Discovery of Biomarkers for Amyotrophic Lateral Sclerosis from Human Cerebrospinal Fluid Using Mass-Spectrometry-Based Proteomics
Abstract
1. Introduction
2. Methods
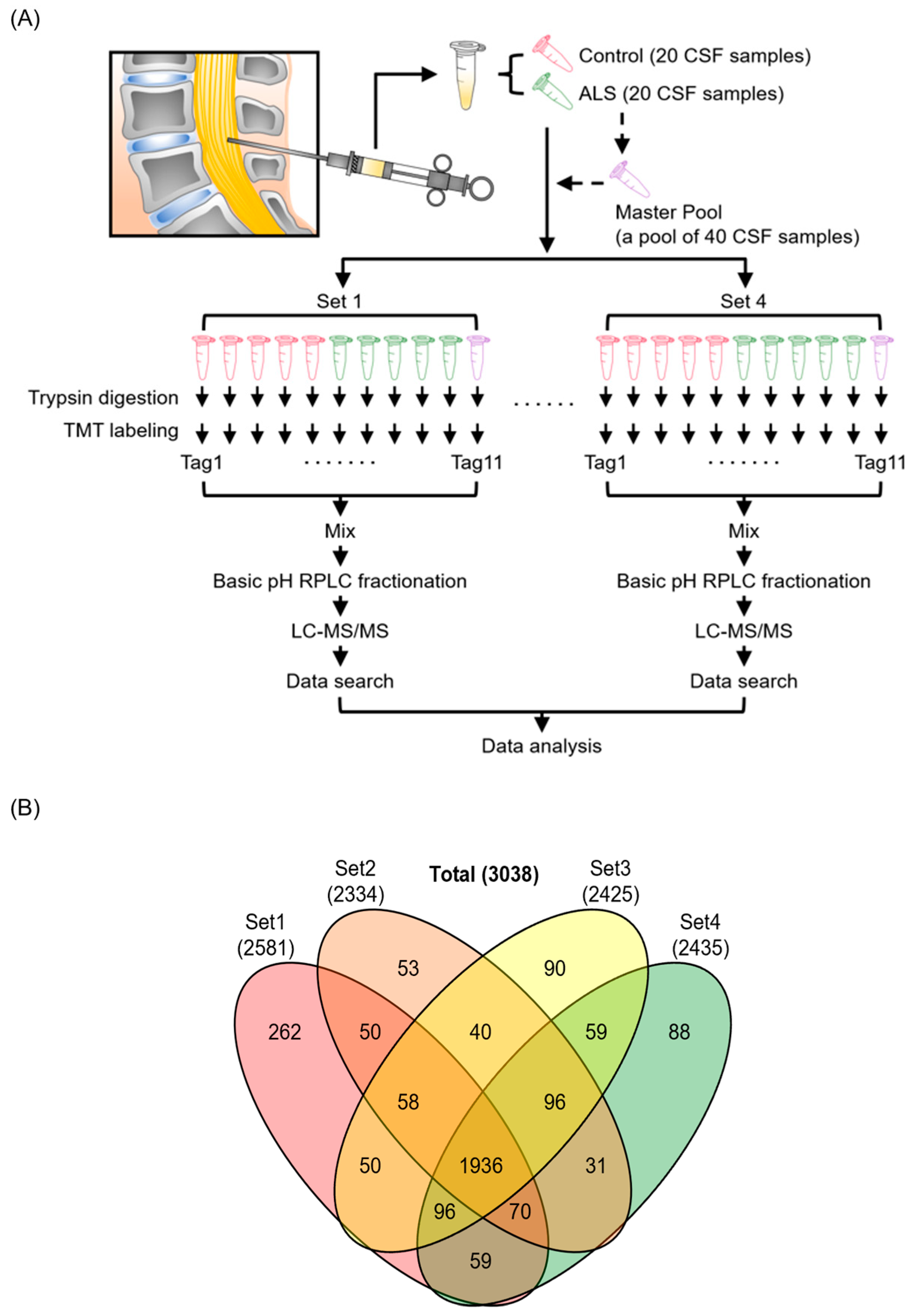
2.1. Collection of CSF Samples
2.2. Sample Preparation and Trypsin Digestion for Discovery Experiments
2.3. LC-MS/MS Analysis for the Discovery of Biomarker Candidates
2.4. Database Searches for Peptide and Protein Identification
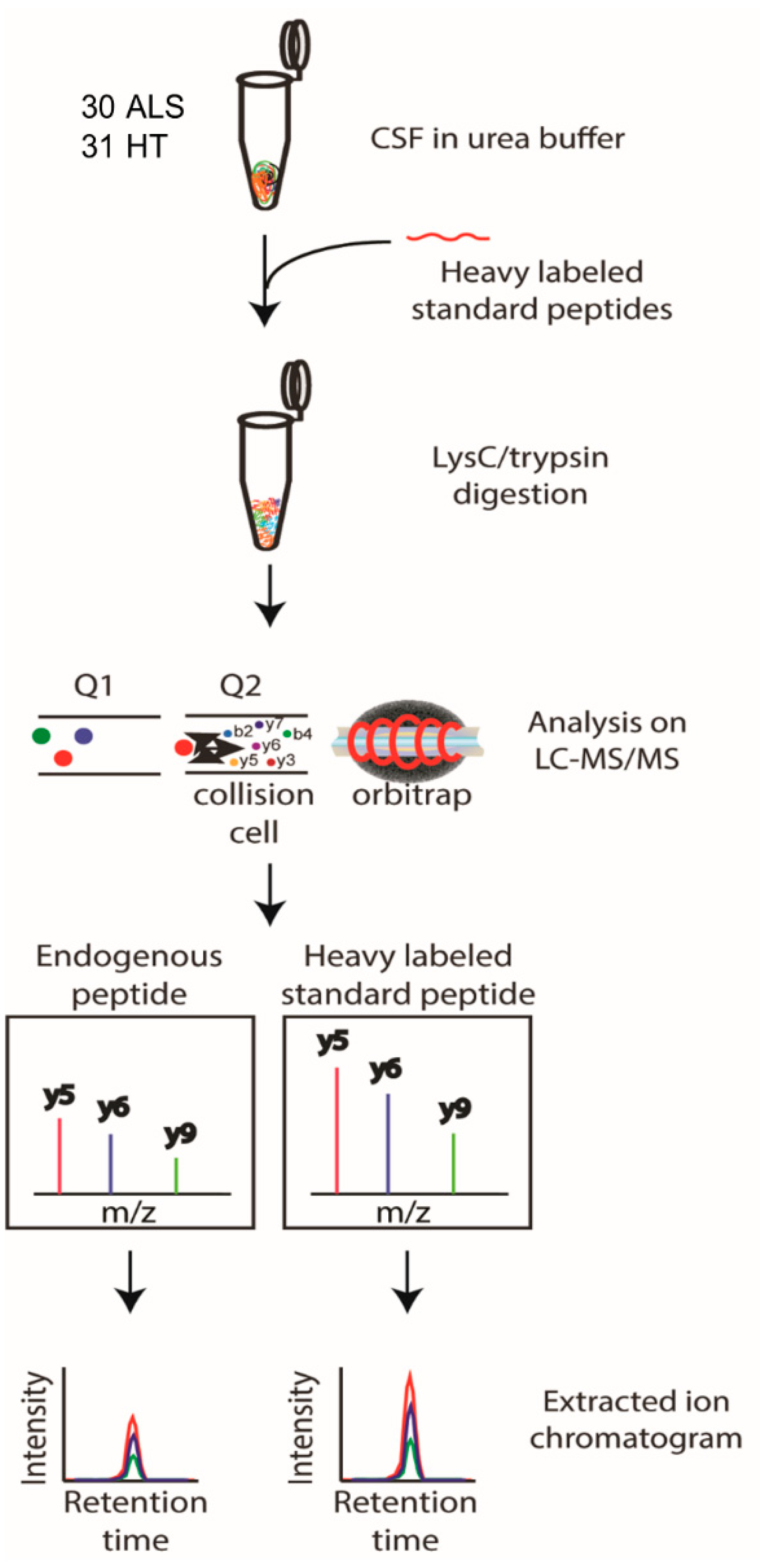
2.5. Preparation of CSF Samples for the PRM Experiments
2.6. LC-MS/MS Analysis for Validation Experiments Using the PRM Method
2.7. Experimental Design and Statistical Rationale
2.8. Disclosure of Previously Published Data
3. Results
3.1. Quantitative Proteome Analysis of CSF Samples
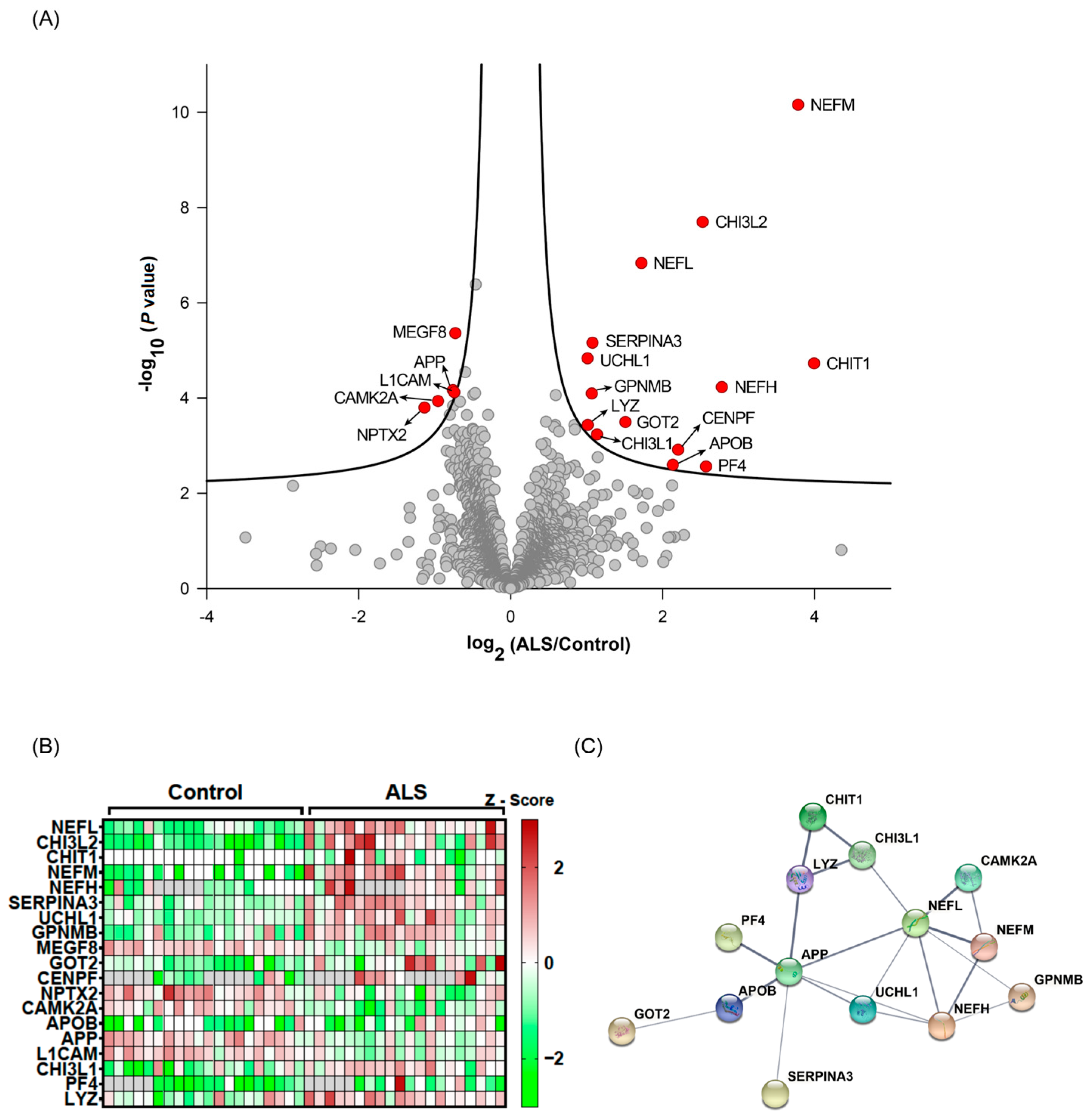
3.2. Selection of Candidate Biomarkers for ALS
3.3. Detectability of Candidate Biomarker Proteins in Targeted PRM Analysis
3.4. Response Curve Test of Candidate Biomarker Proteins in PRM Analysis
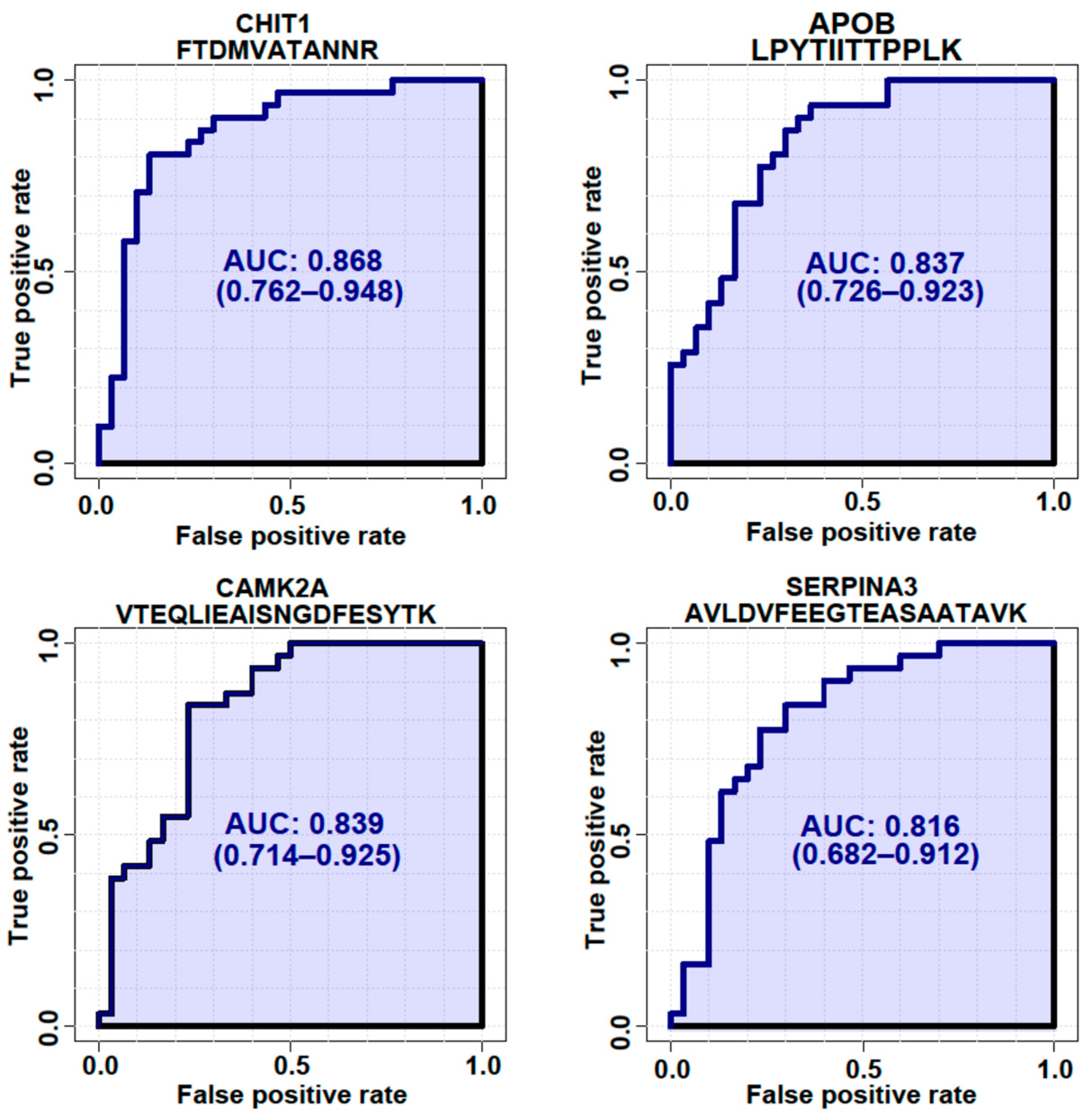
3.5. Targeted Quantification of Candidate ALS Biomarker Peptides in CSF from ALS and HC Individuals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Rowland, L.P.; Shneider, N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001, 344, 1688–1700. [Google Scholar] [CrossRef]
- Mitchell, J.D.; Borasio, G.D. Amyotrophic lateral sclerosis. Lancet 2007, 369, 2031–2041. [Google Scholar] [CrossRef] [PubMed]
- Wijesekera, L.C.; Leigh, P.N. Amyotrophic lateral sclerosis. Orphanet. J. Rare Dis. 2009, 4, 3. [Google Scholar] [CrossRef]
- Kiernan, M.C.; Vucic, S.; Cheah, B.C.; Turner, M.R.; Eisen, A.; Hardiman, O.; Burrell, J.R.; Zoing, M.C. Amyotrophic lateral sclerosis. Lancet 2011, 377, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.T.; Bowser, R. Fluid-Based Biomarkers for Amyotrophic Lateral Sclerosis. Neurotherapeutics 2017, 14, 119–134. [Google Scholar] [CrossRef]
- Collins, M.A.; An, J.; Hood, B.L.; Conrads, T.P.; Bowser, R.P. Label-Free LC-MS/MS Proteomic Analysis of Cerebrospinal Fluid Identifies Protein/Pathway Alterations and Candidate Biomarkers for Amyotrophic Lateral Sclerosis. J. Proteome Res. 2015, 14, 4486–4501. [Google Scholar] [CrossRef]
- Cruz, M.P. Edaravone (Radicava): A Novel Neuroprotective Agent for the Treatment of Amyotrophic Lateral Sclerosis. Pharm. Ther. 2018, 43, 25–28. [Google Scholar]
- Mehta, P.; Kaye, W.; Raymond, J.; Punjani, R.; Larson, T.; Cohen, J.; Muravov, O.; Horton, K. Prevalence of Amyotrophic Lateral Sclerosis—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.; Orrell, R.W. Pathogenesis of amyotrophic lateral sclerosis. Br. Med. Bull. 2016, 119, 87–98. [Google Scholar] [CrossRef]
- Goutman, S.A.; Hardiman, O.; Al-Chalabi, A.; Chio, A.; Savelieff, M.G.; Kiernan, M.C.; Feldman, E.L. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 2022, 21, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; Raymond, J.; Punjani, R.; Han, M.; Larson, T.; Kaye, W.; Nelson, L.M.; Topol, B.; Muravov, O.; Genson, C.; et al. Prevalence of amyotrophic lateral sclerosis in the United States using established and novel methodologies, 2017. Amyotroph. Lateral Scler. Frontotemporal. Degener. 2022, 24, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Balendra, R.; Isaacs, A.M. C9orf72-mediated ALS and FTD: Multiple pathways to disease. Nat. Rev. Neurol. 2018, 14, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Pansarasa, O.; Bordoni, M.; Diamanti, L.; Sproviero, D.; Gagliardi, S.; Cereda, C. SOD1 in Amyotrophic Lateral Sclerosis: “Ambivalent” Behavior Connected to the Disease. Int. J. Mol. Sci. 2018, 19, 1345. [Google Scholar] [CrossRef] [PubMed]
