Impacts of Mir146a Genotypes on Bladder Cancer Risk in Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment of Bladder Cancer Patients and Healthy Controls
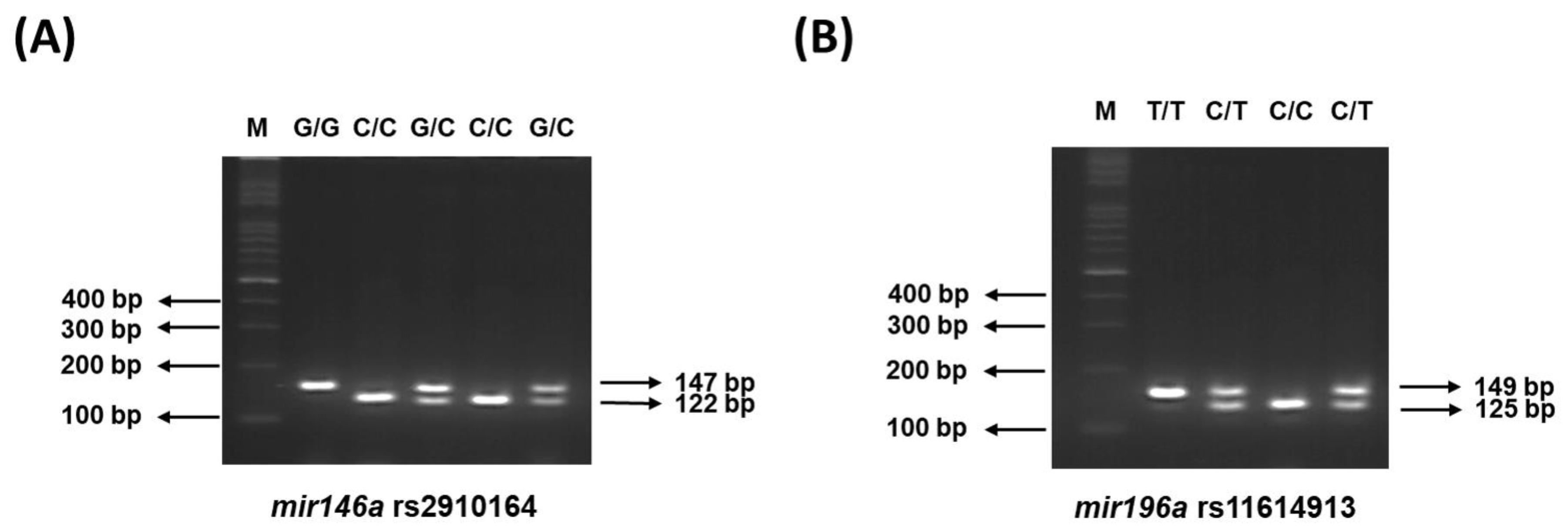
2.2. Genotyping Methodology of Mir146a and Mir196a SNPs and Quality Control
2.3. Quantitative Reverse Transcription Polymerase Chain Reaction for Measuring Mir146a Expression
2.4. Statistical Methodology
3. Results
3.1. Demographic and Clinical Characteristics of Cases and Controls
3.2. Associations of Mir146a and Mir196a Genotypes with Bladder Cancer Risk
3.3. Associations of Mir146a rs2910164 and Mir196a rs11614913 Alleles with BLCA Risk
3.4. Stratified Analyses of Mir146a Genotypes by Age, Gender, Smoking, and Alcohol Drinking Status
3.5. Serum Expression Level of mir146a and Its Correlation with mir146a Genotypes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence. Available online: https://www.hpa.gov.tw/Pages/List.aspx?nodeid=269 (accessed on 27 March 2023).
