PDE3A and GSK3B as Atrial Fibrillation Susceptibility Genes in the Chinese Population via Bioinformatics and Genome-Wide Association Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of DEGs and Susceptibility Genes
2.2. Identification of Potential Therapeutic Targets
2.3. Pathway Enrichment Analysis
2.4. Construction of Pathway Gene Network
2.5. GWAS Study Design and Subjects
2.6. Genotyping
2.7. Statistical Analysis
3. Results
3.1. Identification of DEGs and Susceptibility Genes
3.2. Identification of Potential Therapeutic Targets
3.3. Pathway Enrichment Analysis
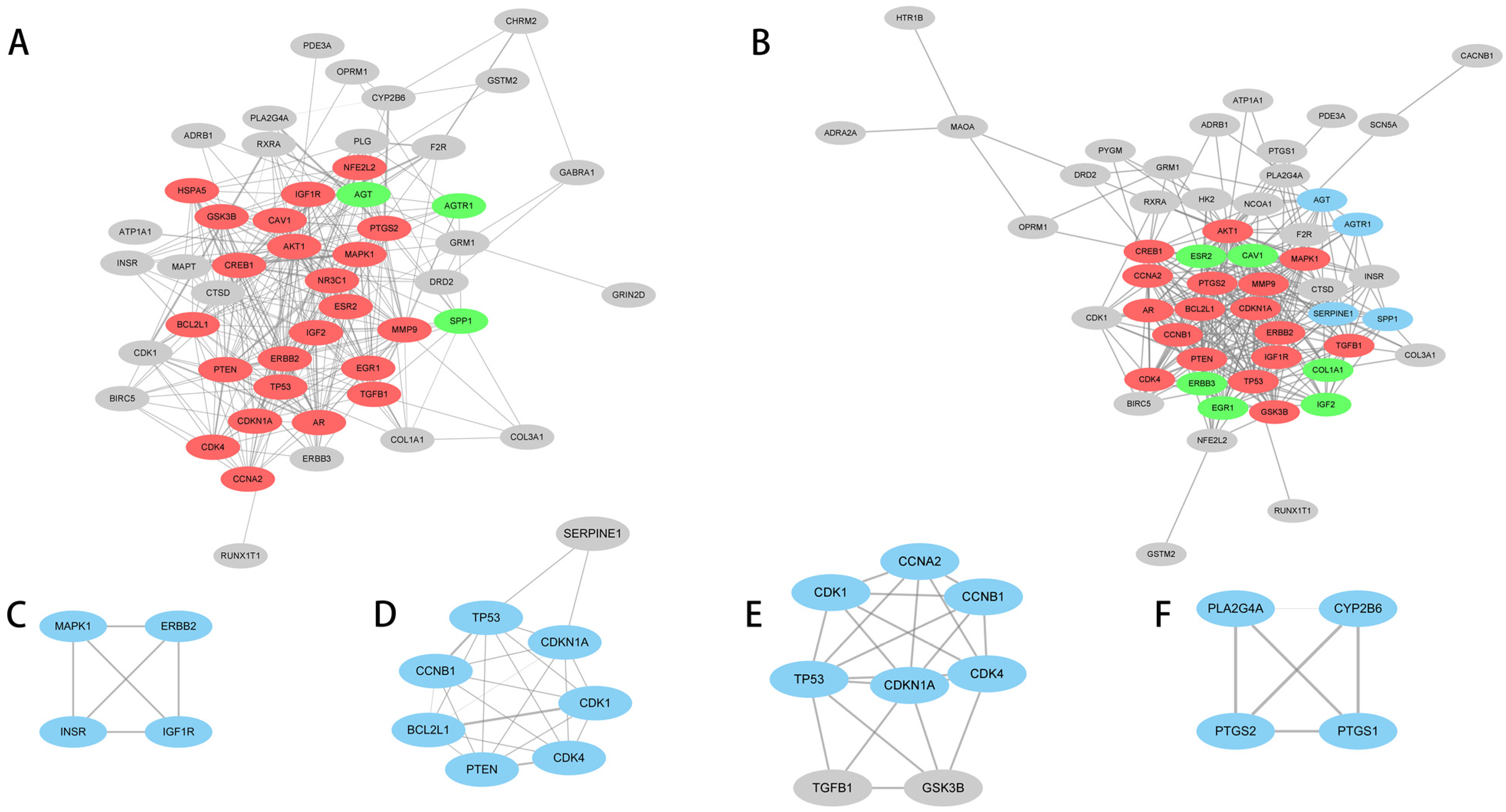
3.4. Construction of Pathway Gene Network
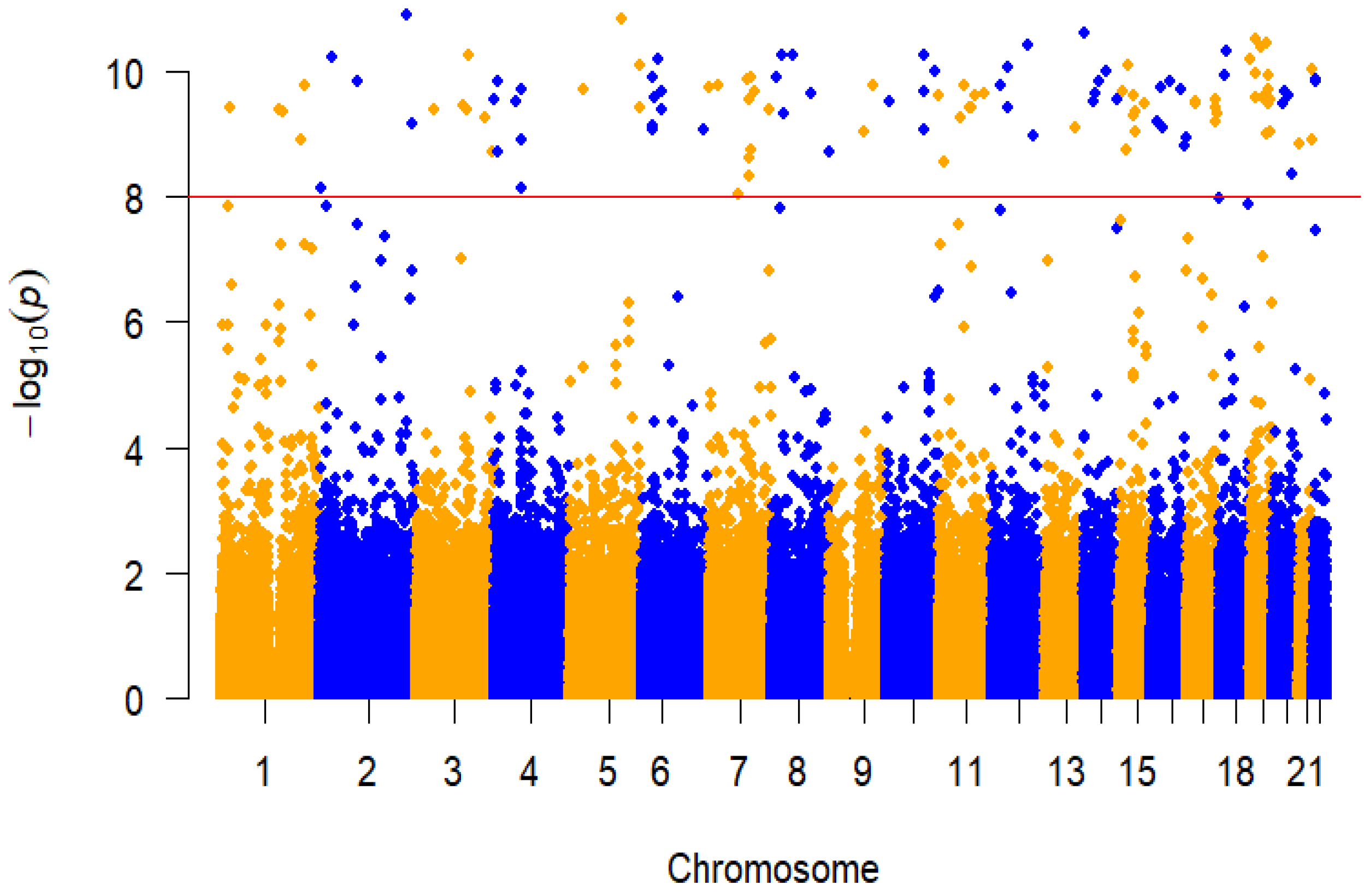
3.5. GWAS Analysis
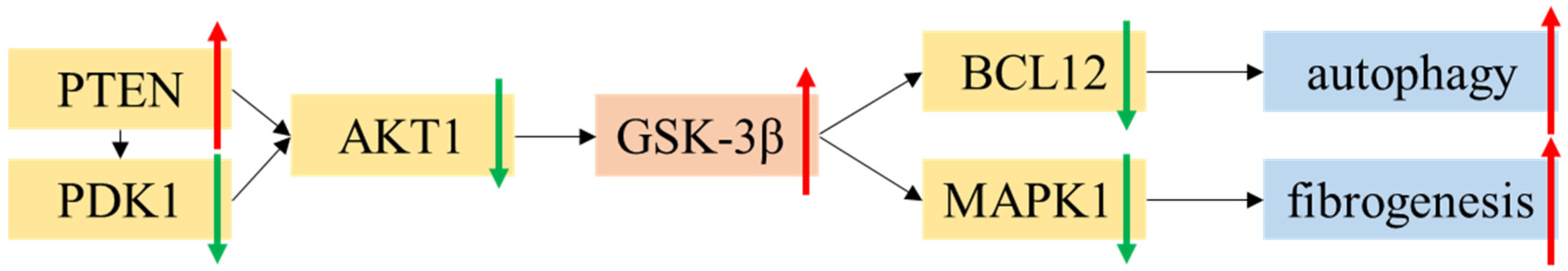
3.6. Bioinformatic Results of PDE3A and GSK-3β
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hindricks, G.; Potpara, T.; Dagres, N.; Arbelo, E.; Bax, J.J.; Blomström-Lundqvist, C.; Boriani, G.; Castella, M.; Dan, G.-A.; Dilaveris, P.E.; et al. Corrigendum to: 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): The Task Force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur. Heart J. 2021, 42, 4194. [Google Scholar] [CrossRef] [PubMed]
- Killu, A.M.; Granger, C.B.; Gersh, B.J. Risk stratification for stroke in atrial fibrillation: A critique. Eur. Heart J. 2019, 40, 1294–1302. [Google Scholar] [CrossRef]
- Xun, A.; True, H.M.; Kuipers, M.F.; YH, L.G.; de Groot Natasja, M.S. Atrial fibrillation. Nat. Rev. Dis. Prim. 2022, 8, 21. [Google Scholar]
- Zhang, J.; Johnsen, S.P.; Guo, Y.; Lip, G.Y. Epidemiology of Atrial Fibrillation: Geographic/Ecological Risk Factors, Age, Sex, Genetics. Card. Electrophysiol. Clin. 2021, 13, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Kornej, J.; Börschel, C.S.; Benjamin, E.J.; Schnabel, R.B. Epidemiology of Atrial Fibrillation in the 21st Century: Novel Methods and New Insights. Circ. Res. 2020, 127, 4–20. [Google Scholar] [CrossRef]
- Peyronnet, R.; Ravens, U. Atria-selective antiarrhythmic drugs in need of alliance partners. Pharmacol. Res. 2019, 145, 104262. [Google Scholar] [CrossRef]
- Roselli, C.; Rienstra, M.; Ellinor, P.T. Genetics of Atrial Fibrillation in 2020: GWAS, Genome Sequencing, Polygenic Risk, and Beyond. Circ. Res. 2020, 127, 21–33. [Google Scholar] [CrossRef]
- Roselli, C.; Chaffin, M.D.; Weng, L.C.; Aeschbacher, S.; Ahlberg, G.; Albert, C.M.; Almgren, P.; Alonso, A.; Anderson, C.D.; Aragam, K.G.; et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 2018, 50, 1225–1233. [Google Scholar] [CrossRef]
- Sutanto, H.; Dobrev, D.; Heijman, J. Genome-wide association studies of atrial fibrillation: Finding meaning in the life of risk loci. IJC Heart Vasc. 2019, 24, 100397. [Google Scholar] [CrossRef]
- Natsume, Y.; Oaku, K.; Takahashi, K.; Nakamura, W.; Oono, A.; Hamada, S.; Yamazoe, M.; Ihara, K.; Sasaki, T.; Goya, M.; et al. Combined Analysis of Human and Experimental Murine Samples Identified Novel Circulating MicroRNAs as Biomarkers for Atrial Fibrillation. Circ. J. 2018, 82, 965–973. [Google Scholar] [CrossRef]