- Suk, T.R.; Rousseaux, M.W.C. The role of TDP-43 mislocalization in amyotrophic lateral sclerosis. Mol. Neurodegener. 2020, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Huang, E.J. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain. Res. 2016, 1647, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Mead, R.J.; Shan, N.; Reiser, H.J.; Marshall, F.; Shaw, P.J. Amyotrophic lateral sclerosis: A neurodegenerative disorder poised for successful therapeutic translation. Nat. Rev. Drug Discov. 2023, 22, 185–212. [Google Scholar] [CrossRef]
- Chang, E.; Ghosh, N.; Yanni, D.; Lee, S.; Alexandru, D.; Mozaffar, T. A Review of Spasticity Treatments: Pharmacological and Interventional Approaches. Crit. Rev. Phys. Rehabil. Med. 2013, 25, 11–22. [Google Scholar] [CrossRef]
- Barschke, P.; Oeckl, P.; Steinacker, P.; Ludolph, A.; Otto, M. Proteomic studies in the discovery of cerebrospinal fluid biomarkers for amyotrophic lateral sclerosis. Expert Rev. Proteom. 2017, 14, 769–777. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, X.H.; Wu, J.J.; Ren, H.M.; Wang, J.; Ding, Z.T.; Jiang, Y.P. Proteomic analysis of cerebrospinal fluid in amyotrophic lateral sclerosis. Exp. Ther. Med. 2016, 11, 2095–2106. [Google Scholar] [CrossRef]
- Thompson, A.G.; Gray, E.; Thezenas, M.L.; Charles, P.D.; Evetts, S.; Hu, M.T.; Talbot, K.; Fischer, R.; Kessler, B.M.; Turner, M.R. Cerebrospinal fluid macrophage biomarkers in amyotrophic lateral sclerosis. Ann. Neurol. 2018, 83, 258–268. [Google Scholar] [CrossRef]
- Kruger, T.; Lautenschlager, J.; Grosskreutz, J.; Rhode, H. Proteome analysis of body fluids for amyotrophic lateral sclerosis biomarker discovery. Proteom. Clin. Appl. 2013, 7, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Bowser, R.; Turner, M.R.; Shefner, J. Biomarkers in amyotrophic lateral sclerosis: Opportunities and limitations. Nat. Rev. Neurol. 2011, 7, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Oeckl, P.; Weydt, P.; Thal, D.R.; Weishaupt, J.H.; Ludolph, A.C.; Otto, M. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol. 2020, 139, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Gaur, N.; Perner, C.; Witte, O.W.; Grosskreutz, J. The Chitinases as Biomarkers for Amyotrophic Lateral Sclerosis: Signals From the CNS and Beyond. Front. Neurol. 2020, 11, 377. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, D.; Meneri, M.; Saccomanno, D.; Bresolin, N.; Comi, G.P.; Corti, S. Diagnostic and Prognostic Role of Blood and Cerebrospinal Fluid and Blood Neurofilaments in Amyotrophic Lateral Sclerosis: A Review of the Literature. Int. J. Mol. Sci. 2019, 20, 4152. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.M.; Sharma, A.; Mishra, P.; Vijayalakshmi, K.; Harsha, H.C.; Sathyaprabha, T.N.; Bharath, S.M.; Nalini, A.; Alladi, P.A.; Raju, T.R. Chitotriosidase—A putative biomarker for sporadic amyotrophic lateral sclerosis. Clin. Proteom. 2013, 10, 19. [Google Scholar] [CrossRef]
- Meeter, L.H.H.; Vijverberg, E.G.; Del Campo, M.; Rozemuller, A.J.M.; Donker Kaat, L.; de Jong, F.J.; van der Flier, W.M.; Teunissen, C.E.; van Swieten, J.C.; Pijnenburg, Y.A.L. Clinical value of neurofilament and phospho-tau/tau ratio in the frontotemporal dementia spectrum. Neurology 2018, 90, e1231–e1239. [Google Scholar] [CrossRef]