- Hung, C.F.; Yang, C.K.; Ou, Y.C. Urologic cancer in Taiwan. Jpn. J. Clin. Oncol. 2016, 46, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Yeh, H.L.; Hsu, S.W.; Chang, Y.C.; Chan, T.C.; Tsou, H.C.; Chang, Y.C.; Chiang, P.H. Spatial Analysis of Ambient PM2.5 Exposure and Bladder Cancer Mortality in Taiwan. Int. J. Environ. Res. Public Health 2017, 14, 508. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.F.; Chen, B.K.; Yang, C.Y. Parity, Age at First Birth, and Risk of Death from Bladder Cancer: A Population-Based Cohort Study in Taiwan. Int. J. Environ. Res. Public Health 2016, 13, 1197. [Google Scholar] [CrossRef] [PubMed]
- Aveta, A.; Cacciapuoti, C.; Barone, B.; Di Zazzo, E.; Del Giudice, F.; Maggi, M.; Ferro, M.; Terracciano, D.; Busetto, G.M.; Lucarelli, G.; et al. The Impact of Meat Intake on Bladder Cancer Incidence: Is It Really a Relevant Risk? Cancers 2022, 14, 4775. [Google Scholar] [CrossRef]
- Ng, K.; Stenzl, A.; Sharma, A.; Vasdev, N. Urinary biomarkers in bladder cancer: A review of the current landscape and future directions. Urol. Oncol. 2021, 39, 41–51. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Ye, R.; Zhang, D.; Li, Q.; An, D.; Fang, L.; Lin, Y.; Hou, Y.; Xu, A.; et al. An epigenetic biomarker combination of PCDH17 and POU4F2 detects bladder cancer accurately by methylation analyses of urine sediment DNA in Han Chinese. Oncotarget 2016, 7, 2754–2764. [Google Scholar] [CrossRef]
- O’Sullivan, P.; Sharples, K.; Dalphin, M.; Davidson, P.; Gilling, P.; Cambridge, L.; Harvey, J.; Toro, T.; Giles, N.; Luxmanan, C.; et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J. Urol. 2012, 188, 741–747. [Google Scholar] [CrossRef]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Lu, J.; Mercer, K.L.; Golub, T.R.; Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007, 39, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Kwak, P.B.; Iwasaki, S.; Tomari, Y. The microRNA pathway and cancer. Cancer Sci. 2010, 101, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Gilad, S.; Meiri, E.; Yogev, Y.; Benjamin, S.; Lebanony, D.; Yerushalmi, N.; Benjamin, H.; Kushnir, M.; Cholakh, H.; Melamed, N.; et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 2008, 3, e3148. [Google Scholar] [CrossRef]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef]
- Lin, J.T.; Tsai, K.W. Circulating miRNAs Act as Diagnostic Biomarkers for Bladder Cancer in Urine. Int. J. Mol. Sci. 2021, 22, 4278. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, K.K.; Gu, J.; Yang, H.; Lin, J.; Ajani, J.A.; Wu, X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. 2008, 1, 460–469. [Google Scholar] [CrossRef]
- Mittal, R.D.; Gangwar, R.; George, G.P.; Mittal, T.; Kapoor, R. Investigative role of pre-microRNAs in bladder cancer patients: A case-control study in North India. DNA Cell Biol. 2011, 30, 401–406. [Google Scholar] [CrossRef]