- da Silva, A.M.G.; de Araújo, J.N.G.; de Oliveira, K.M.; Novaes, A.E.M.; Lopes, M.B.; de Sousa, J.C.V.; Filho, A.A.D.A.; Luchessi, A.D.; de Rezende, A.A.; Hirata, M.H.; et al. Circulating miRNAs in acute new-onset atrial fibrillation and their target mRNA network. J. Cardiovasc. Electrophysiol. 2018, 29, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Chiang, D.Y.; Zhang, M.; Voigt, N.; Alsina, K.M.; Jakob, H.; Martin, J.F.; Dobrev, D.; Wehrens, X.H.; Li, N. Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation. Int. J. Cardiol. 2015, 184, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. In Statistical Genomics; Springer: New York, NY, USA, 2016; pp. 93–110. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic. Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic. Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Kuhn, M.; Szklarczyk, D.; Franceschini, A.; Campillos, M.; von Mering, C.; Jensen, L.J.; Beyer, A.; Bork, P. STITCH 2: An interaction network database for small molecules and proteins. Nucleic Acids Res. 2010, 38, D552–D556. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Uniprot, C. UniProt: The universal protein knowledgebase in 2021. Nucleic. Acids Res. 2021, 49, D480–D489. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and func-tional characterization of user-uploaded gene/measurement sets. Nucleic. Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Tang, X.; Huidong, D.; Chen, D.; Zhan, S.; Zhang, Z.; Dou, H. The Fangshan/Family-based Ischemic Stroke Study in China (FISSIC) protocol. BMC Med Genet. 2007, 8, 60. [Google Scholar] [CrossRef]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010, 5, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Ercu, M.; Klussmann, E. Roles of A-Kinase Anchoring Proteins and Phosphodiesterases in the Cardiovascular System. J. Cardiovasc. Dev. Dis. 2018, 5, 14. [Google Scholar] [CrossRef]
- Choi, Y.H.; Ekholm, D.; Krall, J.; Ahmad, F.; Degerman, E.; Manganiello, V.C.; Movsesian, M.A. Identification of a novel isoform of the cyclic-nucleotide phosphodiesterase PDE3A ex-pressed in vascular smooth-muscle myocytes. Biochem. J. 2001, 353, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Dolce, B.; Christ, T.; Pavlidou, N.G.; Yildirim, Y.; Reichenspurner, H.; Eschenhagen, T.; Nikolaev, V.O.; Kaumann, A.J.; Molina, E.C. Impact of phosphodiesterases PDE3 and PDE4 on 5-hydroxytryptamine receptor4-mediated increase of cAMP in human atrial fibrillation. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 394, 291–298. [Google Scholar] [CrossRef]
- Ding, B.; Abe, J.I.; Wei, H.; Huang, Q.; Walsh, R.A.; Molina, C.A.; Zhao, A.; Sadoshima, J.; Blaxall, B.C.; Berk, B.C.; et al. Functional role of phosphodiesterase 3 in cardiomyocyte apoptosis: Implication in heart failure. Circulation 2005, 111, 2469–2476. [Google Scholar] [CrossRef]
- Ercu, M.; Mücke, M.B.; Pallien, T.; Markó, L.; Sholokh, A.; Schächterle, C.; Aydin, A.; Kidd, A.; Walter, S.; Esmati, Y.; et al. Mutant Phosphodiesterase 3A Protects from Hypertension-Induced Cardiac Damage. Circulation 2022, 146, 1758–1778. [Google Scholar] [CrossRef]
- Wang, Z.; Greenbaum, J.; Qiu, C.; Li, K.; Wang, Q.; Tang, S.-Y.; Deng, H.-W. Identification of pleiotropic genes between risk factors of stroke by multivariate metaCCA analysis. Mol. Genet. Genom. 2020, 295, 1173–1185. [Google Scholar] [CrossRef]
- Sucharov, C.C.; Nakano, S.J.; Slavov, D.; Schwisow, J.A.; Rodriguez, E.; Nunley, K.; Medway, A.; Stafford, N.; Nelson, P.; McKinsey, T.A.; et al. A PDE3A Promoter Polymorphism Regulates cAMP-Induced Transcriptional Ac-tivity in Failing Human Myocardium. J. Am. Coll Cardiol. 2019, 73, 1173–1184. [Google Scholar] [CrossRef]
- Uzbekova, S.; Salhab, M.; Perreau, C.; Mermillod, P.; Dupont, J. Glycogen synthase kinase 3B in bovine oocytes and granulosa cells: Possible in-volvement in meiosis during in vitro maturation. Reproduction 2009, 138, 235–246. [Google Scholar] [CrossRef]
