Anionic Phospholipids Shift the Conformational Equilibrium of the Selectivity Filter in the KcsA Channel to the Conductive Conformation: Predicted Consequences on Inactivation
Abstract
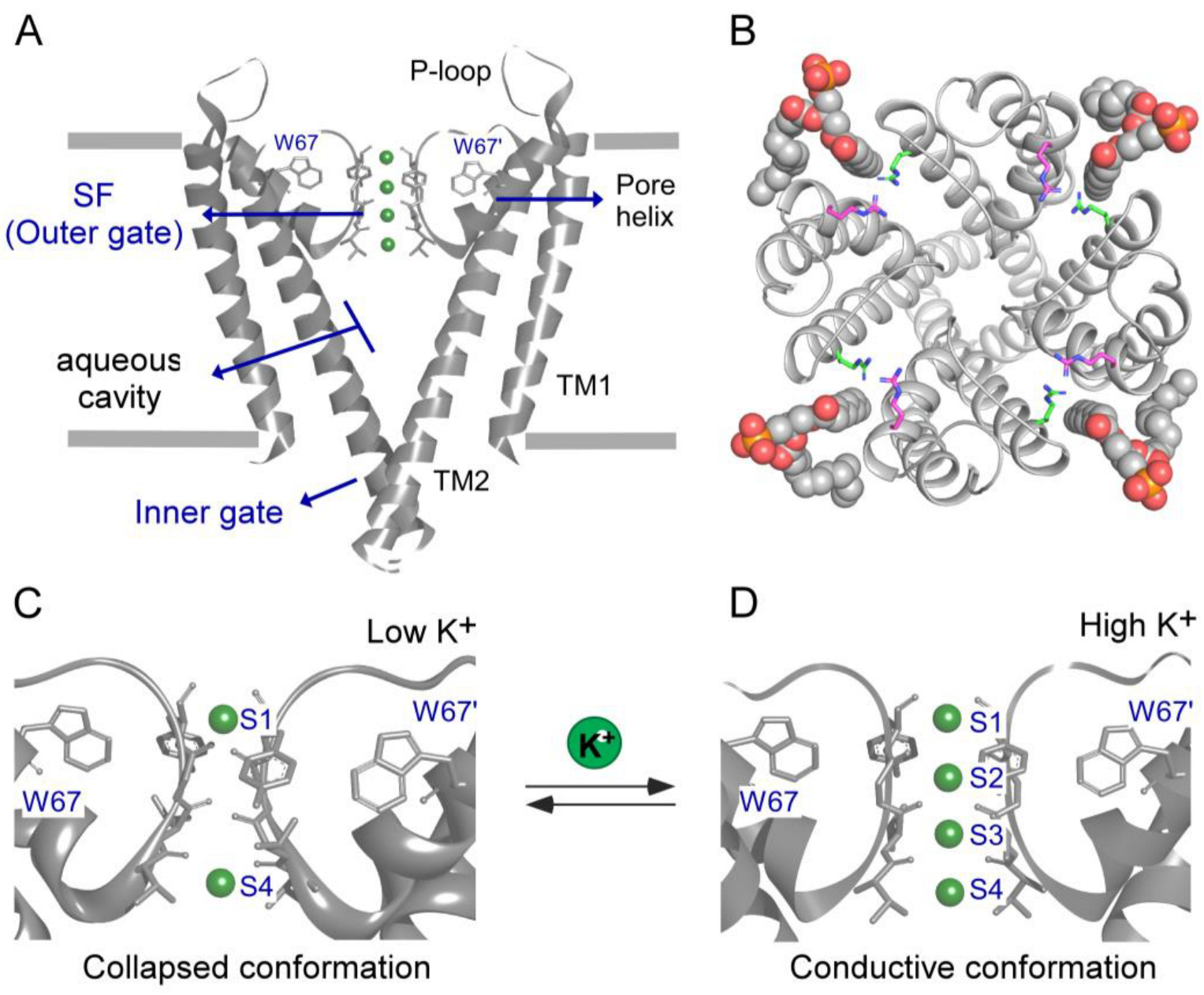
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. KcsA Heterologous Expression and Purification
2.3. Fluorescence Monitoring of Cation Binding through Thermal Denaturation
2.4. Steady-State and Time-Resolved Fluorescence Measurements
2.5. Calculation of the K+ Binding Affinity to KcsA from Changes in the Steady-State Anisotropy Values
2.6. DLS Measurements
2.7. Statistics
3. Results
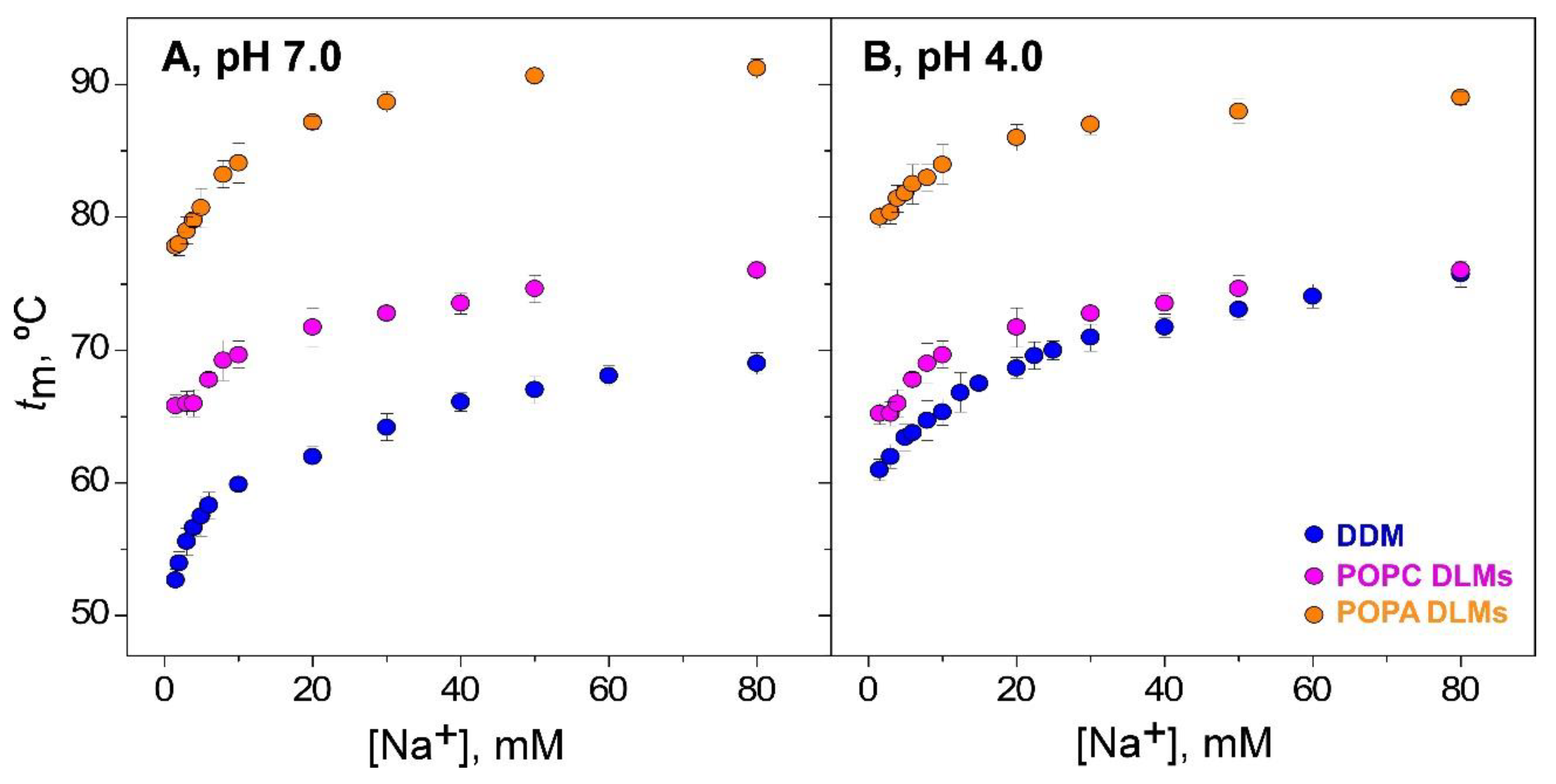
3.1. Thermal Denaturation of WT-KcsA in Mixed Detergent–Lipid Micelles (DLMs)
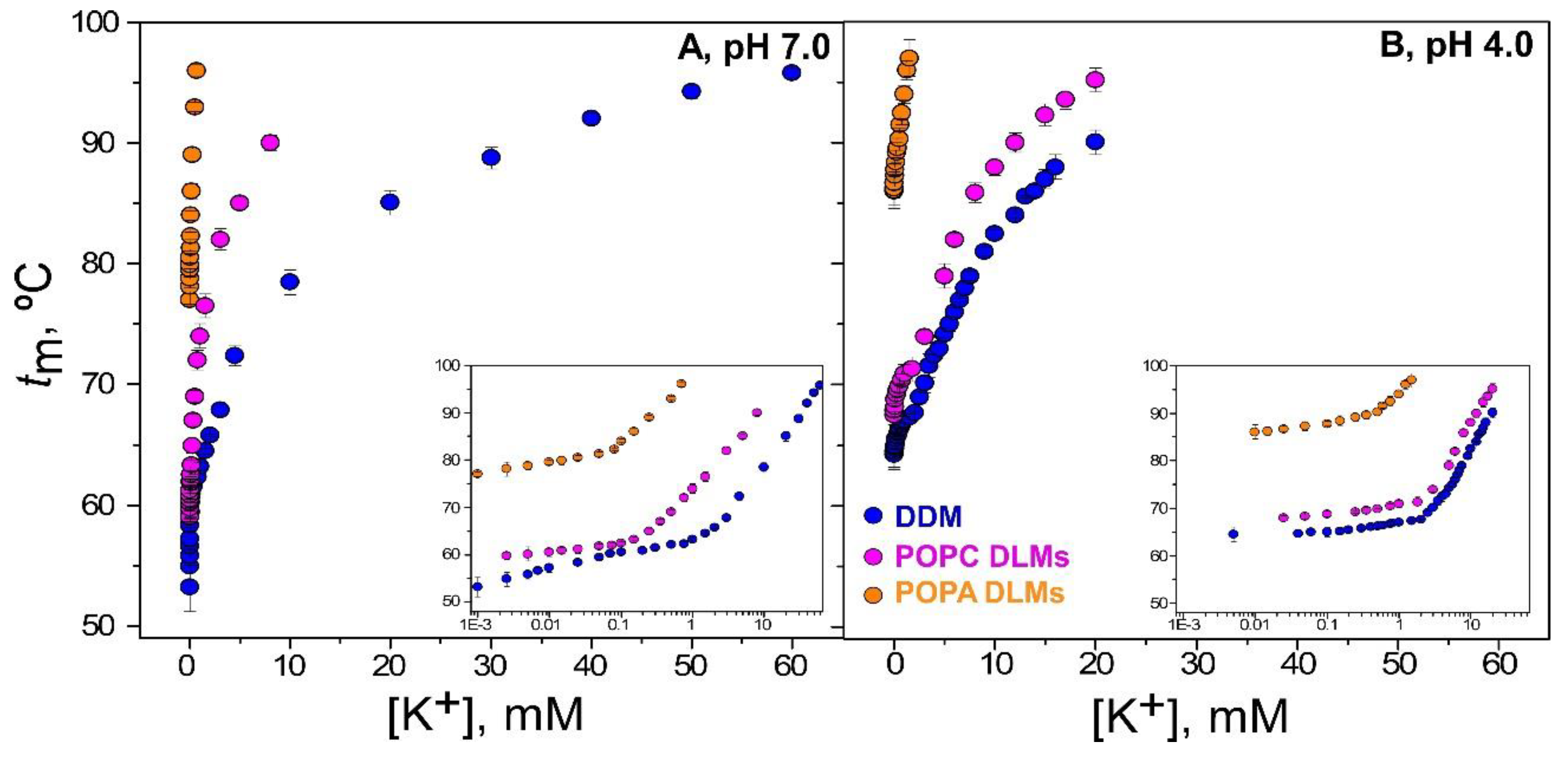
3.2. Binding of K+ to the W67-KcsA Mutant in DLMs Followed by Fluorescence Anisotropy
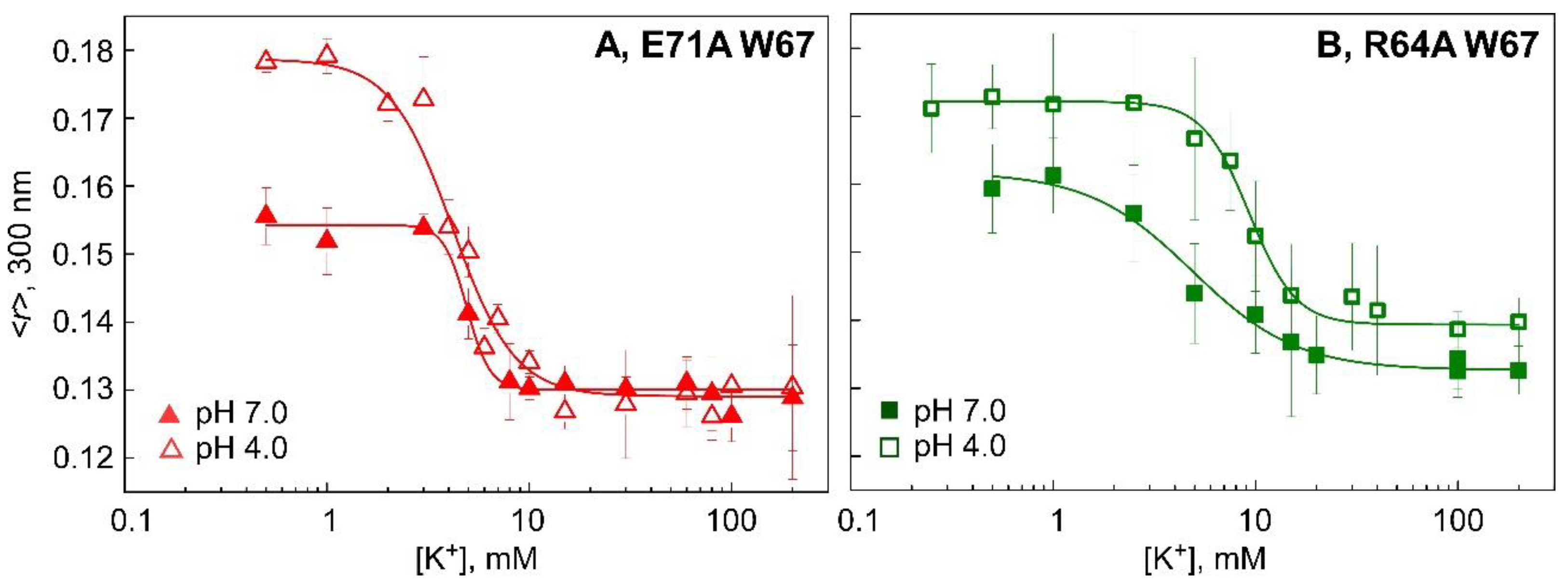
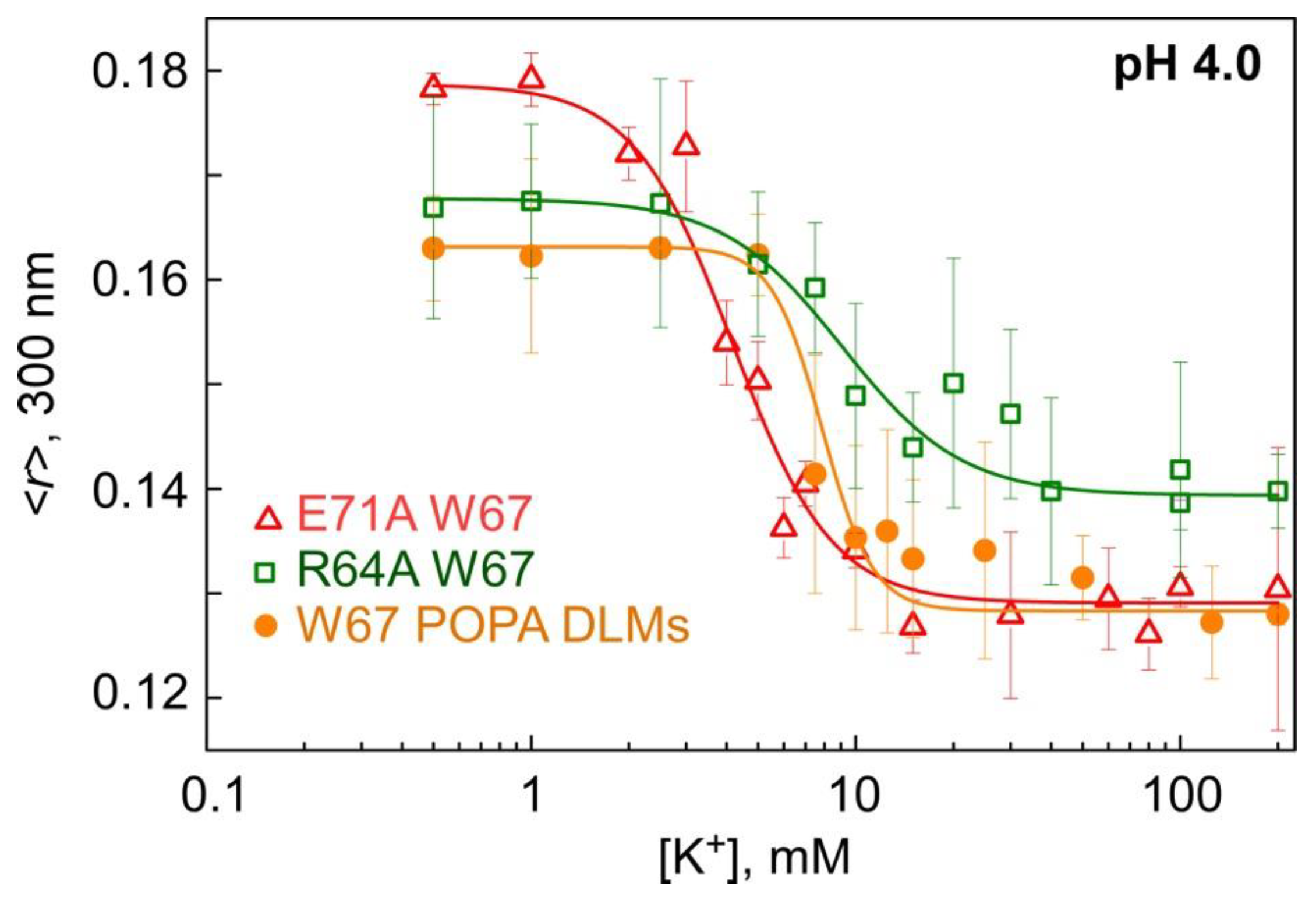
3.3. Binding of K+ to Model Non-Inactivating W67 KcsA Mutants Followed by Fluorescence Anisotropy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, O.S.; Koeppe, R.E. Bilayer Thickness and Membrane Protein Function: An Energetic Perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.G. How Lipids Affect the Activities of Integral Membrane Proteins. Biochim. Biophys. Acta Biomembr. 2004, 1666, 62–87. [Google Scholar]
- Marsh, D. Protein Modulation of Lipids, and Vice-Versa, in Membranes. Biochim. Biophys. Acta Biomembr. 2008, 1778, 1545–1575. [Google Scholar]
- Poveda, J.A.; Giudici, A.M.; Renart, M.L.; Molina, M.L.; Montoya, E.; Fernández-Carvajal, A.; Fernández-Ballester, G.; Encinar, J.A.; González-Ros, J.M. Lipid Modulation of Ion Channels through Specific Binding Sites. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1560–1567. [Google Scholar]
- Poveda, J.A.; Marcela Giudici, A.; Lourdes Renart, M.; Morales, A.; González-Ros, J.M. Towards Understanding the Molecular Basis of Ion Channel Modulation by Lipids: Mechanistic Models and Current Paradigms. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1507–1516. [Google Scholar] [CrossRef]
- Levental, I.; Lyman, E. Regulation of Membrane Protein Structure and Function by Their Lipid Nano-Environment. Nat. Rev. Mol. Cell Biol. 2023, 24, 107–122. [Google Scholar] [CrossRef]
- Uysal, S.; Vasquez, V.; Tereshko, V.; Esaki, K.; Fellouse, F.A.; Sidhu, S.S.; Koide, S.; Perozo, E.; Kossiakoff, A. Crystal Structure of Full-Length KcsA in Its Closed Conformation. Proc. Natl. Acad. Sci. USA 2009, 106, 16644–16649. [Google Scholar] [CrossRef]