- Wang, M.; Chu, H.; Li, P.; Yuan, L.; Fu, G.; Ma, L.; Shi, D.; Zhong, D.; Tong, N.; Qin, C.; et al. Genetic variants in miRNAs predict bladder cancer risk and recurrence. Cancer Res. 2012, 72, 6173–6182. [Google Scholar] [CrossRef]
- Yang, H.; Dinney, C.P.; Ye, Y.; Zhu, Y.; Grossman, H.B.; Wu, X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008, 68, 2530–2537. [Google Scholar] [CrossRef]
- Deng, S.; Wang, W.; Li, X.; Zhang, P. Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J. Surg. Oncol. 2015, 13, 297. [Google Scholar] [CrossRef]
- Liao, C.H.; Tsai, C.W.; Chang, W.S.; Wang, Z.H.; Gong, C.L.; Wu, H.C.; Wang, B.R.; Hsu, S.W.; Huang, W.C.; Shen, T.C.; et al. Association of Matrix Metalloproteinase-1 Genotypes with Bladder Cancer Risk. In Vivo 2021, 35, 2535–2540. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Liu, L.C.; Hsiao, C.L.; Su, C.H.; Wang, H.C.; Ji, H.X.; Tsai, C.W.; Maa, M.C.; Bau, D.T. The contributions of the tissue inhibitor of metalloproteinase-1 genotypes to triple negative breast cancer risk. Biomedicine 2016, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.S.; Chang, W.S.; Hsu, P.C.; Chen, C.C.; Chin, Y.T.; Huang, T.L.; Hsu, Y.N.; Kuo, C.C.; Wang, Y.C.; Tsai, C.W.; et al. Significant Association Between the MiR146a Genotypes and Susceptibility to Childhood Acute Lymphoblastic Leukemia in Taiwan. Cancer Genom. Proteom. 2020, 17, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Hsu, P.C.; Shih, L.C.; Hsu, Y.N.; Kuo, C.C.; Chao, C.Y.; Chang, W.S.; Tsai, C.W.; Bau, D.T.; Pei, J.S. MiR-196a-2 Genotypes Determine the Susceptibility and Early Onset of Childhood Acute Lymphoblastic Leukemia. Anticancer Res. 2020, 40, 4465–4469. [Google Scholar] [CrossRef] [PubMed]
- Dezfuli, N.K.; Alipoor, S.D.; Dalil Roofchayee, N.; Seyfi, S.; Salimi, B.; Adcock, I.M.; Mortaz, E. Evaluation Expression of miR-146a and miR-155 in Non-Small-Cell Lung Cancer Patients. Front. Oncol. 2021, 11, 715677. [Google Scholar] [CrossRef]
- Camargo, J.A.; Lopes, R.E.; Ferreira, G.F.D.; Viana, N.I.; Guimaraes, V.; Leite, K.R.M.; Nahas, W.C.; Srougi, M.; Antunes, A.A.; Reis, S.T. The role of single nucleotide polymorphisms of miRNAs 100 and 146a as prognostic factors for prostate cancer. Int. J. Biol. Markers 2021, 36, 50–56. [Google Scholar] [CrossRef]
- Siasi, E.; Solimani, M. Associations of Single Nucleotide Polymorphism in miR-146a Gene with Susceptibility to Breast Cancer in the Iranian Female. Asian Pac. J. Cancer Prev. 2020, 21, 1585–1593. [Google Scholar] [CrossRef]
- Brincas, H.M.; Augusto, D.G.; Mathias, C.; Cavalli, I.J.; Lima, R.S.; Kuroda, F.; Urban, C.A.; Gradia, D.F.; de Oliveira, J.; de Almeida, R.C.; et al. A genetic variant in microRNA-146a is associated with sporadic breast cancer in a Southern Brazilian Population. Genet. Mol. Biol. 2020, 42, e20190278. [Google Scholar] [CrossRef]