- Smaczniak, C.; Li, N.; Boeren, S.; America, T.; van Dongen, W.; Goerdayal, S.S.; de Vries, S.; Angenent, G.C.; Kaufmann, K. Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protoc. 2012, 7, 2144–2158. [Google Scholar] [CrossRef]
- Scherling, C.S.; Hall, T.; Berisha, F.; Klepac, K.; Karydas, A.; Coppola, G.; Kramer, J.H.; Rabinovici, G.; Ahlijanian, M.; Miller, B.L.; et al. Cerebrospinal fluid neurofilament concentration reflects disease severity in frontotemporal degeneration. Ann. Neurol. 2014, 75, 116–126. [Google Scholar] [CrossRef]
- Jang, Y.; Pletnikova, O.; Troncoso, J.C.; Pantelyat, A.Y.; Dawson, T.M.; Rosenthal, L.S.; Na, C.H. Mass Spectrometry-Based Proteomics Analysis of Human Substantia Nigra From Parkinson’s Disease Patients Identifies Multiple Pathways Potentially Involved in the Disease. Mol. Cell Proteom. 2023, 22, 100452. [Google Scholar] [CrossRef]
- Jang, Y.; Thuraisamy, T.; Redding-Ochoa, J.; Pletnikova, O.; Troncoso, J.C.; Rosenthal, L.S.; Dawson, T.M.; Pantelyat, A.Y.; Na, C.H. Mass spectrometry-based proteomics analysis of human globus pallidus from progressive supranuclear palsy patients discovers multiple disease pathways. Clin. Transl. Med. 2022, 12, e1076. [Google Scholar] [CrossRef]
- Wang, H.; Dey, K.K.; Chen, P.C.; Li, Y.; Niu, M.; Cho, J.H.; Wang, X.; Bai, B.; Jiao, Y.; Chepyala, S.R.; et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 43. [Google Scholar] [CrossRef]
- Yang, G.; Xu, Z.; Lu, W.; Li, X.; Sun, C.; Guo, J.; Xue, P.; Guan, F. Quantitative Analysis of Differential Proteome Expression in Bladder Cancer vs. Normal Bladder Cells Using SILAC Method. PLoS ONE 2015, 10, e0134727. [Google Scholar] [CrossRef] [PubMed]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
- Westerhaus, A.; Joseph, T.; Meyers, A.J.; Jang, Y.; Na, C.H.; Cave, C.; Sockanathan, S. The distribution and function of GDE2, a regulator of spinal motor neuron survival, are disrupted in Amyotrophic Lateral Sclerosis. Acta Neuropathol. Commun. 2022, 10, 73. [Google Scholar] [CrossRef]
- Gaiani, A.; Martinelli, I.; Bello, L.; Querin, G.; Puthenparampil, M.; Ruggero, S.; Toffanin, E.; Cagnin, A.; Briani, C.; Pegoraro, E.; et al. Diagnostic and Prognostic Biomarkers in Amyotrophic Lateral Sclerosis: Neurofilament Light Chain Levels in Definite Subtypes of Disease. JAMA Neurol. 2017, 74, 525–532. [Google Scholar] [CrossRef]
- Mariosa, D.; Hammar, N.; Malmstrom, H.; Ingre, C.; Jungner, I.; Ye, W.; Fang, F.; Walldius, G. Blood biomarkers of carbohydrate, lipid, and apolipoprotein metabolisms and risk of amyotrophic lateral sclerosis: A more than 20-year follow-up of the Swedish AMORIS cohort. Ann. Neurol. 2017, 81, 718–728. [Google Scholar] [CrossRef]
- Barschke, P.; Oeckl, P.; Steinacker, P.; Al Shweiki, M.R.; Weishaupt, J.H.; Landwehrmeyer, G.B.; Anderl-Straub, S.; Weydt, P.; Diehl-Schmid, J.; Danek, A.; et al. Different CSF protein profiles in amyotrophic lateral sclerosis and frontotemporal dementia with C9orf72 hexanucleotide repeat expansion. J. Neurol. Neurosurg. Psychiatry 2020, 91, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Raghunathan, R.; Turajane, K.; Wong, L.C. Biomarkers in Neurodegenerative Diseases: Proteomics Spotlight on ALS and Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 9299. [Google Scholar] [CrossRef]