- Zhou, Y.; Morais-Cabral, J.H.; Kaufman, A.; Mackinnon, R. Chemistry of Ion Coordination and Hydration Revealed by a K+ Channel-Fab Complex at 2.0 Å Resolution. Nature 2001, 414, 43–48. [Google Scholar] [CrossRef]
- Cheng, W.W.L.; McCoy, J.G.; Thompson, A.N.; Nichols, C.G.; Nimigean, C.M. Mechanism for Selectivity-Inactivation Coupling in KcsA Potassium Channels. Proc. Natl. Acad. Sci. USA 2011, 108, 5272–5277. [Google Scholar] [CrossRef]
- Chill, J.H. NMR Study of the Tetrameric KcsA Potassium Channel in Detergent Micelles. Protein Sci. 2006, 15, 684–698. [Google Scholar] [CrossRef]
- Cordero-Morales, J.F.; Cuello, L.G.; Zhao, Y.; Jogini, V.; Cortes, D.M.; Roux, B.; Perozo, E. Molecular Determinants of Gating at the Potassium-Channel Selectivity Filter. Nat. Struct. Mol. Biol. 2006, 13, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Morais-Cabral, J.H.; Zhou, Y.; MacKinnon, R. Energetic Optimization of Ion Conduction Rate by the K+ Selectivity Filter. Nature 2001, 414, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; MacKinnon, R. The Occupancy of Ions in the K+ Selectivity Filter: Charge Balance and Coupling of Ion Binding to a Protein Conformational Change Underlie High Conduction Rates. J. Mol. Biol. 2003, 333, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Ader, C.; Schneider, R.; Hornig, S.; Velisetty, P.; Vardanyan, V.; Giller, K.; Ohmert, I.; Becker, S.; Pongs, O.; Baldus, M. Coupling of Activation and Inactivation Gate in a K+-Channel: Potassium and Ligand Sensitivity. EMBO J. 2009, 28, 2825–2834. [Google Scholar] [PubMed]
- Baker, K.A.; Tzitzilonis, C.; Kwiatkowski, W.; Choe, S.; Riek, R. Conformational Dynamics of the KcsA Potassium Channel Governs Gating Properties. Nat. Struct. Mol. Biol. 2007, 14, 1089–1095. [Google Scholar] [CrossRef]
- Bhate, M.P.; Wylie, B.J.; Tian, L.; McDermott, A.E. Conformational Dynamics in the Selectivity Filter of KcsA in Response to Potassium Ion Concentration. J. Mol. Biol. 2010, 401, 155–166. [Google Scholar] [CrossRef]
- Imai, S.; Osawa, M.; Takeuchi, K.; Shimada, I. Structural Basis Underlying the Dual Gate Properties of KcsA. Proc. Natl. Acad. Sci. USA 2010, 107, 6216–6221. [Google Scholar] [CrossRef]
- Wylie, B.J.; Bhate, M.P.; McDermott, A.E. Transmembrane Allosteric Coupling of the Gates in a Potassium Channel. Proc. Natl. Acad. Sci. USA 2014, 111, 185–190. [Google Scholar] [CrossRef]
- Lockless, S.W.; Zhou, M.; MacKinnon, R. Structural and Thermodynamic Properties of Selective Ion Binding in a K+ Channel. PLoS Biol. 2007, 5, 1079–1088. [Google Scholar] [CrossRef]
- Poveda, J.A.; Giudici, A.M.; Renart, M.L.; Millet, O.; Morales, A.; González-Ros, J.M.; Oakes, V.; Furini, S.; Domene, C. Modulation of the Potassium Channel KcsA by Anionic Phospholipids: Role of Arginines at the Non-Annular Lipid Binding Sites. Biochim. Biophys. Acta Biomembr. 2019, 1861, 183029–183043. [Google Scholar] [CrossRef]