- Wang, Y.H.; Hu, H.N.; Weng, H.; Chen, H.; Luo, C.L.; Ji, J.; Yin, C.Q.; Yuan, C.H.; Wang, F.B. Association between Polymorphisms in MicroRNAs and Risk of Urological Cancer: A Meta-Analysis Based on 17,019 Subjects. Front. Physiol. 2017, 8, 325. [Google Scholar] [CrossRef]
- Sasaki, H.; Yoshiike, M.; Nozawa, S.; Usuba, W.; Katsuoka, Y.; Aida, K.; Kitajima, K.; Kudo, H.; Hoshikawa, M.; Yoshioka, Y.; et al. Expression Level of Urinary MicroRNA-146a-5p Is Increased in Patients with Bladder Cancer and Decreased in Those After Transurethral Resection. Clin. Genitourin. Cancer 2016, 14, e493–e499. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Q.; Ji, L.; Wang, G.; Niu, X.; Sun, S. lncRNA MORT Regulates Bladder Cancer Behaviors by Downregulating MicroRNA-146a-5p. Nephron 2020, 144, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Liao, Y.; Qiu, Y.; Liu, H.; Tan, D.; Wu, T.; Tang, M.; Zhang, S.; Wang, H. miRNA 146a promotes chemotherapy resistance in lung cancer cells by targeting DNA damage inducible transcript 3 (CHOP). Cancer Lett. 2018, 428, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Sundaravinayagam, D.; Kim, H.R.; Wu, T.; Kim, H.H.; Lee, H.S.; Jun, S.; Cha, J.H.; Kee, Y.; You, H.J.; Lee, J.H. miR146a-mediated targeting of FANCM during inflammation compromises genome integrity. Oncotarget 2016, 7, 45976–45994. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, E.I.; Meza-Sosa, K.F.; Lopez-Sevilla, Y.; Camacho-Concha, N.; Sanchez, N.C.; Perez-Martinez, L.; Pedraza-Alva, G. Merlin negative regulation by miR-146a promotes cell transformation. Biochem. Biophys. Res. Commun. 2015, 468, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, N.C.; Medrano-Jimenez, E.; Aguilar-Leon, D.; Perez-Martinez, L.; Pedraza-Alva, G. Tumor Necrosis Factor-Induced miR-146a Upregulation Promotes Human Lung Adenocarcinoma Metastasis by Targeting Merlin. DNA Cell Biol. 2020, 39, 484–497. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, Q.; Jiang, X.Q.; Li, D.Q.; Jiang, X.C.; Wu, X.B.; Cao, Y.L. miR-146a promoted breast cancer proliferation and invasion by regulating NM23-H1. J. Biochem. 2020, 167, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Zhu, E.D.; Li, N.; Lu, D.S.; Li, W.; Li, B.S.; Zhao, Y.L.; Mao, X.H.; Guo, G.; Yu, P.W.; et al. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol. Rep. 2012, 27, 559–566. [Google Scholar]
- Pu, W.; Shang, Y.; Shao, Q.; Yuan, X. miR-146a promotes cell migration and invasion in melanoma by directly targeting SMAD4. Oncol. Lett. 2018, 15, 7111–7117. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Li, T.; Wang, L.; Lv, W.; Wang, S.; Ma, D.; Zang, Y.; Zhu, X.; Xu, Y.; et al. MicroRNA-146a promotes proliferation, migration, and invasion of HepG2 via regulating FLAP. Cancer Cell Int. 2022, 22, 149. [Google Scholar] [CrossRef]