- Yuan, A.; Nixon, R.A. Specialized roles of neurofilament proteins in synapses: Relevance to neuropsychiatric disorders. Brain Res. Bull. 2016, 126, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Day, I.N.; Thompson, R.J. UCHL1 (PGP 9.5): Neuronal biomarker and ubiquitin system protein. Prog. Neurobiol. 2010, 90, 327–362. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wuolikainen, A.; Wu, J.; Ohman, A.; Wingsle, G.; Moritz, T.; Andersen, P.M.; Forsgren, L.; Trupp, M. Targeted Multiple Reaction Monitoring Analysis of CSF Identifies UCHL1 and GPNMB as Candidate Biomarkers for ALS. J. Mol. Neurosci. 2019, 69, 643–657. [Google Scholar] [CrossRef]
- Gille, B.; De Schaepdryver, M.; Dedeene, L.; Goossens, J.; Claeys, K.G.; Van Den Bosch, L.; Tournoy, J.; Van Damme, P.; Poesen, K. Inflammatory markers in cerebrospinal fluid: Independent prognostic biomarkers in amyotrophic lateral sclerosis? J. Neurol. Neurosurg. Psychiatry 2019, 90, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.G.; Gray, E.; Bampton, A.; Raciborska, D.; Talbot, K.; Turner, M.R. CSF chitinase proteins in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2019, 90, 1215–1220. [Google Scholar] [CrossRef]
- Steinacker, P.; Verde, F.; Fang, L.; Feneberg, E.; Oeckl, P.; Roeber, S.; Anderl-Straub, S.; Danek, A.; Diehl-Schmid, J.; Fassbender, K.; et al. Chitotriosidase (CHIT1) is increased in microglia and macrophages in spinal cord of amyotrophic lateral sclerosis and cerebrospinal fluid levels correlate with disease severity and progression. J. Neurol. Neurosurg. Psychiatry 2018, 89, 239–247. [Google Scholar] [CrossRef]
- Picard, C.; Nilsson, N.; Labonte, A.; Auld, D.; Rosa-Neto, P.; Alzheimer’s Disease Neuroimaging, I.; Ashton, N.J.; Zetterberg, H.; Blennow, K.; Breitner, J.C.B.; et al. Apolipoprotein B is a novel marker for early tau pathology in Alzheimer’s disease. Alzheimers Dement. 2022, 18, 875–887. [Google Scholar] [CrossRef]
- Nguyen, K.V. beta-Amyloid precursor protein (APP) and the human diseases. AIMS Neurosci. 2019, 6, 273–281. [Google Scholar] [CrossRef]
- Calingasan, N.Y.; Chen, J.; Kiaei, M.; Beal, M.F. Beta-amyloid 42 accumulation in the lumbar spinal cord motor neurons of amyotrophic lateral sclerosis patients. Neurobiol. Dis. 2005, 19, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Liu, F.; Lin, J.; Li, Y.L.; Fang, Z.H.; Zhou, C.; Li, C.J.; Shen, J.F. Activation of the N-methyl-D-aspartate receptor contributes to orofacial neuropathic and inflammatory allodynia by facilitating calcium-calmodulin-dependent protein kinase II phosphorylation in mice. Brain Res. Bull. 2022, 185, 174–192. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Giese, K.P. Calcium/calmodulin-dependent kinase II and Alzheimer’s disease. Mol. Brain 2015, 8, 78. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, Y.; Chen, F.; Tong, H.; Reddy, M.V.; Luo, L.; Seshadrinathan, S.; Zhang, L.; Holthauzen, L.M.; Craig, A.M.; et al. Calsyntenin-3 molecular architecture and interaction with neurexin 1alpha. J. Biol. Chem. 2014, 289, 34530–34542. [Google Scholar] [CrossRef]
- Paladini, F.; Fiorillo, M.T.; Tedeschi, V.; Mattorre, B.; Sorrentino, R. The Multifaceted Nature of Aminopeptidases ERAP1, ERAP2, and LNPEP: From Evolution to Disease. Front. Immunol. 2020, 11, 1576. [Google Scholar] [CrossRef]