- Hardt, S.E.; Sadoshima, J. Glycogen synthase kinase-3beta: A novel regulator of cardiac hypertrophy and development. Circ. Res. 2002, 90, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Morishima, M.; Li, D.; Takahashi, N.; Saikawa, T.; Nattel, S.; Ono, K. Binge Alcohol Exposure Triggers Atrial Fibrillation Through T-Type Ca(2+) Channel Upregulation via Protein Kinase C (PKC)/Glycogen Synthesis Kinase 3beta (GSK3beta)/Nuclear Factor of Activated T-Cells (NFAT) Signaling- An Experimental Account of Holiday Heart Syndrome. Circ. J. 2020, 84, 1931–1940. [Google Scholar] [PubMed]
- Hamstra, S.I.; Braun, J.L.; Chelko, S.P.; Fajardo, V.A. GSK3-inhibition improves maximal SERCA activity in a murine model of Ar-rhythmogenic cardiomyopathy. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166536. [Google Scholar] [CrossRef] [PubMed]
- Sardu, C.; Santulli, G.; Guerra, G.; Trotta, M.C.; Santamaria, M.; Sacra, C.; Testa, N.; Ducceschi, V.; Gatta, G.; Amico, M.D.; et al. Modulation of SERCA in Patients with Persistent Atrial Fibrillation Treated by Epicardial Thoracoscopic Ablation: The CAMAF Study. J. Clin. Med. 2020, 9, 544. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bertero, E.; Maack, C. Calcium Signaling and Reactive Oxygen Species in Mitochondria. Circ. Res. 2018, 122, 1460–1478. [Google Scholar] [CrossRef]
- Liu, D.; Yang, M.; Yao, Y.; He, S.; Wang, Y.; Cao, Z.; Chen, H.; Fu, Y.; Liu, H.; Zhao, Q. Cardiac Fibroblasts Promote Ferroptosis in Atrial Fibrillation by Secreting Exo-miR-23a-3p Targeting SLC7A11. Oxid. Med. Cell. Longev. 2022, 2022, 3961495. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, H.; Wang, J. Metabolism and Chronic Inflammation: The Links Between Chronic Heart Failure and Comorbidities. Front. Cardiovasc. Med. 2021, 8, 650278. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, R.; Wang, X.; Li, J.; Yuan, M.; Liu, E.; Liu, T.; Li, G. Attenuation of atrial remodeling by aliskiren via affecting oxidative stress, inflammation and PI3K/Akt signaling pathway. Cardiovasc. Drugs Ther. 2021, 35, 587–598. [Google Scholar] [CrossRef]
- Zeng, B.; Liao, X.; Liu, L.; Zhang, C.; Ruan, H.; Yang, B. Thyroid hormone mediates cardioprotection against postinfarction remodeling and dysfunction through the IGF-1/PI3K/AKT signaling pathway. Life Sci. 2021, 267, 118977. [Google Scholar] [CrossRef]
- Wen, L.; Yang, Q.H.; Ma, X.L.; Li, T.; Xiao, S.; Sun, C.F. Inhibition of TNFAIP1 ameliorates the oxidative stress and inflammatory injury in myocardial ischemia/reperfusion injury through modulation of Akt/GSK-3beta/Nrf2 pathway. Int. Immunopharmacol. 2021, 99, 107993. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Kumar, A.; Sahu, M.; Sharma, G.; Datusalia, A.K.; Rajput, S.K. Exercise preconditioning and low dose copper nanoparticles exhibits cardioprotection through targeting GSK-3beta phosphorylation in ischemia/reperfusion induced myocardial infarction. Microvasc. Res. 2018, 120, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liu, Y.; Xu, Q.Q.; Xian, Y.F.; Lin, Z.X. Sulforaphene Ameliorates Neuroinflammation and Hyperphosphorylated Tau Protein via Regulating the PI3K/Akt/GSK-3beta Pathway in Experimental Models of Alzheimer’s Disease. Oxid. Med. Cell. Longev. 2020, 2020, 4754195. [Google Scholar] [CrossRef] [PubMed]
- Smit, M.D.; Maass, A.H.; De Jong, A.M.; Muller Kobold, A.C.; Van Veldhuisen, D.J.; Van Gelder, I.C. Role of inflammation in early atrial fibrillation recurrence. Europace 2012, 14, 810–817. [Google Scholar] [CrossRef]
- Sardu, C.; Santulli, G.; Santamaria, M.; Barbieri, M.; Sacra, C.; Paolisso, P.; D’Amico, F.; Testa, N.; Caporaso, I.; Paolisso, G.; et al. Effects of Alpha Lipoic Acid on Multiple Cytokines and Biomarkers and Recurrence of Atrial Fibrillation Within 1 Year of Catheter Ablation. Am. J. Cardiol. 2017, 119, 1382–1386. [Google Scholar] [CrossRef] [PubMed]