- Wang, L.; Chen, Y.; Yan, Y.; Guo, X.; Fang, Y.; Su, Y.; Wang, L.; Pathak, J.L.; Ge, L. miR-146a Overexpression in Oral Squamous Cell Carcinoma Potentiates Cancer Cell Migration and Invasion Possibly via Targeting HTT. Front. Oncol. 2020, 10, 585976. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, G.; Wang, F.; Li, C.; Wang, X. Hypoxia-Regulated miR-146a Targets Cell Adhesion Molecule 2 to Promote Proliferation, Migration, and Invasion of Clear Cell Renal Cell Carcinoma. Cell Physiol. Biochem. 2018, 49, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.S.; Liu, C.J.; Chou, C.S.; Kao, S.Y.; Yang, C.C.; Chang, K.W.; Chiu, T.H.; Lin, S.C. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS ONE 2013, 8, e79926. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.A.; Akter, T.; Islam, M.S. Effect of miR-196a2 rs11614913 Polymorphism on Cancer Susceptibility: Evidence From an Updated Meta-Analysis. Technol. Cancer Res. Treat. 2022, 21, 15330338221109798. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Zhang, Y.; Chen, L. Correlation between miRNA-196a2 and miRNA-499 polymorphisms and bladder cancer. Int. J. Clin. Exp. Med. 2016, 9, 20484–20488. [Google Scholar]


| Character | Controls (n = 375) | Cases (n = 375) | p-Value | ||||
|---|---|---|---|---|---|---|---|
| n | % | Mean (SD) | n | % | Mean (SD) | ||
| Age (years) | 62.9 (9.8) | 61.4 (10.3) | 0.7315 a | ||||
| Age group (years) | 0.7108 b | ||||||
| ≤55 | 152 | 40.5% | 158 | 42.1% | |||
| >55 | 223 | 59.5% | 217 | 57.9% | |||
| Gender | 0.5525 b | ||||||
| Male | 287 | 76.5% | 279 | 74.4% | |||
| Female | 88 | 23.5% | 96 | 25.6% | |||
| Personal habits | |||||||
| Cigarette smoking | 186 | 49.6% | 201 | 53.6% | 0.3063 b | ||
| Alcohol drinking | 176 | 46.9% | 189 | 50.4% | 0.3807 b | ||
| Stage | |||||||
| Non-muscle-invasive | 235 | 62.7% | |||||
| Muscle-invasive | 140 | 37.3% | |||||
| Grade | |||||||
| Low | 151 | 40.3% | |||||
| High | 224 | 59.7% | |||||
| SNP | Genotype | Cases | Controls | p-Value | OR (95% CI) |
|---|---|---|---|---|---|
| rs2910164 | CC | 82 (21.9%) | 119 (31.7%) | 1.00 (Ref) | |
| CG | 166 (44.3%) | 171 (45.6%) | 0.0695 | 1.41 (0.99–2.01) | |
| GG | 127 (33.8%) | 85 (22.7%) | 0.0002 * | 2.17 (1.46–3.21) | |
| Ptrend | 0.0005 * | ||||
| CG + GG | 293 (78.1%) | 256 (68.3%) | 0.0030 * | 1.66 (1.20–2.30) | |
| PHWE | 0.1193 |
| SNP | Genotype | Cases | Controls | p-Value | OR (95%CI) |
|---|---|---|---|---|---|
| rs11614913 | TT | 125 (33.3%) | 116 (30.9%) | 1.00 (Ref) | |
| CT | 180 (48.0%) | 186 (49.6%) | 0.5722 | 0.90 (0.65–1.24) | |
| CC | 70 (18.7%) | 73 (19.5%) | 0.6549 | 0.89 (0.59–1.35) | |
| Ptrend | 0.7798 | ||||
| CT + CC | 250 (66.7%) | 259 (69.1%) | 0.5316 | 0.90 (0.66–1.22) | |
| PHWE | 0.9195 |
| Allele | Cases | Controls | p-Value | OR (95% CI) |
|---|---|---|---|---|
| mir146a rs2910164 | ||||
| C | 330 (44.0%) | 409 (54.5%) | 1.00 (Ref) | |
| G | 420 (56.0%) | 341 (45.5%) | 0.0001 * | 1.53 (1.25–1.87) |