- Ziff, O.J.; Taha, D.M.; Crerar, H.; Clarke, B.E.; Chakrabarti, A.M.; Kelly, G.; Neeves, J.; Tyzack, G.E.; Luscombe, N.M.; Patani, R. Reactive astrocytes in ALS display diminished intron retention. Nucleic Acids Res. 2021, 49, 3168–3184. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Fujikawa, A.; Komatsu, Y.; Kuboyama, K.; Tanga, N.; Noda, M. Enhanced extinction of aversive memories in mice lacking SPARC-related protein containing immunoglobulin domains 1 (SPIG1/FSTL4). Neurobiol. Learn Mem. 2018, 152, 61–70. [Google Scholar] [CrossRef]
- Bhown, A.S.; Mole, J.E.; Bennett, J.C. Primary structure of human J chain: Isolation and characterization of tryptic and chymotryptic peptides of human J chain. Biochemistry 1977, 16, 3501–3507. [Google Scholar] [CrossRef]
- van der Lienden, M.J.C.; Gaspar, P.; Boot, R.; Aerts, J.; van Eijk, M. Glycoprotein Non-Metastatic Protein B: An Emerging Biomarker for Lysosomal Dysfunction in Macrophages. Int. J. Mol. Sci. 2018, 20, 66. [Google Scholar] [CrossRef]
- Genc, B.; Jara, J.H.; Schultz, M.C.; Manuel, M.; Stanford, M.J.; Gautam, M.; Klessner, J.L.; Sekerkova, G.; Heller, D.B.; Cox, G.A.; et al. Absence of UCHL 1 function leads to selective motor neuropathy. Ann. Clin. Transl. Neurol. 2016, 3, 331–345. [Google Scholar] [CrossRef]
- Dahme, M.; Bartsch, U.; Martini, R.; Anliker, B.; Schachner, M.; Mantei, N. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 1997, 17, 346–349. [Google Scholar] [CrossRef]
- Linneberg, C.; Toft, C.L.F.; Kjaer-Sorensen, K.; Laursen, L.S. L1cam-mediated developmental processes of the nervous system are differentially regulated by proteolytic processing. Sci. Rep. 2019, 9, 3716. [Google Scholar] [CrossRef] [PubMed]
- Chapman, G.; Shanmugalingam, U.; Smith, P.D. The Role of Neuronal Pentraxin 2 (NP2) in Regulating Glutamatergic Signaling and Neuropathology. Front. Cell Neurosci. 2019, 13, 575. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.F.; Xu, D.; Craig, M.T.; Pelkey, K.A.; Chien, C.C.; Shi, Y.; Zhang, J.; Resnick, S.; Pletnikova, O.; Salmon, D.; et al. NPTX2 and cognitive dysfunction in Alzheimer’s Disease. eLife 2017, 6, e23798. [Google Scholar] [CrossRef] [PubMed]
- Ebbert, M.T.W.; Ross, C.A.; Pregent, L.J.; Lank, R.J.; Zhang, C.; Katzman, R.B.; Jansen-West, K.; Song, Y.; da Rocha, E.L.; Palmucci, C.; et al. Conserved DNA methylation combined with differential frontal cortex and cerebellar expression distinguishes C9orf72-associated and sporadic ALS, and implicates SERPINA1 in disease. Acta Neuropathol. 2017, 134, 715–728. [Google Scholar] [CrossRef]
- Gollin, P.A.; Kalaria, R.N.; Eikelenboom, P.; Rozemuller, A.; Perry, G. Alpha 1-antitrypsin and alpha 1-antichymotrypsin are in the lesions of Alzheimer’s disease. Neuroreport 1992, 3, 201–203. [Google Scholar] [CrossRef]
- Abu-Rumeileh, S.; Halbgebauer, S.; Steinacker, P.; Anderl-Straub, S.; Polischi, B.; Ludolph, A.C.; Capellari, S.; Parchi, P.; Otto, M. CSF SerpinA1 in Creutzfeldt-Jakob disease and frontotemporal lobar degeneration. Ann. Clin. Transl. Neurol. 2020, 7, 191–199. [Google Scholar] [CrossRef]
- Zattoni, M.; Mearelli, M.; Vanni, S.; Colini Baldeschi, A.; Tran, T.H.; Ferracin, C.; Catania, M.; Moda, F.; Di Fede, G.; Giaccone, G.; et al. Serpin Signatures in Prion and Alzheimer’s Diseases. Mol. Neurobiol. 2022, 59, 3778–3799. [Google Scholar] [CrossRef]
- Fissolo, N.; Matute-Blanch, C.; Osman, M.; Costa, C.; Pinteac, R.; Miro, B.; Sanchez, A.; Brito, V.; Dujmovic, I.; Voortman, M.; et al. CSF SERPINA3 Levels Are Elevated in Patients With Progressive MS. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e941. [Google Scholar] [CrossRef]
- Vizcaino, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef]