- Weingarth, M.; Prokofyev, A.; Van Der Cruijsen, E.A.W.; Nand, D.; Bonvin, A.M.J.J.; Pongs, O.; Baldus, M. Structural Determinants of Specific Lipid Binding to Potassium Channels. J. Am. Chem. Soc. 2013, 135, 3983–3988. [Google Scholar] [CrossRef]
- Marius, P.; De Planque, M.R.R.; Williamson, P.T.F. Probing the Interaction of Lipids with the Non-Annular Binding Sites of the Potassium Channel KcsA by Magic-Angle Spinning NMR. Biochim. Biophys. Acta 2012, 1818, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Renart, M.L.; Barrera, F.N.; Molina, M.L.; Encinar, J.A.; Poveda, J.A.; Fernández, A.M.; Gómez, J.; González-Ros, J.M. Effects of Conducting and Blocking Ions on the Structure and Stability of the Potassium Channel KcsA. J. Biol. Chem. 2006, 281, 29905–29915. [Google Scholar] [CrossRef] [PubMed]
- Renart, M.L.; Montoya, E.; Giudici, A.M.; Poveda, J.A.; Fernández, A.M.; Morales, A.; González-Ros, J.M. Selective Exclusion and Selective Binding Both Contribute to Ion Selectivity in KcsA, a Model Potassium Channel. J. Biol. Chem. 2017, 292, 15552–15560. [Google Scholar] [CrossRef] [PubMed]
- Montoya, E.; Lourdes Renart, M.; Marcela Giudici, A.; Poveda, J.A.; Fernández, A.M.; Morales, A.; González-Ros, J.M. Differential Binding of Monovalent Cations to KcsA: Deciphering the Mechanisms of Potassium Channel Selectivity. Biochim. Biophys. Acta Biomembr. 2017, 1859, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Renart, M.L.; Triano, I.; Poveda, J.A.; Encinar, J.A.; Fernández, A.M.; Ferrer-Montiel, A.V.; Gómez, J.; González Ros, J.M. Ion Binding to KcsA: Implications in Ion Selectivity and Channel Gating. Biochemistry 2010, 49, 9480–9487. [Google Scholar] [CrossRef]
- Renart, M.L.; Giudici, A.M.; Poveda, J.A.; Fedorov, A.; Berberan-Santos, M.N.; Prieto, M.; Díaz-García, C.; González-Ros, J.M.; Coutinho, A. Conformational Plasticity in the KcsA Potassium Channel Pore Helix Revealed by Homo-FRET Studies. Sci. Rep. 2019, 9, 6215–6228. [Google Scholar] [CrossRef]
- Doyle, D.A.; Cabral, J.M.; Pfuetzner, R.A.; Kuo, A.; Gulbis, J.M.; Cohen, S.L.; Chait, B.T.; MacKinnon, R. The Structure of the Potassium Channel: Molecular Basis of K+ Conduction and Selectivity. Science 1998, 280, 69–77. [Google Scholar] [CrossRef]
- Shealy, R.T.; Murphy, A.D.; Ramarathnam, R.; Jakobsson, E.; Subramaniam, S. Sequence-Function Analysis of the K+-Selective Family of Ion Channels Using a Comprehensive Alignment and the KcsA Channel Structure. Biophys. J. 2003, 84, 2929–2942. [Google Scholar] [CrossRef]