- Sardu, C.; Santamaria, M.; Paolisso, G.; Marfella, R. microRNA expression changes after atrial fibrillation catheter ablation. Pharmacogenomics 2015, 16, 1863–1877. [Google Scholar] [CrossRef]
- Shen, N.N.; Zhang, Z.L.; Li, Z.; Zhang, C.; Li, H.; Wang, J.L.; Wang, J.; Gu, Z.C. Identification of microRNA biomarkers in atrial fibrillation: A protocol for systematic review and bioinformatics analysis. Medicine 2019, 98, e16538. [Google Scholar] [CrossRef]
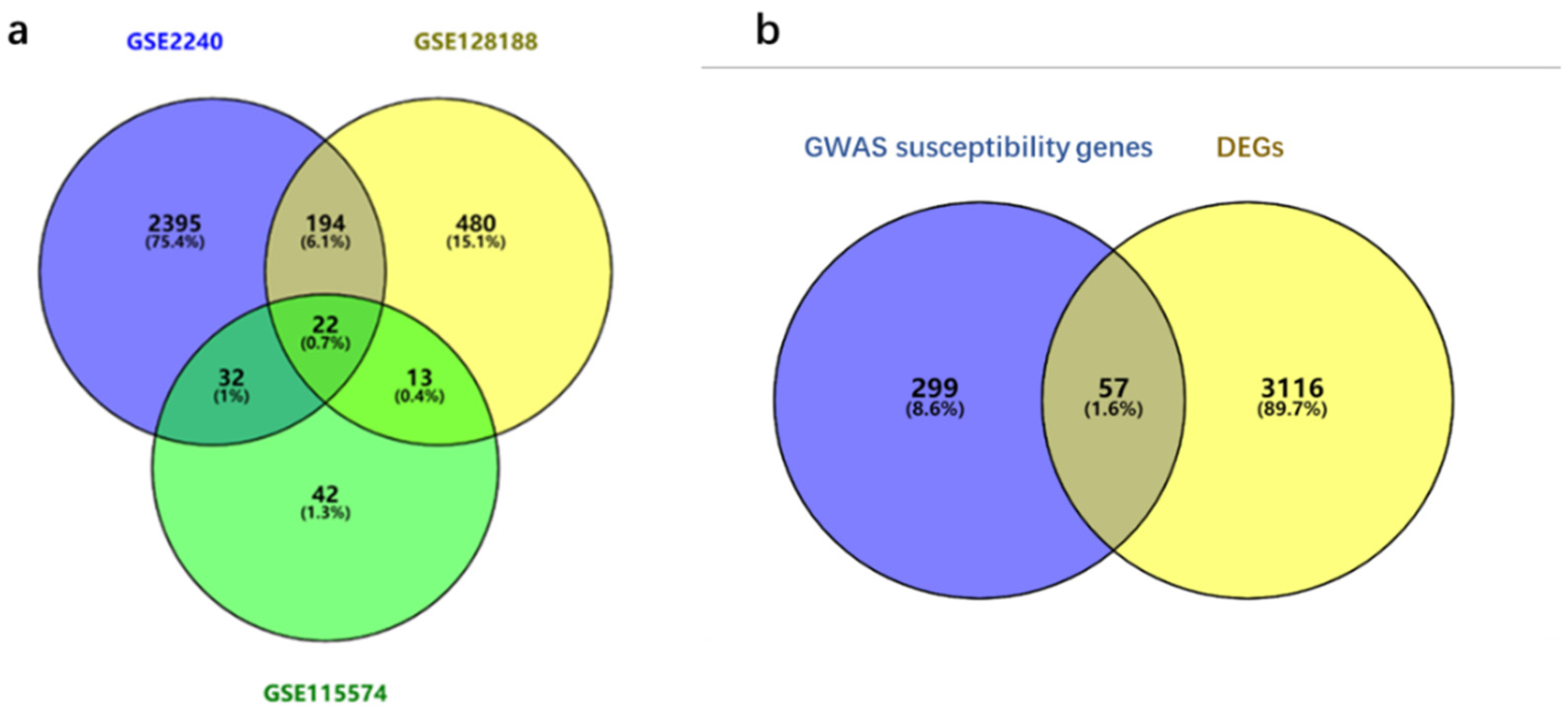
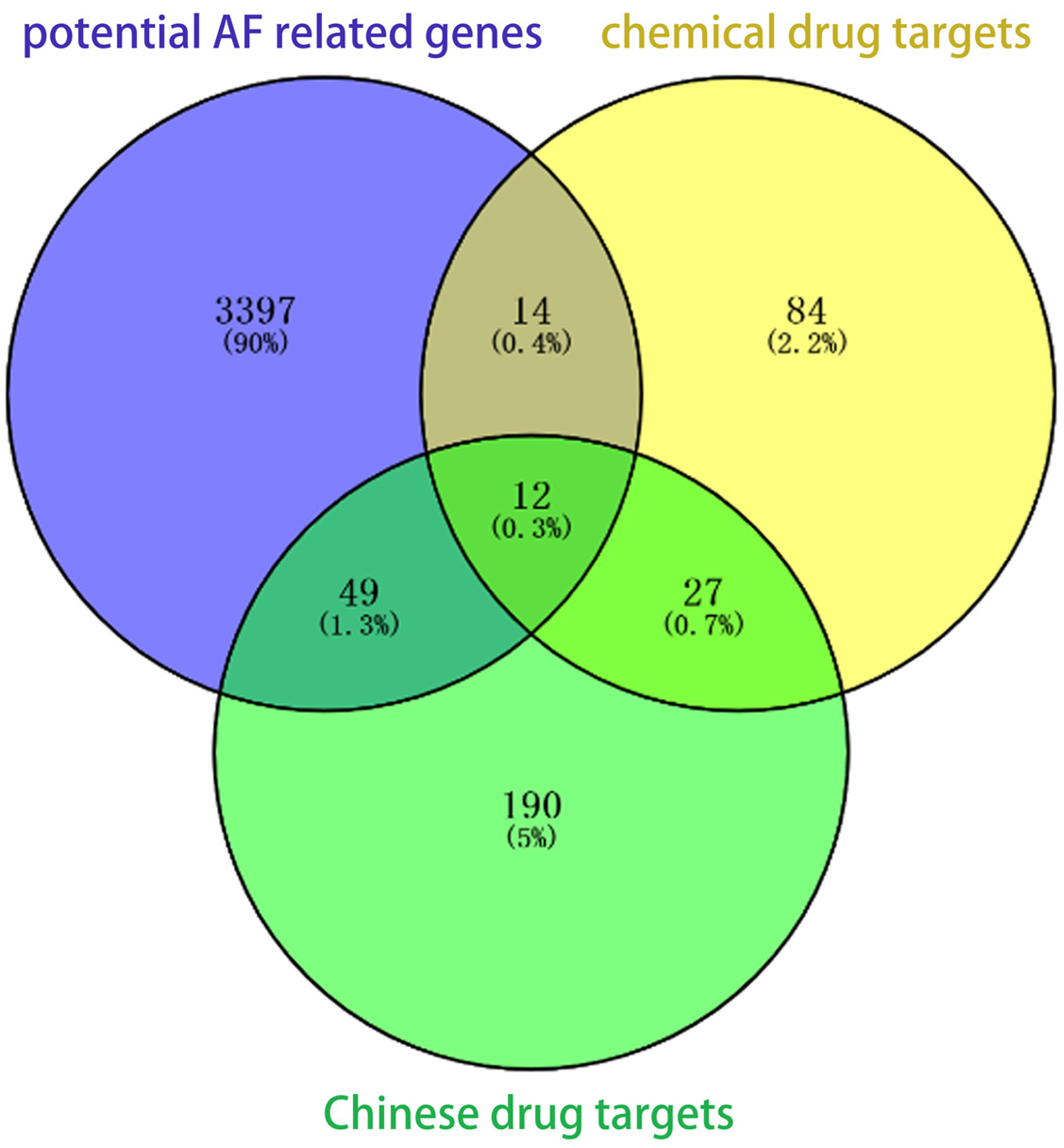
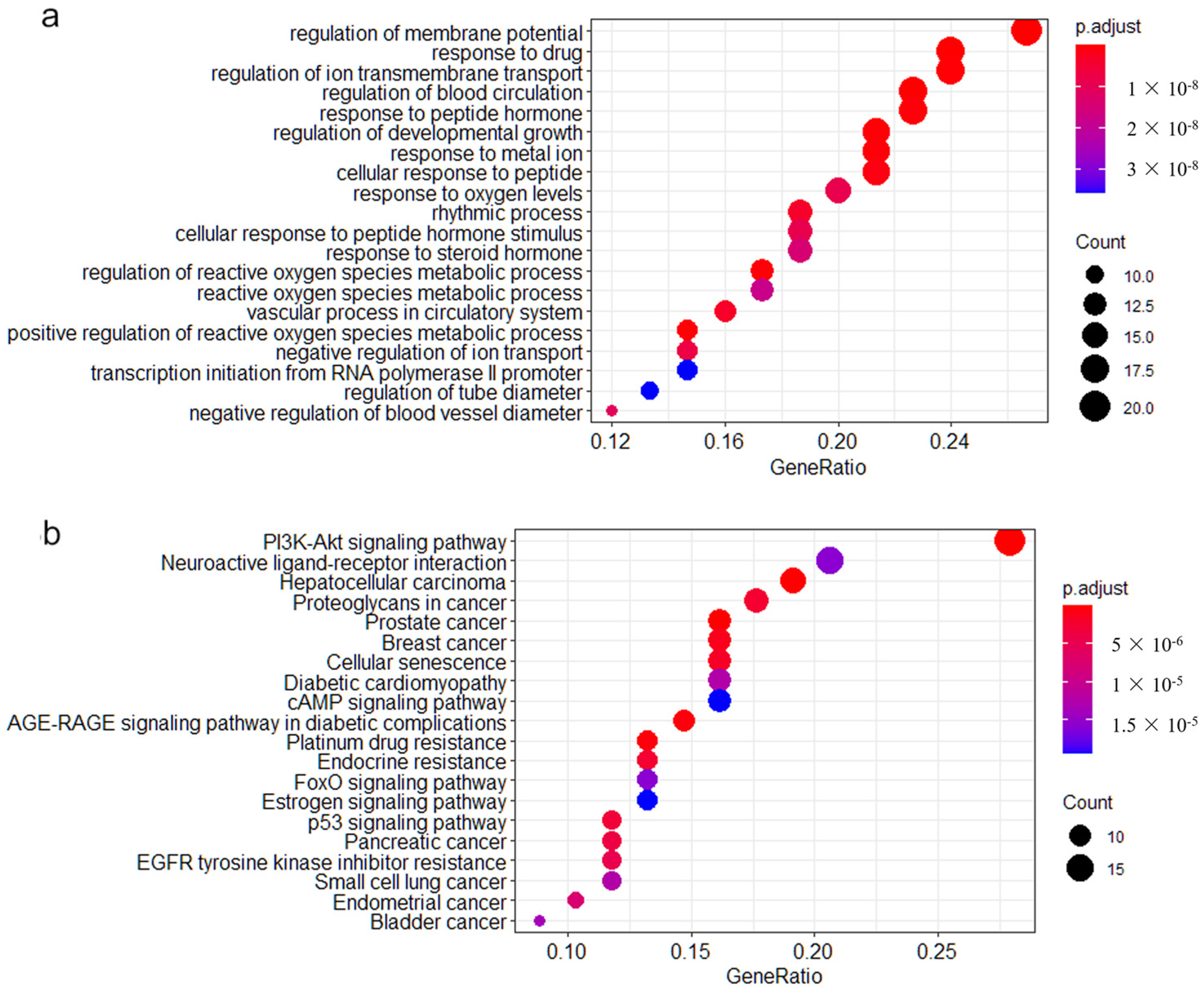
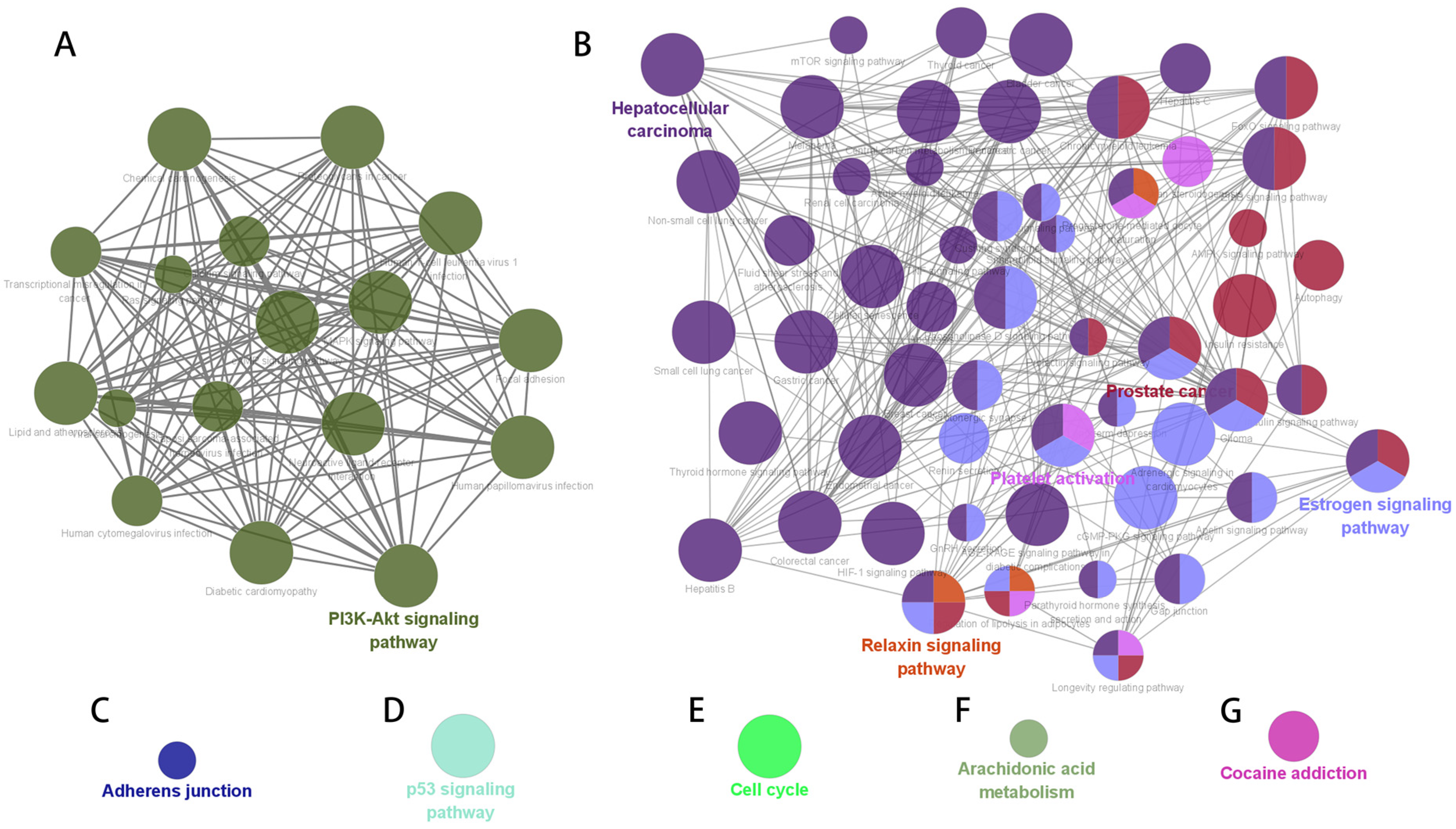
| Data Sets | Tissues | Chips (Batches) | Number of DEGs | Up-Regulated Genes | Down-Regulated Genes |
|---|---|---|---|---|---|
| GSE2240 | atrium | GPL96 | 1819 | 1029 | 790 |
| GPL97 | 508 | 230 | 278 | ||
| GPL96 + GPL97 | 885 | 443 | 442 | ||
| GSE128188 | left auricle | GPL18573 | 13 | 3 | 10 |
| right auricle | GPL18573 | 1 | 0 | 1 | |
| auricle | GPL18573 | 708 | 350 | 358 | |
| GSE115574 | left atrium | GPL570 | 1 | 0 | 1 |
| right atrium | GPL570 | 1 | 1 | 0 | |
| atrium | GPL570 | 111 | 64 | 47 | |
| Total | 3173 | 1757 | 1432 |
| Targets | Entrez ID | Genes |
|---|---|---|
| Prostaglandin G/H synthase 2 | 5743 | PTGS2 |
| Prostaglandin G/H synthase 1 | 5742 | PTGS1 |
| C-reactive protein | 1401 | CRP |
| Cellular tumor antigen p53 | 7157 | TP53 |
| Matrix metalloproteinase-9 | 4318 | MMP9 |
| Potassium voltage-gated channel subfamily H member 2 | 3757 | KCNH2 |
| Angiotensinogen | 183 | AGT |
| Beta-1 adrenergic receptor | 153 | ADRB1 |
| Lactotransferrin | 4057 | LTF |
| Microtubule-associated protein tau | 4137 | MAPT |
| Type-1 angiotensin II receptor | 185 | AGTR1 |
| RAC-alpha serine/threonine-protein kinase | 207 | AKT1 |
| Alpha-2A adrenergic receptor | 150 | ADRA2A |
| Cytochrome P450 2B6 | 1555 | CYP2B6 |
| Sodium/potassium-transporting ATPase subunit alpha-1 | 476 | ATP1A1 |
| Early growth response protein 1 | 1958 | EGR1 |
| Sodium channel protein type 5 subunit alpha | 6331 | SCN5A |
| Triadin | 10345 | TRDN |
| Sodium channel protein type 10 subunit alpha | 6336 | SCN10A |
| Sodium channel protein type 8 subunit alpha | 6334 | SCN8A |
| Sodium channel protein type 3 subunit alpha | 6328 | SCN3A |
| Heparin cofactor 2 | 3053 | SERPIND1 |
| Plasminogen | 5340 | PLG |
| Voltage-dependent L-type calcium channel subunit beta-1 | 782 | CACNB1 |
| 5-hydroxytryptamine 1B receptor | 3351 | HTR1B |
| Potassium channel subfamily K member 17 | 89822 | KCNK17 |
| 78 kDa glucose-regulated protein | 3309 | HSPA5 |
| Amine oxidase [flavin-containing] A | 4128 | MAOA |
| Androgen receptor | 367 | AR |
| Baculoviral IAP repeat-containing protein 5 | 332 | BIRC5 |
| Bcl-2-like protein 1 | 598 | BCL2L1 |
| Carbonic anhydrase II | 760 | CA2 |
| Cathepsin D | 1509 | CTSD |
| Caveolin-1 | 857 | CAV1 |
| Cell division control protein 2 homolog | 983 | CDK1 |
| Cell division protein kinase 4 | 1019 | CDK4 |
| CGMP-inhibited 3’,5’-cyclic phosphodiesterase A | 5139 | PDE3A |
| Collagen alpha-1(I) chain | 1277 | COL1A1 |
| Collagen alpha-1(III) chain | 1281 | COL3A1 |
| Cyclic AMP-responsive element-binding protein 1 | 1385 | CREB1 |
| Cyclin-A2 | 890 | CCNA2 |
| Cyclin-dependent kinase inhibitor 1 | 1026 | CDKN1A |
| Cytosolic phospholipase A2 | 5321 | PLA2G4A |
| D(2) dopamine receptor | 1813 | DRD2 |
| DNA topoisomerase 2-alpha | 7153 | TOP2A |
| Estrogen receptor beta | 2100 | ESR2 |