| Subject ID | Sample Type | Group | Gender | Race | Age | Application of the Samples in This Study |
|---|---|---|---|---|---|---|
| ALS01 | CSF | ALSFRS 40 | Male | White | 62 | Discovery and Validation |
| ALS02 | CSF | ALSFRS 36 | Female | White | 68 | Discovery and Validation |
| ALS03 | CSF | ALSFRS 44 | Male | White | 54 | Discovery and Validation |
| ALS04 | CSF | ALSFRS 33 | Female | White | 55 | Discovery and Validation |
| ALS05 | CSF | ALSFRS 37 | Female | White | 44 | Discovery and Validation |
| ALS06 | CSF | ALSFRS 42 | Female | White | 51 | Discovery and Validation |
| ALS07 | CSF | ALSFRS 42 | Male | White | 64 | Discovery and Validation |
| ALS08 | CSF | ALSFRS 33 | Female | White | 52 | Discovery and Validation |
| ALS09 | CSF | ALSFRS 35 | Male | White | 68 | Discovery and Validation |
| ALS10 | CSF | ALSFRS 34 | Male | White | 55 | Discovery and Validation |
| ALS11 | CSF | ALSFRS 34 | Male | White | 61 | Discovery and Validation |
| ALS12 | CSF | ALSFRS 35 | Male | White | 79 | Discovery and Validation |
| ALS13 | CSF | ALSFRS 42 | Male | White | 63 | Discovery and Validation |
| ALS14 | CSF | ALSFRS 40 | Male | White | 59 | Discovery and Validation |
| ALS15 | CSF | ALSFRS 44 | Female | White | 66 | Discovery and Validation |
| ALS16 | CSF | ALSFRS 43 | Male | White | 73 | Discovery and Validation |
| ALS17 | CSF | ALSFRS 41 | Male | White | 55 | Discovery and Validation |
| ALS18 | CSF | ALSFRS 48 | Male | White | 65 | Discovery and Validation |
| ALS19 | CSF | ALSFRS 46 | Male | White | 40 | Discovery and Validation |
| ALS20 | CSF | ALSFRS 34 | Female | White | 53 | Discovery and Validation |
| ALS21 | CSF | No score | Male | White | 64 | Validation |
| ALS22 | CSF | ALSFRS 32 | Male | White | 58 | Validation |
| ALS23 | CSF | ALSFRS 42 | Male | White | 45 | Validation |
| ALS24 | CSF | ALSFRS 40 | Female | White | 41 | Validation |
| ALS25 | CSF | ALSFRS 47 | Male | White | 44 | Validation |
| ALS26 | CSF | ALSFRS 41 | Male | White | 51 | Validation |
| ALS27 | CSF | ALSFRS 40 | Male | White | 68 | Validation |
| ALS28 | CSF | ALSFRS 27 | Female | White | 44 | Validation |
| ALS29 | CSF | ALSFRS 48 | Male | White | 57 | Validation |
| ALS30 | CSF | ALSFRS 15 | Female | White | 63 | Validation |
| HC01 | CSF | Healthy Control | Male | White | 38 | Discovery and Validation |
| HC02 | CSF | Healthy Control | Male | White | 58 | Discovery and Validation |
| HC03 | CSF | Healthy Control | Female | White | 23 | Discovery and Validation |
| HC04 | CSF | Healthy Control | Female | White | 58 | Discovery and Validation |
| HC05 | CSF | Healthy Control | Female | Asian | 43 | Discovery and Validation |
| HC06 | CSF | Healthy Control | Male | White | 35 | Discovery and Validation |
| HC07 | CSF | Healthy Control | Male | White | 57 | Discovery and Validation |
| HC08 | CSF | Healthy Control | Female | White | 46 | Discovery and Validation |
| HC09 | CSF | Healthy Control | Male | White | 76 | Discovery and Validation |
| HC10 | CSF | Healthy Control | Female | White | 44 | Discovery and Validation |
| HC11 | CSF | Healthy Control | Female | White | 47 | Discovery and Validation |
| HC12 | CSF | Healthy Control | Male | White | 70 | Discovery and Validation |
| HC13 | CSF | Healthy Control | Female | White | 62 | Discovery and Validation |
| HC14 | CSF | Healthy Control | Male | White | 74 | Discovery and Validation |
| HC15 | CSF | Healthy Control | Male | White | 66 | Discovery and Validation |