- Renart, M.L.; Montoya, E.; Fernández, A.M.; Molina, M.L.; Poveda, J.A.; Encinar, J.A.; Ayala, J.L.; Ferrer-Montiel, A.V.; Gómez, J.; Morales, A.; et al. Contribution of Ion Binding Affinity to Ion Selectivity and Permeation in KcsA, a Model Potassium Channel. Biochemistry 2012, 51, 3891–3900. [Google Scholar] [CrossRef]
- Giudici, A.; Díaz-García, C.; Renart, M.; Coutinho, A.; Prieto, M.; González-Ros, J.; Poveda, J. Tetraoctylammonium, a Long Chain Quaternary Ammonium Blocker, Promotes a Noncollapsed, Resting-Like Inactivated State in KcsA. Int. J. Mol. Sci. 2021, 22, 490. [Google Scholar] [CrossRef]
- Cuello, L.G.; Cortes, D.M.; Jogini, V.; Sompornpisut, A.; Perozo, E. A Molecular Mechanism for Proton-Dependent Gating in KcsA. FEBS Lett. 2010, 584, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Onishi, Y.; Yanagida, T.; Ide, T. Role of the KcsA Channel Cytoplasmic Domain in PH-Dependent Gating. Biophys. J. 2011, 101, 2157–2162. [Google Scholar] [CrossRef] [PubMed]
- Posson, D.J.; Thompson, A.N.; McCoy, J.G.; Nimigean, C.M. Molecular Interactions Involved in Proton-Dependent Gating in KcsA Potassium Channels. J. Gen. Physiol. 2013, 142, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Mi, X.; Paajanen, V.; Wang, K.; Fan, Z. Activation-Coupled Inactivation in the Bacterial Potassium Channel KcsA. Proc. Natl. Acad. Sci. USA 2005, 102, 17630–17635. [Google Scholar] [CrossRef]
- Li, J.; Ostmeyer, J.; Cuello, L.G.; Perozo, E.; Roux, B. Rapid Constriction of the Selectivity Filter Underlies C-Type Inactivation in the KcsA Potassium Channel. J. Gen. Physiol. 2018, 150, 1408–1420. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Morales, J.F.; Jogini, V.; Lewis, A.; Vasquez, V.; Cortes, D.M.; Roux, B.; Perozo, E. Molecular Driving Forces Determining Potassium Channel Slow Inactivation. Nat. Struct. Mol. Biol. 2007, 14, 1062–1069. [Google Scholar] [CrossRef]
- Rotem, D.; Mason, A.; Bayley, H. Inactivation of the KcsA Potassium Channel Explored with Heterotetramers. J. Gen. Physiol. 2010, 135, 29–42. [Google Scholar] [CrossRef]
- Cordero-Morales, J.F.; Jogini, V.; Chakrapani, S.; Perozo, E. A Multipoint Hydrogen-Bond Network Underlying KcsA C-Type Inactivation. Biophys. J. 2011, 100, 2387–2393. [Google Scholar] [CrossRef] [PubMed]
- Cuello, L.G.; Cortes, D.M.; Perozo, E. The Gating Cycle of a K+ Channel at Atomic Resolution. eLife 2017, 6, e28032. [Google Scholar] [CrossRef]
- Valiyaveetil, F.I.; Zhou, Y.; MacKinnon, R. Lipids in the Structure, Folding, and Function of the KcsA K+ Channel. Biochemistry 2002, 41, 10771–10777. [Google Scholar] [CrossRef] [PubMed]