| G2/mitotic-specific cyclin-B1 | 891 | CCNB1 |
| Gamma-aminobutyric acid receptor subunit alpha-1 | 2554 | GABRA1 |
| Glucocorticoid receptor | 2908 | NR3C1 |
| Glutamate [NMDA] receptor subunit epsilon-4 | 2906 | GRIN2D |
| Glutathione S-transferase Mu 2 | 2946 | GSTM2 |
| Glycogen phosphorylase, muscle form | 5837 | PYGM |
| Glycogen synthase kinase-3 beta | 2932 | GSK-3β |
| Hexokinase-2 | 3099 | HK2 |
| Insulin-like growth factor 1 receptor | 3480 | IGF1R |
| Insulin-like growth factor II | 3481 | IGF2 |
| Insulin receptor | 3643 | INSR |
| Metabotropic glutamate receptor 1 | 2911 | GRM1 |
| Mitogen-activated protein kinase 1 | 5594 | MAPK1 |
| Mu-type opioid receptor | 4988 | OPRM1 |
| Muscarinic acetylcholine receptor M2 | 1129 | CHRM2 |
| Nuclear factor erythroid 2-related factor 2 | 4780 | NFE2L2 |
| Nuclear receptor coactivator 1 | 8648 | NCOA1 |
| Osteopontin | 6696 | SPP1 |
| Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | 5728 | PTEN |
| Plasminogen activator inhibitor 1 | 5054 | SERPINE1 |
| Protein CBFA2T1 | 862 | RUNX1T1 |
| Receptor tyrosine-protein kinase erbB-2 | 2064 | ERBB2 |
| Receptor tyrosine-protein kinase erbB-3 | 2065 | ERBB3 |
| Retinoic acid receptor RXR-alpha | 6256 | RXRA |
| Serum paraoxonase/arylesterase 1 | 5444 | PON1 |
| Thrombin | 2149 | F2R |
| Transforming growth factor beta-1 | 7040 | TGFB1 |
| Type I iodothyronine deiodinase | 1733 | DIO1 |
| Beta-secretase 2 | 25825 | BACE2 |
| Subnetworks | Clusters | MCODE Scores | Nodes | Lines | Hub Genes |
|---|---|---|---|---|---|
| A | 1 | 16.818 | 23 | 185 | NR3C1, TP53, ESR2, EGR1, HSPA5, CREB1, CAV1, AKT1, TGFB1, GSK-3β, MAPK1, NFE2L2, CCNA2, IGF2, BCL2L1, CDKN1A, PTGS2, PTEN, CDK4, ERBB2, MMP9, AR, IGF1R |
| A | 2 | 3 | 3 | 3 | AGT, SPP1, AGTR1 |
| B | 1 | 15.25 | 17 | 122 | TP53, CREB1, AKT1, TGFB1, GSK-3β, MAPK1, CCNA2, BCL2L1, CDKN1A, PTGS2, PTEN, CCNB1, CDK4, ERBB2, MMP9, AR, IGF1R |
| B | 2 | 3.333 | 4 | 5 | SERPINE1, AGT, SPP1, AGTR1 |
| B | 3 | 3.2 | 6 | 8 | ESR2, ERBB3, COL1A1, CAV1, IGF2, EGR1 |
| C | 1 | 4 | 4 | 6 | MAPK1, ERBB2, INSR, IGF1R |
| D | 1 | 7 | 7 | 21 | PTEN, CDKN1A, CDK1, CCNB1, BCL2L1, TP53, CDK4 |
| E | 1 | 6 | 6 | 15 | CDKN1A, CCNB1, CCNA2, CDK1, TP53, CDK4 |
| F | 1 | 4 | 4 | 6 | PTGS2, PTGS1, PLA2G4A, CYP2B6 |
| G | - | - | - | - | - |
| Gene | SNP | Chr:position | Allele1 | Allele0 | MAF | β | SE | p |
|---|---|---|---|---|---|---|---|---|
| NBPF3 | rs147300495 | 1:21777939 | T | C | 0.02 | 0.878 | 0.139 | 3.71 × 10−10 |
| PDE4DIP | rs1628310 | 1:144868170 | C | T | 0.019 | 0.881 | 0.14 | 4.11 × 10−10 |
| LOC101929703 | rs974690619 | 1:155538990 | G | A | 0.019 | 0.881 | 0.14 | 4.54 × 10−10 |
| NR5A2 | rs7546336 | 1:199994841 | C | T | 0.02 | 0.855 | 0.14 | 1.21 × 10−9 |
| SYT14 | rs76437946 | 1:210335855 | T | C | 0.019 | 0.901 | 0.14 | 1.73 × 10−10 |
| EIPR1 | rs199623295 | 2:3359563 | C | T | 0.019 | 0.604 | 0.104 | 7.33 × 10−9 |
| NLRC4 | rs1408931915 | 2:32468262 | G | A | 0.019 | 0.916 | 0.139 | 6.05 × 10−11 |
| ANKRD36C | rs5005869 | 2:96521297 | C | T | 0.019 | 0.903 | 0.14 | 1.50 × 10−10 |
| USP37 | rs182055303 | 2:219384724 | G | A | 0.019 | 0.955 | 0.14 | 1.26 × 10−11 |
| UGT1A8 | rs1042591 | 2:234526794 | G | T | 0.019 | 0.871 | 0.14 | 6.87 × 10−10 |
| MAP4 | rs137991644 | 3:48118180 | C | A | 0.02 | 0.877 | 0.139 | 4.16 × 10−10 |
| GSK3B | rs796944992 | 3:119787193 | G | A | 0.019 | 0.888 | 0.14 | 3.47 × 10−10 |
| LINC01565 | rs141327567 | 3:128293330 | C | A | 0.02 | 0.876 | 0.139 | 4.07 × 10−10 |
| CPNE4 | rs139775204 | 3:131493768 | T | C | 0.012 | 0.259 | 0.039 | 5.43 × 10−11 |
