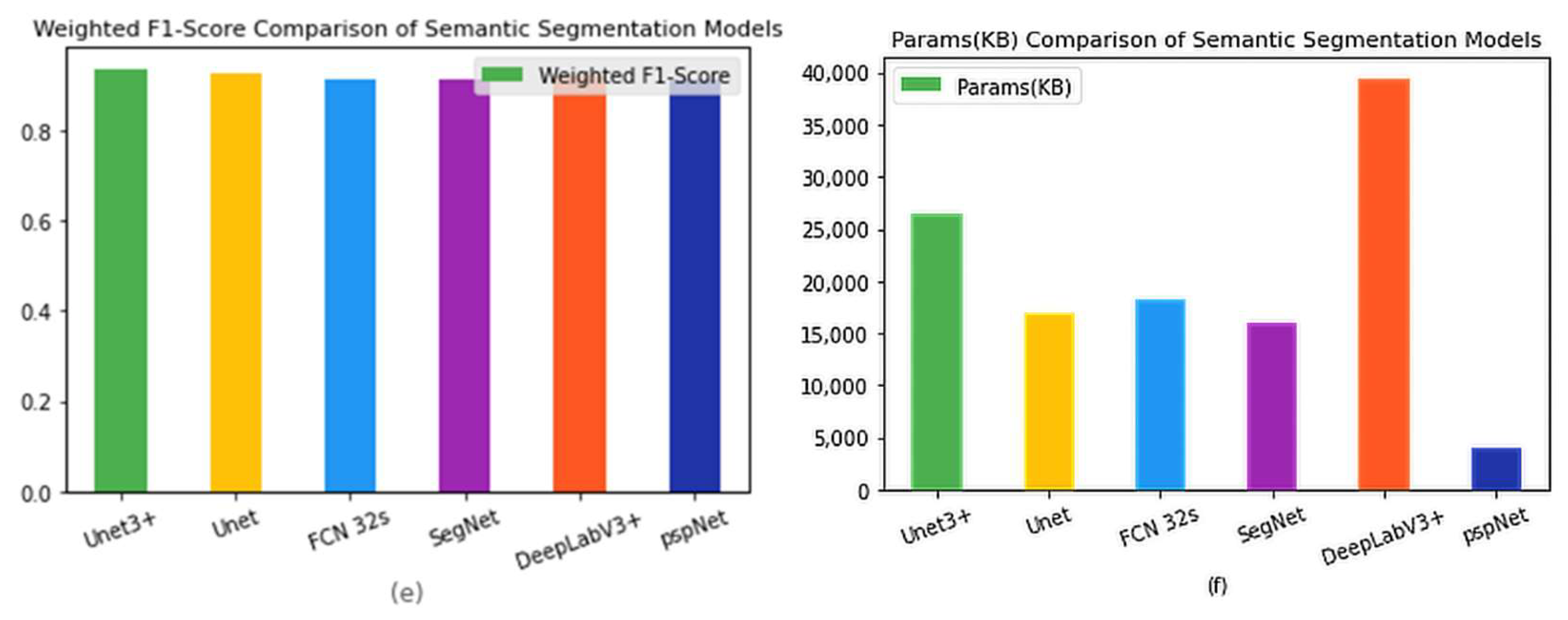
Figure 3 shows the semantic segmentation performance (Metrics) of the six models used in this study, including Unet3+, Unet, FCN-32s, SegNet, pspNet and DeeplabV3+. From
Figure 3, we can see that Unet3+ had the highest global accuracy with better mean accuracy, mean IoU and weighted F1 score. Specifically, Unet3+ achieved 0.90 for mean accuracy, 0.82 for mean IoU and 0.52 for mean weighted IoU, while its weighted F1 score was found to be 0.83 with superior Parmas (KB) compared to other models. Although DeepLabV3+ had a higher global accuracy of 0.91 than other models, our analysis showed that Unet3+ outperformed it in terms of overall performance, based on metrics such as mean accuracy, mean IoU, weighted F1-score and Parmas (KB).
Figure 4 shows the results presented separately for each model and indicator item, to facilitate comparison between models.
Table 2 lists the original data.
5. Discussion
In this study, we employed the Unet3+ deep learning architecture to accurately segment the malignant BI-RADS lexicon feature of breast cancer in medical images. Our approach successfully addressed the challenge of identifying the small and irregularly shaped malignant features on a tumour, which is a difficult task in medical image analysis. The segmentation map clearly demonstrates that our use of learning networks and fully connected networks resulted in an improved BI-RADS lexical recognition compared to other models such as Unet, FCN 32s, SegNet, DeepLabV3+, and pspNet. Notably, the Unet3+ model outperformed all other models tested when applied to the test set.