- Demmers, J.A.; van Dalen, A.; de Kruijff, B.; Heck, A.J.; Killian, J.A. Interaction of the K+ Channel KcsA with Membrane Phospholipids as Studied by ESI Mass Spectrometry. FEBS Lett. 2003, 541, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Deol, S.S.; Domene, C.; Bond, P.J.; Sansom, M.S.P. Anionic Phospholipid Interactions with the Potassium Channel KcsA: Simulation Studies. Biophys. J. 2006, 90, 822–830. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.G. Lipid-Protein Interactions in Biological Membranes: A Structural Perspective. Biochim. Biophys. Acta Biomembr. 2003, 1612, 1–40. [Google Scholar] [CrossRef]
- Alvis, S.J.; Williamson, I.M.; East, J.M.; Lee, A.G. Interactions of Anionic Phospholipids and Phosphatidylethanolamine with the Potassium Channel KcsA. Biophys. J. 2003, 85, 3828–3838. [Google Scholar] [CrossRef]
- Marius, P.; Alvis, S.J.; East, J.M.; Lee, A.G. The Interfacial Lipid Binding Site on the Potassium Channel KcsA Is Specific for Anionic Phospholipids. Biophys. J. 2005, 89, 4081–4089. [Google Scholar] [CrossRef]
- Triano, I.; Barrera, F.N.; Renart, M.L.; Molina, M.L.; Fernandez-Ballester, G.; Poveda, J.A.; Fernandez, A.M.; Encinar, J.A.; Ferrer-Montiel, A.V.; Otzen, D.; et al. Occupancy of Nonannular Lipid Binding Sites on KcsA Greatly Increases the Stability of the Tetrameric Protein. Biochemistry 2010, 49, 5397–5404. [Google Scholar] [CrossRef] [PubMed]
- Opekarová, M.; Tanner, W. Specific Lipid Requirements of Membrane Proteins—A Putative Bottleneck in Heterologous Expression. Biochim. Biophys. Acta Biomembr. 2003, 1610, 11–22. [Google Scholar] [CrossRef]
- Barrera, F.N.; Renart, M.L.; Molina, M.L.; Poveda, J.A.; Encinar, J.A.; Fernández, A.M.; Neira, J.L.; González-Ros, J.M. Unfolding and Refolding in Vitro of a Tetrameric, Alpha-Helical Membrane Protein: The Prokaryotic Potassium Channel KcsA. Biochemistry 2005, 44, 14344–14352. [Google Scholar] [CrossRef]
- Barrera, F.N.; Renart, M.L.; Poveda, J.A.; De Kruijff, B.; Killian, J.A.; González-Ros, J.M. Protein Self-Assembly and Lipid Binding in the Folding of the Potassium Channel KcsA. Biochemistry 2008, 47, 2123–2133. [Google Scholar] [CrossRef]
- Renart, M.L.; Giudici, A.M.; Díaz-García, C.; Molina, M.L.; Morales, A.; González-Ros, J.M.; Poveda, J.A. Modulation of Function, Structure and Clustering of K+ Channels by Lipids: Lessons Learnt from KcsA. Int. J. Mol. Sci. 2020, 21, 2554. [Google Scholar] [CrossRef] [PubMed]