| TBL1XR1 | rs148786696 | 3:176879400 | G | A | 0.019 | 0.875 | 0.14 | 5.42 × 10−10 |
| MUC4 | rs74500246 | 3:195513563 | C | T | 0.02 | 0.842 | 0.139 | 1.92 × 10−9 |
| ZNF141 | rs1303526299 | 4:382920 | C | T | 0.02 | 0.891 | 0.14 | 2.78 × 10−10 |
| non-coding | rs184180522 | 4:8930460 | A | G | 0.02 | 0.839 | 0.139 | 1.91 × 10−9 |
| non-coding | rs1394588518 | 4:9274164 | C | A | 0.02 | 0.894 | 0.139 | 1.49 × 10−10 |
| non-coding | rs1015522933 | 4:55067908 | G | A | 0.02 | 0.881 | 0.139 | 3.05 × 10−10 |
| UGT2B15 | rs4148260 | 4:69531574 | A | G | 0.02 | 0.895 | 0.139 | 1.91 × 10−10 |
| UGT2B15 | rs3862051 | 4:69534405 | C | T | 0.02 | 0.857 | 0.14 | 1.20 × 10−9 |
| UGT2B7 | rs6600887 | 4:69969788 | C | T | 0.016 | 0.514 | 0.088 | 7.23 × 10−9 |
| SKP2 | rs763496236 | 5:36183898 | G | A | 0.02 | 0.895 | 0.139 | 1.91 × 10−10 |
| IL4 | rs376951889 | 5:132008827 | C | T | 0.02 | 0.699 | 0.103 | 1.44 × 10−11 |
| non-coding | rs1081806 | 5:176198317 | G | A | 0.019 | 0.881 | 0.14 | 3.90 × 10−10 |
| ADAMTS2 | rs1213209228 | 5:178551750 | G | A | 0.019 | 0.918 | 0.14 | 7.73 × 10−11 |
| MUC21 | rs767391626 | 6:30954375 | D | I | 0.019 | 0.91 | 0.14 | 1.29 × 10−10 |
| non-coding | rs1261299467 | 6:31030655 | G | A | 0.019 | 0.867 | 0.14 | 7.53 × 10−10 |
| HLA-B | rs12697943 | 6:31324057 | A | C | 0.019 | 0.833 | 0.135 | 8.27 × 10−10 |
| TNXB | rs200135227 | 6:32029369 | T | C | 0.02 | 0.884 | 0.139 | 2.60 × 10−10 |
| CUL7 | rs201406974 | 6:43014042 | G | A | 0.019 | 0.923 | 0.14 | 6.24 × 10−11 |
| GSTA1 | rs2894804 | 6:52668546 | G | A | 0.019 | 0.88 | 0.14 | 4.01 × 10−10 |
| GSTA1 | rs9296692 | 6:52668943 | T | C | 0.019 | 0.855 | 0.134 | 2.16 × 10−10 |
| TULP4 | rs113382463 | 6:158847210 | G | A | 0.02 | 0.858 | 0.139 | 8.69 × 10−10 |
| non-coding | rs1443575050 | 7:2919444 | G | A | 0.019 | 0.901 | 0.14 | 1.78 × 10−10 |
| IGF2BP3 | rs118111412 | 7:23416772 | C | T | 0.02 | 0.893 | 0.139 | 1.67 × 10−10 |
| non-coding | rs1158528647 | 7:75813735 | C | T | 0.016 | 0.51 | 0.088 | 9.18 × 10−9 |
| CYP3A5 | rs76293380 | 7:99250394 | D | I | 0.019 | 0.908 | 0.14 | 1.39 × 10−10 |
| MUC3A | rs73163748 | 7:100549573 | T | G | 0.019 | 0.892 | 0.14 | 2.83 × 10−10 |
| MUC3A | rs74197937 | 7:100550133 | G | A | 0.017 | 0.706 | 0.12 | 4.75 × 10−9 |
| MUC12 | rs75466554 | 7:100615387 | C | T | 0.02 | 0.837 | 0.139 | 2.39 × 10−9 |
| KMT2E | rs149680168 | 7:104698096 | C | T | 0.02 | 0.902 | 0.139 | 1.25 × 10−10 |
| LAMB1 | rs6959803 | 7:107621620 | A | G | 0.02 | 0.845 | 0.139 | 1.75 × 10−9 |
| CFTR | rs34517638 | 7:117146425 | C | T | 0.02 | 0.891 | 0.139 | 2.14 × 10−10 |
| KMT2C | rs74977767 | 7:151932876 | T | G | 0.019 | 0.849 | 0.135 | 4.14 × 10−10 |
| non-coding | rs5004426 | 8:13679081 | C | T | 0.02 | 0.896 | 0.138 | 1.28 × 10−10 |
| non-coding | rs375571715 | 8:24929402 | A | G | 0.02 | 0.917 | 0.139 | 5.65 × 10−11 |
| non-coding | rs149373333 | 8:29332974 | T | C | 0.02 | 0.875 | 0.139 | 4.84 × 10−10 |
| RP1 | rs759385909 | 8:55533903 | T | C | 0.02 | 0.922 | 0.139 | 5.41 × 10−11 |
| RIDA | rs187976179 | 8:99119874 | A | G | 0.02 | 0.867 | 0.136 | 2.33 × 10−10 |
| ARHGAP39 | rs113552609 | 8:145790524 | C | T | 0.02 | 0.842 | 0.139 | 1.93 × 10−9 |
| SLC28A3 | rs112132735 | 9:86980920 | G | A | 0.02 | 0.86 | 0.139 | 9.18 × 10−10 |
| LOC105376205 | rs12553896 | 9:110375131 | C | A | 0.019 | 0.905 | 0.141 | 1.73 × 10−10 |
| non-coding | rs190890438 | 10:11501497 | C | T | 0.019 | 0.89 | 0.14 | 2.97 × 10−10 |
| CYP2C19 | rs540418228 | 10:96561899 | G | A | 0.02 | 0.89 | 0.139 | 2.17 × 10−10 |
| CYP2C19 | rs113934938 | 10:96602752 | G | A | 0.019 | 0.917 | 0.139 | 5.70 × 10−11 |