Figure 5 shows the results of semantic segmentation of breast ultrasound images using the model used in this study. In
Figure 5, we simultaneously allowed six models to perform semantic segmentation on ten randomly selected images from the image dataset and provided their ground truth as a comparison for the segmentation results. FCN-32 and Unet produced good segmentation results, while SegNet performed poorly in comparison. These findings suggest that the classification or recognition performance of a well-trained deep learning model, may decline to some extent when applied to datasets with varied sizes or characteristics. In comparing FCN-32s and deepLab3+ with ground truth, DeepLabV3+ showed improved recognition of shadowing features. Similarly, in comparing SegNet and pspNet with ground truth, pspNet demonstrated an enhanced recognition of shadowing features due to its emphasis on obtaining multi-scale contextual information through the pyramid pooling module. This allowed for effective integration of information at multiple scales and established a thorough comprehension of complex scenes and objects, resulting in more accurate segmentation findings aligned with the ground truth.
DeepLabV3+ also demonstrated the competitive performance when it came to matching the ground truth. DeepLabV3+ was able to capture multi-scale information without downsampling, maintaining spatial features. The decoder module, in conjunction with skip connections, aided in the recovery of lost spatial data, which enhanched segmentation accuracy even further.
The Unet architecture adequately reflected the ground reality and demonstrated ease of use and good performance in semantic segmentation tasks. The incorporation of skip links between the encoder and decoder facilitated the merging of low-level and high-level characteristics, resulting in a more precise object localization. While Unet’s performance was not as strong as that of Unet3+, pspNet, or DeepLabV3+, it still produced segmentations that closely approximated the ground truth.
The pspNet framework, despite incorporating the improved Unet3+ module, did not perform satisfactorily. It encountered issues with pixel category classification, leading to significant deviations from the ground truth in twenty-two segments. In contrast, the Unet3+ model utilized the redesigned skip connections and combined multiple scale features through multi-scale deep supervision. This theoretically enhances its ability to acquire and fuse image features while computing pixels for better segmentation results than the Unet model. However, experimental outcomes failed to demonstrate any improvement over the Unet model, as there was a noticeable loss of detail observed in the test set images [
36,
37,
38].
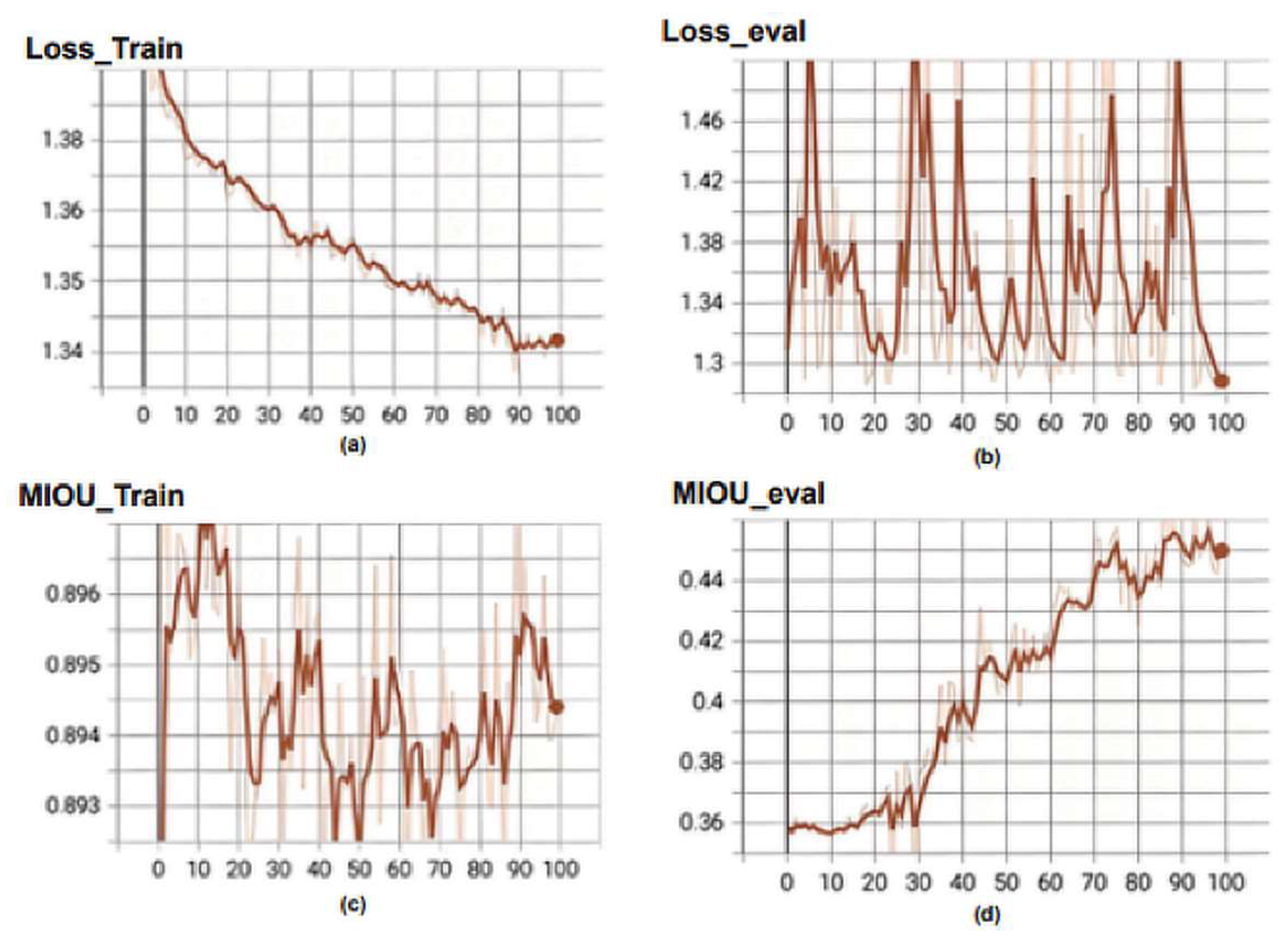