- Marius, P.; Zagnoni, M.; Sandison, M.E.; Malcolm East, J.; Morgan, H.; Lee, A.G. Binding of Anionic Lipids to at Least Three Nonannular Sites on the Potassium Channel KcsA Is Required for Channel Opening. Biophys. J. 2008, 94, 1689–1698. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Oiki, S. Amphipathic Antenna of an Inward Rectifier K+ Channel Responds to Changes in the Inner Membrane Leaflet. Proc. Natl. Acad. Sci. USA 2013, 110, 749–754. [Google Scholar] [CrossRef]
- Cosseddu, S.M.; Choe, E.J.; Khovanov, I.A. Unraveling of a Strongly Correlated Dynamical Network of Residues Controlling the Permeation of Potassium in KcsA Ion Channel. Entropy 2021, 23, 72. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.L.; Encinar, J.A.; Barrera, F.N.; Fernandez-Ballester, G.; Riquelme, G.; Gonzalez-Ros, J.M. Influence of C-Terminal Protein Domains and Protein-Lipid Interactions on Tetramerization and Stability of the Potassium Channel KcsA. Biochemistry 2004, 43, 14924–14931. [Google Scholar] [CrossRef]
- Thompson, A.N.; Kim, I.; Panosian, T.D.; Iverson, T.M.; Allen, T.W.; Nimigean, C.M. Mechanism of Potassium-Channel Selectivity Revealed by Na(+) and Li(+) Binding Sites within the KcsA Pore. Nat. Struct. Mol. Biol. 2009, 16, 1317–1324. [Google Scholar] [CrossRef]
- Giudici, A.M.; Renart, M.L.; Díaz-García, C.; Morales, A.; Poveda, J.A.; González-Ros, J.M. Accessibility of Cations to the Selectivity Filter of KcsA in the Inactivated State: An Equilibrium Binding Study. Int. J. Mol. Sci. 2019, 20, 689. [Google Scholar] [CrossRef]
- Rasmusson, R.L.; Morales, M.J.; Wang, S.; Liu, S.; Campbell, D.L.; Brahmajothi, M.V.; Strauss, H.C. Inactivation of Voltage-Gated Cardiac K+ Channels. Circ. Res. 1998, 82, 739–750. [Google Scholar] [CrossRef]
- Swenson, R.P.; Armstrong, C.M. K+channels Close More Slowly in the Presence of External K+and Rb+. Nature 1981, 291, 427–429. [Google Scholar] [CrossRef]
- Santos, J.S.; Syeda, R.; Montal, M. Stabilization of the Conductive Conformation of a Voltage-Gated K+ (Kv) Channel: The Lid Mechanism. J. Biol. Chem. 2013, 288, 16619–16628. [Google Scholar] [CrossRef]
- Matulef, K.; Komarov, A.G.; Costantino, C.A.; Valiyaveetil, F.I. Using Protein Backbone Mutagenesis to Dissect the Link between Ion Occupancy and C-Type Inactivation in K+ Channels. Proc. Natl. Acad. Sci. USA 2013, 110, 17886–17891. [Google Scholar] [CrossRef] [PubMed]
- Cuello, L.G.; Jogini, V.; Cortes, D.M.; Perozo, E. Structural Mechanism of C-Type Inactivation in K(+) Channels. Nature 2010, 466, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Matulef, K.; Annen, A.W.; Nix, J.C.; Valiyaveetil, F.I. Individual Ion Binding Sites in the K(+) Channel Play Distinct Roles in C-Type Inactivation and in Recovery from Inactivation. Structure 2016, 24, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Kiya, T.; Takeshita, K.; Kawanabe, A.; Fujiwara, Y. Intermolecular Functional Coupling between Phosphoinositides and the Potassium Channel KcsA. J. Biol. Chem. 2022, 298, 102257. [Google Scholar] [CrossRef] [PubMed]