| CYP2C9 | rs774607211 | 10:96701973 | G | A | 0.019 | 0.865 | 0.14 | 8.46 × 10−10 |
| non-coding | rs199864119 | 10:127201227 | A | G | 0.02 | 0.905 | 0.139 | 1.01 × 10−10 |
| MUC6 | rs200695483 | 11:1016957 | G | T | 0.019 | 0.897 | 0.141 | 2.51 × 10−10 |
| BTBD10 | rs1399839941 | 11:13436715 | T | C | 0.017 | 0.715 | 0.119 | 2.76 × 10−9 |
| OR9G1 | rs78036340 | 11:56467819 | G | A | 0.02 | 0.875 | 0.14 | 5.37 × 10−10 |
| non-coding | rs188394047 | 11:62423576 | A | G | 0.02 | 0.889 | 0.138 | 1.69 × 10−10 |
| non-coding | rs191739873 | 11:79947826 | C | A | 0.019 | 0.883 | 0.14 | 3.87 × 10−10 |
| non-coding | rs375180756 | 11:81527364 | T | C | 0.02 | 0.876 | 0.139 | 3.85 × 10−10 |
| non-coding | rs200877836 | 11:89331149 | G | A | 0.02 | 0.89 | 0.14 | 2.50 × 10−10 |
| non-coding | rs76168892 | 11:113838928 | C | T | 0.019 | 0.896 | 0.14 | 2.24 × 10−10 |
| PDE3A | rs11613698 | 12:20743447 | A | G | 0.019 | 0.898 | 0.14 | 1.72 × 10−10 |
| non-coding | rs1158482043 | 12:38739656 | A | G | 0.019 | 0.918 | 0.14 | 8.31 × 10−11 |
| non-coding | rs796791101 | 12:38739890 | C | T | 0.019 | 0.882 | 0.14 | 3.88 × 10−10 |
| non-coding | rs10777371 | 12:92375102 | G | A | 0.019 | 0.931 | 0.14 | 3.95 × 10−11 |
| TDG | rs372823872 | 12:104359779 | C | T | 0.02 | 0.857 | 0.14 | 1.10 × 10−9 |
| HS6ST3 | rs117321325 | 13:97247454 | T | C | 0.02 | 0.86 | 0.139 | 7.71 × 10−10 |
| AP1G2 | rs199900328 | 14:24031771 | G | A | 0.02 | 0.943 | 0.14 | 2.43 × 10−11 |
| non-coding | rs915392393 | 14:46302004 | T | C | 0.02 | 0.879 | 0.138 | 3.02 × 10−10 |
| LINC01599 | rs56246200 | 14:50523448 | G | A | 0.019 | 0.899 | 0.141 | 2.25 × 10−10 |
| non-coding | rs141410596 | 14:59276647 | C | A | 0.02 | 0.897 | 0.139 | 1.42 × 10−10 |
| NRXN3 | rs117501552 | 14:79223516 | T | C | 0.02 | 0.904 | 0.139 | 1.02 × 10−10 |
| non-coding | rs2919632 | 14:105325224 | G | A | 0.019 | 0.893 | 0.14 | 2.78 × 10−10 |
| LOC100288637 | rs12902692 | 15:31042778 | C | T | 0.019 | 0.898 | 0.14 | 2.03 × 10−10 |
| non-coding | rs181979589 | 15:38936842 | C | T | 0.02 | 0.838 | 0.138 | 1.76 × 10−9 |
| SORD2P | rs2462045 | 15:45138196 | C | T | 0.019 | 0.917 | 0.14 | 8.08 × 10−11 |
| ALDH1A2 | rs151009495 | 15:58318627 | A | G | 0.02 | 0.872 | 0.139 | 5.21 × 10−10 |
| AQP9 | rs59710194 | 15:58470240 | A | G | 0.02 | 0.887 | 0.139 | 2.51 × 10−10 |
| GOLGA2P11 | rs1188201439 | 15:62538744 | G | A | 0.019 | 0.863 | 0.14 | 9.46 × 10−10 |
| SLC24A1 | rs182375570 | 15:65920491 | A | C | 0.02 | 0.868 | 0.138 | 4.54 × 10−10 |
| non-coding | rs76141469 | 15:85841674 | A | G | 0.019 | 0.88 | 0.139 | 3.23 × 10−10 |
| ABCC1 | rs779233080 | 16:16117841 | C | T | 0.02 | 0.871 | 0.14 | 6.38 × 10−10 |
| non-coding | rs651252 | 16:26733084 | C | T | 0.019 | 0.901 | 0.14 | 1.77 × 10−10 |
| SH2B1 | rs79172792 | 16:28872240 | C | T | 0.02 | 0.866 | 0.14 | 7.85 × 10−10 |
| ABCC12 | rs9302750 | 16:48149467 | G | A | 0.019 | 0.896 | 0.139 | 1.44 × 10−10 |
| non-coding | rs147492577 | 16:78128820 | C | T | 0.019 | 0.899 | 0.14 | 1.96 × 10−10 |
| ZCCHC14-DT | rs1027308723 | 16:87538248 | C | T | 0.017 | 0.592 | 0.097 | 1.57 × 10−9 |
| ANKRD11 | rs1345338837 | 16:89530633 | T | C | 0.019 | 0.852 | 0.139 | 1.18 × 10−9 |
| MAP2K3 | rs56369732 | 17:21215552 | T | C | 0.02 | 0.886 | 0.14 | 3.35 × 10−10 |
| KCNJ12 | rs76267885 | 17:21319743 | G | A | 0.02 | 0.887 | 0.14 | 2.95 × 10−10 |
| non-coding | rs2620043 | 17:71833095 | G | A | 0.019 | 0.887 | 0.14 | 3.74 × 10−10 |
| non-coding | rs1229451307 | 17:73434022 | G | A | 0.019 | 0.848 | 0.136 | 6.57 × 10−10 |
| UNC13D | rs145811063 | 17:73833216 | G | A | 0.02 | 0.883 | 0.139 | 2.81 × 10−10 |