Figure 6 illustrates the loss function curve in training process of the Unet3+ model. The following metrics are shown: (a) Loss_Train, which represents the loss function during training; (b) Loss_eval, which is the value of the loss function computed on the validation set during model evaluation; (c) MIOU_Train, which is Mean Intersection over Union (MIOU) computed on the training set during model training; and (d) MIOU_eval, which refers to Mean Intersection over Union (MI-OU) computed on the validation set during model evaluation. The loss curve for the training set gradually decreases with an increase in the number of epochs. It is important to note that Unet3+ belongs to the deep supervision model and has five outputs calculating the loss, which requires dividing by five to obtain a loss of 0.269 from 1.347. However, there is significant fluctuation in the loss function of the validation set, reaching a value of 0.252 at epoch = 100 from 1.281/5 = 0.252. There exists a considerable difference between MIOU_train (0.894) and MIOU_eval (0.453), representing training and validation sets, respectively, in terms of numerical values. During the training phase, the Unet3+ model’s loss and MIOU curves exhibit volatility with significant numerical disparities between the training and validation sets.
6. Conclusions
This study demonstrates the effectiveness of the Unet3+ model, a modified version of the Unet architecture, in accurately segmenting breast tumours from mammogram images. By incorporating dense skip connections and attention gates, this model can capture multi-scale features and focus on important regions of the image. These improvements lead to more accurate tumour segmentation and highlight the potential for deep learning models to improve breast cancer diagnosis and treatment. The Unet3+ model provides a promising platform for further research in this area and, after refinement and optimisation, has the potential to be a reliable algorithm for radiologists and clinicians in the detection and diagnosis of breast cancer.
To fully utilize the potential of the model in clinical practice, future work should focus on enhancing and optimizing it. This can be achieved by investigating the application of transfer learning to adapt the model to various medical imaging modalities, including clinical data that can improve segmentation accuracy. Additionally, verifying the performance of the model on larger datasets with a wider range of images is necessary. Integrating the Unet3+ model with decision support systems would provide doctors access to more thorough analyses of medical imaging. This integration will enhance breast cancer detection and management capabilities.
Author Contributions
T.A.: conceptualization, methodology, writing—original draft preparation, validation. W.-C.S.: conceptualization, investigation, data curation, methodology, resources, writing—review and editing, project administration, supervision, funding acquisition. F.-R.H.: project administration, resources, supervision. T.H.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Department of Research, Changhua Christian Hospital, Taiwan. Grant number: 110-CCH-IRP-012, 112-CCH-IRP-117 and by Ministry of Science and Technology Grant number MOST 111-2221-E-035-015.