| mir196a rs11614913 | ||||
| T | 430 (57.3%) | 418 (55.7%) | 1.00 (Ref) | |
| C | 320 (42.7%) | 332 (44.3%) | 0.5667 | 0.94 (0.76–1.15) |
| Genotype | Controls | Cases | OR (95% CI) a | aOR (95% CI) b | p-Value |
|---|---|---|---|---|---|
| Age | |||||
| ≤55 years old | |||||
| CC | 47 | 33 | 1.00 (ref) | 1.00 (ref) | |
| CG | 66 | 68 | 1.47 (0.84–2.57) | 1.55 (0.87–2.34) | 0.2283 |
| GG | 39 | 57 | 2.08 (1.14–3.81) | 2.27 (1.18–3.56) | 0.0248 * |
| >55 years old | |||||
| CC | 72 | 49 | 1.00 (ref) | 1.00 (ref) | |
| CG | 105 | 98 | 1.37 (0.87–2.16) | 1.49 (0.89–2.03) | 0.2130 |
| GG | 46 | 70 | 2.24 (1.33–3.76) | 2.58 (1.45–3.46) | 0.0034 * |
| Gender | |||||
| Males | |||||
| CC | 89 | 60 | 1.00 (ref) | 1.00 (ref) | |
| CG | 134 | 122 | 1.35 (0.90–2.03) | 1.39 (0.87–1.96) | 0.1810 |
| GG | 64 | 97 | 2.25 (1.43–3.54) | 2.36 (1.66–3.31) | 0.0007 * |
| Females | |||||
| CC | 30 | 22 | 1.00 (ref) | 1.00 (ref) | |
| CG | 37 | 44 | 1.62 (0.80–3.27) | 1.59 (0.77–3.18) | 0.2402 |
| GG | 21 | 30 | 1.95 (0.89–4.26) | 2.01 (0.92–3.95) | 0.1391 |
| Smoking behaviors | |||||
| Non-smokers | |||||
| CC | 61 | 37 | 1.00 (ref) | 1.00 (ref) | |
| CG | 84 | 80 | 1.57 (0.94–2.62) | 1.65 (0.97–2.47) | 0.1077 |
| GG | 44 | 57 | 2.14 (1.21–3.77) | 2.31 (1.33–2.98) | 0.0125 * |
| Smokers | |||||
| CC | 58 | 45 | 1.00 (ref) | 1.00 (ref) | |
| CG | 87 | 86 | 1.27 (0.78–2.08) | 1.35 (0.83–2.03) | 0.3985 |
| GG | 41 | 70 | 2.20 (1.27–3.81) | 2.26 (1.32–3.74) | 0.0069 * |
| Alcohol drinking behaviors | |||||
| Non-drinkers | |||||
| CC | 61 | 41 | 1.00 (ref) | 1.00 (ref) | |
| CG | 95 | 79 | 1.24 (0.75–2.03) | 1.32 (0.84–2.01) | 0.4738 |
| GG | 43 | 66 | 2.28 (1.32–3.96) | 2.34 (1.37–4.04) | 0.0048 * |
| Drinkers | |||||
| CC | 58 | 41 | 1.00 (ref) | 1.00 (ref) | |
| CG | 76 | 87 | 1.62 (0.98–2.68) | 1.69 (0.97–2.54) | 0.0801 |
| GG | 42 | 61 | 2.05 (1.17–3.60) | 2.11 (1.24–3.52) | 0.0168 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.-R.; Chang, W.-S.; Liao, C.-H.; Wang, Y.-C.; Gu, J.; Bau, D.-T.; Tsai, C.-W. Impacts of Mir146a Genotypes on Bladder Cancer Risk in Taiwan. Biomedicines 2023, 11, 1396. https://doi.org/10.3390/biomedicines11051396
Wang B-R, Chang W-S, Liao C-H, Wang Y-C, Gu J, Bau D-T, Tsai C-W. Impacts of Mir146a Genotypes on Bladder Cancer Risk in Taiwan. Biomedicines. 2023; 11(5):1396. https://doi.org/10.3390/biomedicines11051396
Chicago/Turabian StyleWang, Bo-Ren, Wen-Shin Chang, Cheng-Hsi Liao, Yun-Chi Wang, Jian Gu, Da-Tian Bau, and Chia-Wen Tsai. 2023. "Impacts of Mir146a Genotypes on Bladder Cancer Risk in Taiwan" Biomedicines 11, no. 5: 1396. https://doi.org/10.3390/biomedicines11051396
APA StyleWang, B.-R., Chang, W.-S., Liao, C.-H., Wang, Y.-C., Gu, J., Bau, D.-T., & Tsai, C.-W. (2023). Impacts of Mir146a Genotypes on Bladder Cancer Risk in Taiwan. Biomedicines, 11(5), 1396. https://doi.org/10.3390/biomedicines11051396