| non-coding | rs9902558 | 17:76635886 | G | A | 0.019 | 0.881 | 0.14 | 4.59 × 10−10 |
| ANKRD20A5P | rs200431864 | 18:14179412 | G | A | 0.019 | 0.909 | 0.14 | 1.20 × 10−10 |
| OSBPL1A | rs4800569 | 18:21962607 | C | T | 0.02 | 0.915 | 0.138 | 4.84 × 10−11 |
| non-coding | rs140152353 | 19:2379463 | G | A | 0.02 | 0.921 | 0.14 | 6.35 × 10−11 |
| CYP4F12 | rs149121004 | 19:15792430 | G | A | 0.019 | 0.895 | 0.141 | 2.59 × 10−10 |
| CYP4F2 | rs4020346 | 19:15989730 | C | T | 0.02 | 0.908 | 0.14 | 1.11 × 10−10 |
| MED26 | rs75838150 | 19:16691304 | C | T | 0.019 | 0.935 | 0.139 | 2.99 × 10−11 |
| non-coding | rs181407642 | 19:30211111 | C | A | 0.02 | 0.915 | 0.138 | 4.26 × 10−11 |
| CHST8 | rs4805919 | 19:34148544 | C | T | 0.02 | 0.879 | 0.138 | 2.69 × 10−10 |
| CYP2S1 | rs187503524 | 19:41708962 | A | G | 0.019 | 0.854 | 0.139 | 9.88 × 10−10 |
| RELB | rs187976859 | 19:45533628 | C | A | 0.02 | 0.925 | 0.138 | 3.50 × 10−11 |
| non-coding | rs113900465 | 19:46748849 | G | A | 0.019 | 0.893 | 0.14 | 2.79 × 10−10 |
| CCDC9 | rs144604956 | 19:47766123 | G | A | 0.019 | 0.894 | 0.14 | 2.72 × 10−10 |
| SULT2B1 | rs7248627 | 19:49084356 | C | T | 0.019 | 0.89 | 0.14 | 3.25 × 10−10 |
| non-coding | rs73046773 | 19:49582674 | G | A | 0.02 | 0.904 | 0.139 | 1.18 × 10−10 |
| SIGLEC11 | rs375426790 | 19:50451606 | G | A | 0.02 | 0.897 | 0.14 | 1.92 × 10−10 |
| ZNF350 | rs113541493 | 19:52484200 | G | A | 0.02 | 0.886 | 0.139 | 2.74 × 10−10 |
| CACNG8 | rs1280762104 | 19:54478295 | C | T | 0.02 | 0.858 | 0.139 | 9.13 × 10−10 |
| LINC01733 | rs73601743 | 20:25947923 | C | T | 0.019 | 0.888 | 0.14 | 3.28 × 10−10 |
| FRG1BP | rs138922778 | 20:29632727 | D | I | 0.019 | 0.87 | 0.136 | 2.08 × 10−10 |
| ZHX3 | rs1363587961 | 20:39898770 | G | A | 0.019 | 0.894 | 0.14 | 2.35 × 10−10 |
| SNAI1 | rs749239193 | 20:48600541 | C | T | 0.019 | 0.612 | 0.104 | 4.32 × 10−9 |
| non-coding | rs113148353 | 21:14361624 | C | T | 0.019 | 0.834 | 0.137 | 1.40 × 10−9 |
| non-coding | rs79163003 | 21:47475650 | A | G | 0.02 | 0.85 | 0.139 | 1.25 × 10−9 |
| DIP2A | rs368500330 | 21:47986571 | C | T | 0.019 | 0.913 | 0.14 | 9.18 × 10−11 |
| non-coding | rs61731379 | 22:22730619 | A | T | 0.019 | 0.904 | 0.14 | 1.33 × 10−10 |
| non-coding | rs67984407 | 22:24230687 | C | T | 0.019 | 0.907 | 0.14 | 1.45 × 10−10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Wang, Y.; Li, X.; Zhang, Y.; Yuan, L.; Chen, D.; Wang, X. PDE3A and GSK3B as Atrial Fibrillation Susceptibility Genes in the Chinese Population via Bioinformatics and Genome-Wide Association Analysis. Biomedicines 2023, 11, 908. https://doi.org/10.3390/biomedicines11030908
Zhou Z, Wang Y, Li X, Zhang Y, Yuan L, Chen D, Wang X. PDE3A and GSK3B as Atrial Fibrillation Susceptibility Genes in the Chinese Population via Bioinformatics and Genome-Wide Association Analysis. Biomedicines. 2023; 11(3):908. https://doi.org/10.3390/biomedicines11030908
Chicago/Turabian StyleZhou, Zechen, Yu Wang, Xiaoyi Li, Yinan Zhang, Lichuang Yuan, Dafang Chen, and Xuedong Wang. 2023. "PDE3A and GSK3B as Atrial Fibrillation Susceptibility Genes in the Chinese Population via Bioinformatics and Genome-Wide Association Analysis" Biomedicines 11, no. 3: 908. https://doi.org/10.3390/biomedicines11030908
APA StyleZhou, Z., Wang, Y., Li, X., Zhang, Y., Yuan, L., Chen, D., & Wang, X. (2023). PDE3A and GSK3B as Atrial Fibrillation Susceptibility Genes in the Chinese Population via Bioinformatics and Genome-Wide Association Analysis. Biomedicines, 11(3), 908. https://doi.org/10.3390/biomedicines11030908