| HC16 | CSF | Healthy Control | Male | White | 72 | Discovery and Validation |
| HC17 | CSF | Healthy Control | Female | White | 62 | Discovery and Validation |
| HC18 | CSF | Healthy Control | Female | White | 66 | Discovery and Validation |
| HC19 | CSF | Healthy Control | Male | White | 69 | Discovery and Validation |
| HC20 | CSF | Healthy Control | Female | White | 55 | Discovery and Validation |
| HC21 | CSF | Healthy Control | Female | White | 61 | Validation |
| HC22 | CSF | Healthy Control | Male | White | 66 | Validation |
| HC23 | CSF | Healthy Control | Male | White | 67 | Validation |
| HC24 | CSF | Healthy Control | Female | White | 64 | Validation |
| HC25 | CSF | Healthy Control | Female | ND | 59 | Validation |
| HC26 | CSF | Healthy Control | Male | White | 65 | Validation |
| HC27 | CSF | Healthy Control | Male | White | 66 | Validation |
| HC28 | CSF | Healthy Control | Female | White | 58 | Validation |
| HC29 | CSF | Healthy Control | Male | White | 54 | Validation |
| HC30 | CSF | Healthy Control | Female | White | 59 | Validation |
| HC31 | CSF | Healthy Control | Female | ND | 60 | Validation |
| Name | Gene Symbol | p Value | q-Value | Log2 (ALS/HC) | Detectability by PRM | q-Value |
|---|---|---|---|---|---|---|
| Chitinase-3-like protein 2 | CHI3L2 | 2.01 × 10−8 | 0 | 2.527697 | <0.05 | |
| Chitotriosidase-1 | CHIT1 | 1.88 × 10−5 | 0 | 3.99428 | Detectable | |
| Neurofilament light polypeptide | NEFL | 1.47 × 10−7 | 0 | 1.722488 | ||
| Neurofilament medium polypeptide | NEFM | 6.97 × 10−11 | 0 | 3.784436 | ||
| Neurofilament heavy polypeptide | NEFH | 5.91 × 10−5 | 0.001333 | 2.779995 | ||
| Alpha-1-antichymotrypsin | SERPINA3 | 6.94 × 10−6 | 0.008571 | 1.077944 | Detectable | |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | 1.48 × 10−5 | 0.015 | 1.012955 | Detectable | |
| Transmembrane glycoprotein NMB | GPNMB | 8.06 × 10−5 | 0.0244 | 1.069444 | Detectable | |
| Multiple epidermal-growth-factor-like domains protein 8 | MEGF8 | 4.36 × 10−6 | 0.026182 | −0.72796 | Detectable | |
| Aspartate aminotransferase, mitochondrial | GOT2 | 0.00032 | 0.026222 | 1.511867 | ||
| Centromere protein F | CENPF | 0.001221 | 0.028 | 2.205369 | ||
| Neuronal pentraxin-2 | NPTX2 | 0.00016 | 0.028667 | −1.13379 | Detectable | |
| Calcium/calmodulin-dependent protein kinase type II subunit alpha | CAMK2A | 0.000116 | 0.033143 | −0.95518 | Detectable | |
| Apolipoprotein B-100 | APOB | 0.002532 | 0.041053 | 2.13572 | Detectable | |
| Amyloid-beta A4 protein | APP | 6.85 × 10−5 | 0.042444 | −0.75982 | Detectable | |
| Neural cell adhesion molecule L1 | L1CAM | 7.59 × 10−5 | 0.043 | −0.74116 | Detectable | |
| Chitinase-3-like protein 1 | CHI3L1 | 0.000583 | 0.044 | 1.137858 | Detectable | |
| Platelet factor 4 | PF4 | 0.002747 | 0.044533 | 2.573066 | ||
| Lysozyme C | LYZ | 0.000368 | 0.0465 | 1.015916 | ||
| Calsyntenin-1 | CLSTN1 | 0.000146 | 0.051429 | −0.75724 | Detectable | <0.1 |
| Contactin-associated protein-like 2 | CNTNAP2 | 0.000225 | 0.052182 | −0.79877 | Detectable | |
| Macrophage-capping protein | CAPG | 0.00037 | 0.056522 | 0.840849 | ||
| Triosephosphate isomerase | TPI1 | 2.87 × 10−5 | 0.05712 | −0.59761 | Detectable | |
| Microfibril-associated glycoprotein 4 | MFAP4 | 0.00012 | 0.058429 | −0.66703 | Detectable | |
| Endoplasmic reticulum aminopeptidase 2 | ERAP2 | 0.001754 | 0.058897 | 1.147899 | Detectable | |