Institutional Review Board Statement
This study was approved by the Institutional Review Board (IRB) of Changhua Christian Hospital, Changhua, Taiwan (No. 181235). All experimental methods were supervised by the IRB and conducted in accordance with the relevant guidelines and the Declaration of Helsinki.
Informed Consent Statement
Informed consent requirement was waived by the ethics committee because of the retrospective nature of the study.
Data Availability Statement
The datasets produced and analyzed in this study are not publicly accessible due to IRB and institutional limitations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Michael, E.; Ma, H.; Li, H.; Kulwa, F.; Li, J. Breast Cancer Segmentation Methods: Current Status and Future Potentials. BioMed Res. Int. 2021, 2021, 9962109. [Google Scholar] [CrossRef] [PubMed]
- Schoemaker, M.J.; Jones, M.E.; Wright, L.B.; Griffin, J.; McFadden, E.; Ashworth, A.; Swerdlow, A.J. Psychological stress, adverse life events and breast cancer incidence: A cohort investigation in 106,000 women in the United Kingdom. Breast Cancer Res. 2016, 18, 72. [Google Scholar] [CrossRef]
- Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef] [PubMed]
- Shia, W.C.; Lin, L.S.; Chen, D.R. Classification of malignant tumours in breast ultrasound using unsupervised machine learning approaches. Sci. Rep. 2021, 11, 1418. [Google Scholar] [CrossRef]
- Hsieh, Y.H.; Hsu, F.R.; Dai, S.T.; Huang, H.Y.; Chen, D.R.; Shia, W.C. Incorporating the Breast Imaging Reporting and Data System Lexicon with a Fully Convolutional Network for Malignancy Detection on Breast Ultrasound. Diagnostics 2021, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Irfan, R.; Almazroi, A.A.; Rauf, H.T.; Damaševičius, R.; Nasr, E.A.; Abdelgawad, A.E. Dilated Semantic Segmentation for Breast Ultrasonic Lesion Detection Using Parallel Feature Fusion. Diagnostics 2021, 11, 1212. [Google Scholar] [CrossRef]
- Larbi, A.; Fourneret, B.; Lukas, C.; Baron, M.-P.; Molinari, N.; Taourel, P.; Cyteval, C. Prevalence and topographic distribution of spinal inflammation on MR imaging in patients recently diagnosed with axial spondyloarthritis. Diagn. Interv. Imaging 2017, 98, 347–353. [Google Scholar] [CrossRef]
- Bartolotta, T.V.; Taibbi, A.; Picone, D.; Anastasi, A.; Midiri, M.; Lagalla, R. Detection of liver metastases in cancer patients with geographic fatty infiltration of the liver: The added value of contrast-enhanced sonography. Ultrasonography 2017, 36, 160. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Sasikala, S.; Gomathi, S.; Geetha, V.; Anbumani, V. Segmentation and classification of breast cancer using novel deep learning architecture. Neural Comput. Appl. 2022, 34, 16533–16545. [Google Scholar] [CrossRef]
- Hai, J.; Qiao, K.; Chen, J.; Tan, H.; Xu, J.; Zeng, L.; Shi, D.; Yan, B. Fully convolutional densenet with multiscale context for automated breast tumor segmentation. J. Healthc. Eng. 2019, 2019, 8415485. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Civilibal, S.; Cevik, K.K.; Bozkurt, A. A deep learning approach for automatic detection, segmentation and classification of breast lesions from thermal images. Expert Syst. Appl. 2023, 212, 118774. [Google Scholar] [CrossRef]
- Guo, Y.; Duan, X.; Wang, C.; Guo, H. Segmentation and recognition of breast ultrasound images based on an expanded U-Net. PLoS ONE 2021, 16, e0253202. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, R.; Zhao, D.; Yu, F.; Heidari, A.A.; Xu, Z.; Chen, H.; Algarni, A.D.; Elmannai, H.; Xu, S. Multi-level threshold segmentation framework for breast cancer images using enhanced differential evolution. Biomed. Signal Process. Control. 2023, 80, 104373. [Google Scholar] [CrossRef]
- Avcı, H.; Karakaya, J. A Novel Medical Image Enhancement Algorithm for Breast Cancer Detection on Mammography Images Using Machine Learning. Diagnostics 2023, 13, 348. [Google Scholar] [CrossRef]