- Cuello, L.G.; Jogini, V.; Cortes, D.M.; Pan, A.C.; Gagnon, D.G.; Dalmas, O.; Cordero-Morales, J.F.; Chakrapani, S.; Roux, B.; Perozo, E. Structural Basis for the Coupling between Activation and Inactivation Gates in K+channels. Nature 2010, 466, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Focke, P.J.; Matulef, K.; Bian, X.; Moënne-Loccoz, P.; Valiyaveetil, F.I.; Lockless, S.W. Ion-Binding Properties of a K+ Channel Selectivity Filter in Different Conformations. Proc. Natl. Acad. Sci. USA 2015, 112, 15096–15100. [Google Scholar] [CrossRef]
- Devaraneni, P.K.; Komarov, A.G.; Costantino, C.A.; Devereaux, J.J.; Matulef, K.; Valiyaveetil, F.I. Semisynthetic K+ Channels Show That the Constricted Conformation of the Selectivity Filter Is Not the C-Type Inactivated State. Proc. Natl. Acad. Sci. USA 2013, 110, 15698–15703. [Google Scholar] [CrossRef]
- van der Cruijsen, E.A.W.; Prokofyev, A.V.; Pongs, O.; Baldus, M. Probing Conformational Changes during the Gating Cycle of a Potassium Channel in Lipid Bilayers. Biophys. J. 2017, 112, 99–108. [Google Scholar] [CrossRef]
- Xu, Y.; Bhate, M.P.; McDermott, A.E. Transmembrane Allosteric Energetics Characterization for Strong Coupling between Proton and Potassium Ion Binding in the KcsA Channel. Proc. Natl. Acad. Sci. USA 2017, 114, 8788–8793. [Google Scholar] [CrossRef]
- Baukrowitz, T.; Yellen, G. Modulation of K+current by Frequency and External [K+]: A Tale of Two Inactivation Mechanisms. Neuron 1995, 15, 951–960. [Google Scholar] [CrossRef]
- López-Barneo, J.; Hoshi, T.; Heinemann, S.H.; Aldrich, R.W. Effects of External Cations and Mutations in the Pore Region on C-Type Inactivation of Shaker Potassium Channels. Recept. Channels 1993, 1, 61–71. [Google Scholar]
- Chakrapani, S.; Cordero-Morales, J.F.; Perozo, E. A Quantitative Description of KcsA Gating I: Macroscopic Currents. J. Gen. Physiol. 2007, 130, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, T.; Zagotta, W.N.; Aldrich, R.W. Biophysical and Molecular Mechanisms of Shaker Potassium Channel Inactivation. Science 1990, 250, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Fedida, D.; Maruoka, N.D.; Lin, S. Modulation of Slow Inactivation in Human Cardiac Kv1.5 Channels by Extra- and Intracellular Permeant Cations. J. Physiol. 1999, 515 Pt 2, 315–329. [Google Scholar] [CrossRef]
- Rasmusson, R.L.; Morales, M.J.; Castellino, R.C.; Zhang, Y.; Campbell, D.L.; Strauss, H.C. C-Type Inactivation Controls Recovery in a Fast Inactivating Cardiac K+ Channel (Kv1.4) Expressed in Xenopus Oocytes. J. Physiol. 1995, 489 Pt 3, 709–721. [Google Scholar] [CrossRef]