| Gamma-crystallin D | CRYGD | 0.007001 | 0.058963 | −2.86543 | ||
| Syndecan binding protein (Syntenin), isoform CRA_a | SDCBP | 0.000461 | 0.059 | 0.846597 | ||
| Follistatin-related protein 4 | FSTL4 | 0.000542 | 0.059333 | −0.82485 | Detectable | |
| Transport and Golgi organization protein 1 homolog | MIA3 | 4.14 × 10−7 | 0.061231 | −0.45853 | ||
| Neurocan core protein | NCAN | 0.000476 | 0.070545 | −0.73593 | Detectable | |
| Platelet basic protein | PPBP | 0.006893 | 0.072625 | 2.127046 | Detectable | |
| Leukotriene A-4 hydrolase | LTA4H | 8.72 × 10−5 | 0.073677 | 0.593814 | Detectable | |
| Chondroitin sulfate proteoglycan 5 | CSPG5 | 0.001286 | 0.083412 | −0.84017 | Detectable | |
| Band 4.1-like protein 3 | EPB41L3 | 0.004408 | 0.092686 | 1.101059 | ||
| Immunoglobulin heavy variable 2-5 | IGHV2-5 | 0.010187 | 0.096522 | 1.802516 | ||
| Plectin | PLEC | 0.00037 | 0.096681 | 0.586559 | ||
| Myosin light chain kinase, smooth muscle | MYLK | 0.000406 | 0.09688 | 0.572703 | ||
| Calsyntenin-3 | CLSTN3 | 8.53 × 10−5 | 0.097255 | −0.49128 | Detectable | |
| Serine/threonine-protein phosphatase 2A activator | PTPA | 0.000505 | 0.0978 | 0.637236 | ||
| UMP-CMP kinase | CMPK1 | 0.002623 | 0.097956 | 0.843523 | ||
| Neuronal pentraxin-1 | NPTX1 | 0.002511 | 0.098222 | −0.79414 | Detectable | |
| Alpha-1-antitrypsin | SERPINA1 | 0.002667 | 0.098449 | 0.842153 | Detectable | |
| Leucine-rich repeat and immunoglobulin-like domain-containing nogo receptor-interacting protein 1 | LINGO1 | 0.000955 | 0.098462 | −0.65034 | Detectable | |
| Immunoglobulin lambda variable 3-19 | IGLV3-19 | 0.006846 | 0.098537 | 1.408966 | Detectable | |
| Immunoglobulin lambda variable 3-27 | IGLV3-27 | 0.003614 | 0.098566 | 0.886375 | ||
| Alpha-2,8-sialyltransferase 8E | ST8SIA5 | 0.000228 | 0.098974 | −0.5754 | Detectable | |
| Butyrophilin subfamily 3 member A2 | BTN3A2 | 0.005124 | 0.099 | 1.036734 | ||
| Reticulon-4 receptor-like 2 | RTN4RL2 | 0.00064 | 0.099111 | −0.67662 | Detectable | |
| Protein kinase C-binding protein NELL2 | NELL2 | 0.001377 | 0.099135 | −0.78332 | Detectable | |
| Immunoglobulin J chain | JCHAIN | 0.010018 | 0.099167 | 1.661328 | Detectable | |
| Multiple epidermal-growth-factor-like domains protein 9 | MEGF9 | 0.000603 | 0.099263 | −0.66189 | ||
| Glutathione S-transferase Mu 1 | GSTM1 | 0.008847 | 0.09981 | 1.620058 | ||
| Neurofascin | NFASC | 0.00052 | 0.099907 | −0.62704 | Detectable |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, S.; Jang, Y.; Na, C.H. Discovery of Biomarkers for Amyotrophic Lateral Sclerosis from Human Cerebrospinal Fluid Using Mass-Spectrometry-Based Proteomics. Biomedicines 2023, 11, 1250. https://doi.org/10.3390/biomedicines11051250
Oh S, Jang Y, Na CH. Discovery of Biomarkers for Amyotrophic Lateral Sclerosis from Human Cerebrospinal Fluid Using Mass-Spectrometry-Based Proteomics. Biomedicines. 2023; 11(5):1250. https://doi.org/10.3390/biomedicines11051250
Chicago/Turabian StyleOh, Sungtaek, Yura Jang, and Chan Hyun Na. 2023. "Discovery of Biomarkers for Amyotrophic Lateral Sclerosis from Human Cerebrospinal Fluid Using Mass-Spectrometry-Based Proteomics" Biomedicines 11, no. 5: 1250. https://doi.org/10.3390/biomedicines11051250
APA StyleOh, S., Jang, Y., & Na, C. H. (2023). Discovery of Biomarkers for Amyotrophic Lateral Sclerosis from Human Cerebrospinal Fluid Using Mass-Spectrometry-Based Proteomics. Biomedicines, 11(5), 1250. https://doi.org/10.3390/biomedicines11051250