- Zhang, S.; Liao, M.; Wang, J.; Zhu, Y.; Zhang, Y.; Zhang, J.; Zheng, R.; Lv, L.; Zhu, D.; Chen, H. Fully automatic tumor segmentation of breast ultrasound images with deep learning. J. Appl. Clin. Med. Phys. 2023, 24, e13863. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.-W.; Heng, P.-A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Punitha, S.; Amuthan, A.; Joseph, K.S. Benign and malignant breast cancer segmentation using optimized region growing technique. Future Comput. Inform. J. 2018, 3, 348–358. [Google Scholar] [CrossRef]
- Rouhi, R.; Jafari, M.; Kasaei, S.; Keshavarzian, P. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst. Appl. 2015, 42, 990–1002. [Google Scholar] [CrossRef]
- Setiawan, A.S.; Wesley, J.; Purnama, Y. Mammogram classification using law’s texture energy measure and neural networks. Procedia Comput. Sci. 2015, 59, 92–97. [Google Scholar] [CrossRef]
- Varela, C.; Tahoces, P.G.; Mendez, A.J.; Souto, M.; Vidal, J.J. Computerized detection of breast masses in digitized mammograms. Comput. Biol. Med. 2007, 37, 214–226. [Google Scholar] [CrossRef]
- Patel, B.C.; Sinha, G. Mammography feature analysis and mass detection in breast cancer images. In Proceedings of the 2014 International Conference on Electronic Systems, Signal Processing and Computing Technologies, Nagpur, India, 9–11 January 2014; pp. 474–478. [Google Scholar]
- Pereira, D.C.; Ramos, R.P.; Do Nascimento, M.Z. Segmentation and detection of breast cancer in mammograms combining wavelet analysis and genetic algorithm. Comput. Methods Programs Biomed. 2014, 114, 88–101. [Google Scholar] [CrossRef]
- Chen, Z.; Strange, H.; Oliver, A.; Denton, E.R.; Boggis, C.; Zwiggelaar, R. Topological modeling and classification of mammographic microcalcification clusters. IEEE Trans. Biomed. Eng. 2014, 62, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Q.; Celebi, M.E.; García, I.F. Breast mass segmentation using region-based and edge-based methods in a 4-stage multiscale system. Biomed. Signal Process. Control. 2013, 8, 204–214. [Google Scholar] [CrossRef]
- Kashyap, K.L.; Bajpai, M.K.; Khanna, P. Breast cancer detection in digital mammograms. In Proceedings of the 2015 IEEE International Conference on Imaging Systems and Techniques (IST), Macau, China, 16–18 September 2015; pp. 1–6. [Google Scholar]
- Rezk, E.; Awan, Z.; Islam, F.; Jaoua, A.; Al Maadeed, S.; Zhang, N.; Das, G.; Rajpoot, N. Conceptual data sampling for breast cancer histology image classification. Comput. Biol. Med. 2017, 89, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Rakhlin, A.; Shvets, A.; Iglovikov, V.; Kalinin, A.A. Deep convolutional neural networks for breast cancer histology image analysis. In Proceedings of the Image Analysis and Recognition: 15th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, 27–29 June 2018; pp. 737–744. [Google Scholar]
- Li, X.; Monga, V.; Rao, U.A. Analysis-synthesis model learning with shared features: A new framework for histopathological image classification. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 203–206. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Proceedings 4. pp. 3–11. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Piramanayagam, S.; Saber, E.; Schwartzkopf, W.; Koehler, F.W. Supervised classification of multisensor remotely sensed images using a deep learning framework. Remote Sens. 2018, 10, 1429. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, Hawaii, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 5–12 June 2015; pp. 3431–3440. [Google Scholar]
- Yin, X.-X.; Sun, L.; Fu, Y.; Lu, R.; Zhang, Y. U-Net-Based medical image segmentation. J. Healthc. Eng. 2022, 2022, 4189781. [Google Scholar] [CrossRef]
- Soulami, K.B.; Kaabouch, N.; Saidi, M.N.; Tamtaoui, A. Breast cancer: One-stage automated detection, segmentation, and classification of digital mammograms using UNet model based-semantic segmentation. Biomed. Signal Process. Control. 2021, 66, 102481. [Google Scholar] [CrossRef]
- Lu, H.; She, Y.; Tie, J.; Xu, S. Half-UNet: A simplified U-Net architecture for medical image segmentation. Front. Neuroinformatics 2022, 16, 911679. [Google Scholar] [CrossRef]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).