- Lee, A.G. Biological Membranes: The Importance of Molecular Detail. Trends Biochem. Sci. 2011, 36, 493–500. [Google Scholar] [CrossRef]
- Yeagle, P.L. Non-Covalent Binding of Membrane Lipids to Membrane Proteins. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1548–1559. [Google Scholar] [CrossRef]

| pH | Cation | Number of Binding Events Detected | DDM | POPC DLMs | POPA DLMs |
|---|---|---|---|---|---|
| KD (mM) | KD (mM) | KD (mM) | |||
| 7.0 | Na+ | 1 | 3.3 ± 0.5 | 4.5 ± 0.6 | 4.6 ± 0.1 |
| K+ | 2 | (1.9 ± 0.5) × 10−3 | (1.4 ± 1.1) × 10−3 | (2.1 ± 0.2) × 10−3 | |
| 3.5 ± 0.5 | 0.36 ± 0.01 a | 0.17 ± 0.01 a | |||
| 4.0 | Na+ | 1 | 7.7 ± 0.6 | 5.1 ± 0.6 | 5.2 ± 0.1 |
| K+ | 2 | (91 ± 11) × 10−3 | (56 ± 11) × 10−3 | (62 ± 10) × 10−3 | |
| 8.4 ± 0.5 | 4.9 ± 0.5 | 0.26 ± 0.05 a |
| Channel | Micelles | pH 7.0 | pH 4.0 |
|---|---|---|---|
| KD (mM) | KD (mM) | ||
| W67 | DDM | 0.45 ± 0.09 | 46 ± 2 a |
| POPC DLMs | 1.0 ± 0.1 | 51 ± 3 a | |
| POPA DLMs | 0.5 ± 0.1 | 8 ± 2 a | |
| E71A W67 | DDM | 6.0 ± 0.7 | 4.1 ± 0.3 |
| R64A W67 | DDM | 4.9 ± 0.8 | 9.0 ± 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renart, M.L.; Giudici, A.M.; Coll-Díez, C.; González-Ros, J.M.; Poveda, J.A. Anionic Phospholipids Shift the Conformational Equilibrium of the Selectivity Filter in the KcsA Channel to the Conductive Conformation: Predicted Consequences on Inactivation. Biomedicines 2023, 11, 1376. https://doi.org/10.3390/biomedicines11051376
Renart ML, Giudici AM, Coll-Díez C, González-Ros JM, Poveda JA. Anionic Phospholipids Shift the Conformational Equilibrium of the Selectivity Filter in the KcsA Channel to the Conductive Conformation: Predicted Consequences on Inactivation. Biomedicines. 2023; 11(5):1376. https://doi.org/10.3390/biomedicines11051376
Chicago/Turabian StyleRenart, María Lourdes, Ana Marcela Giudici, Carlos Coll-Díez, José M. González-Ros, and José A. Poveda. 2023. "Anionic Phospholipids Shift the Conformational Equilibrium of the Selectivity Filter in the KcsA Channel to the Conductive Conformation: Predicted Consequences on Inactivation" Biomedicines 11, no. 5: 1376. https://doi.org/10.3390/biomedicines11051376
APA StyleRenart, M. L., Giudici, A. M., Coll-Díez, C., González-Ros, J. M., & Poveda, J. A. (2023). Anionic Phospholipids Shift the Conformational Equilibrium of the Selectivity Filter in the KcsA Channel to the Conductive Conformation: Predicted Consequences on Inactivation. Biomedicines, 11(5), 1376. https://doi.org/10.3390/biomedicines11051376









