The Antibacterial Effect of Cannabigerol toward Streptococcus mutans Is Influenced by the Autoinducers 21-CSP and AI-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Strains, Culture Conditions and End Point Planktonic Growth Assay
2.3. Growth Kinetics Studies
2.4. Investigation of AI-2 Production
2.5. RNA Isolation
2.6. Quantitative Real-Time PCR
2.7. Statistical Analysis
3. Results
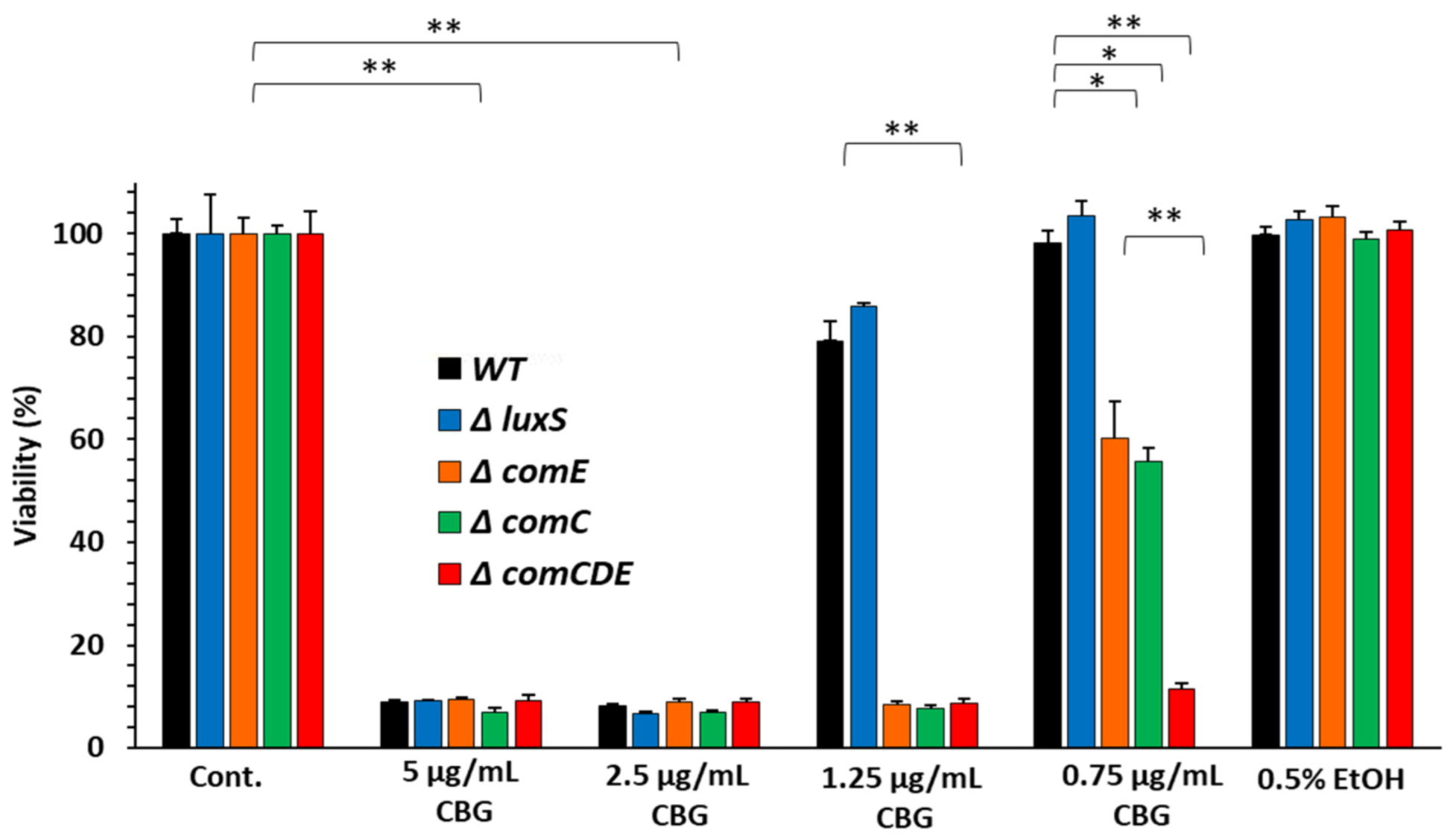
3.1. Streptococcus Mutans Strains Deficient in the comCDE QS System Showed Increased Sensitivity to CBG
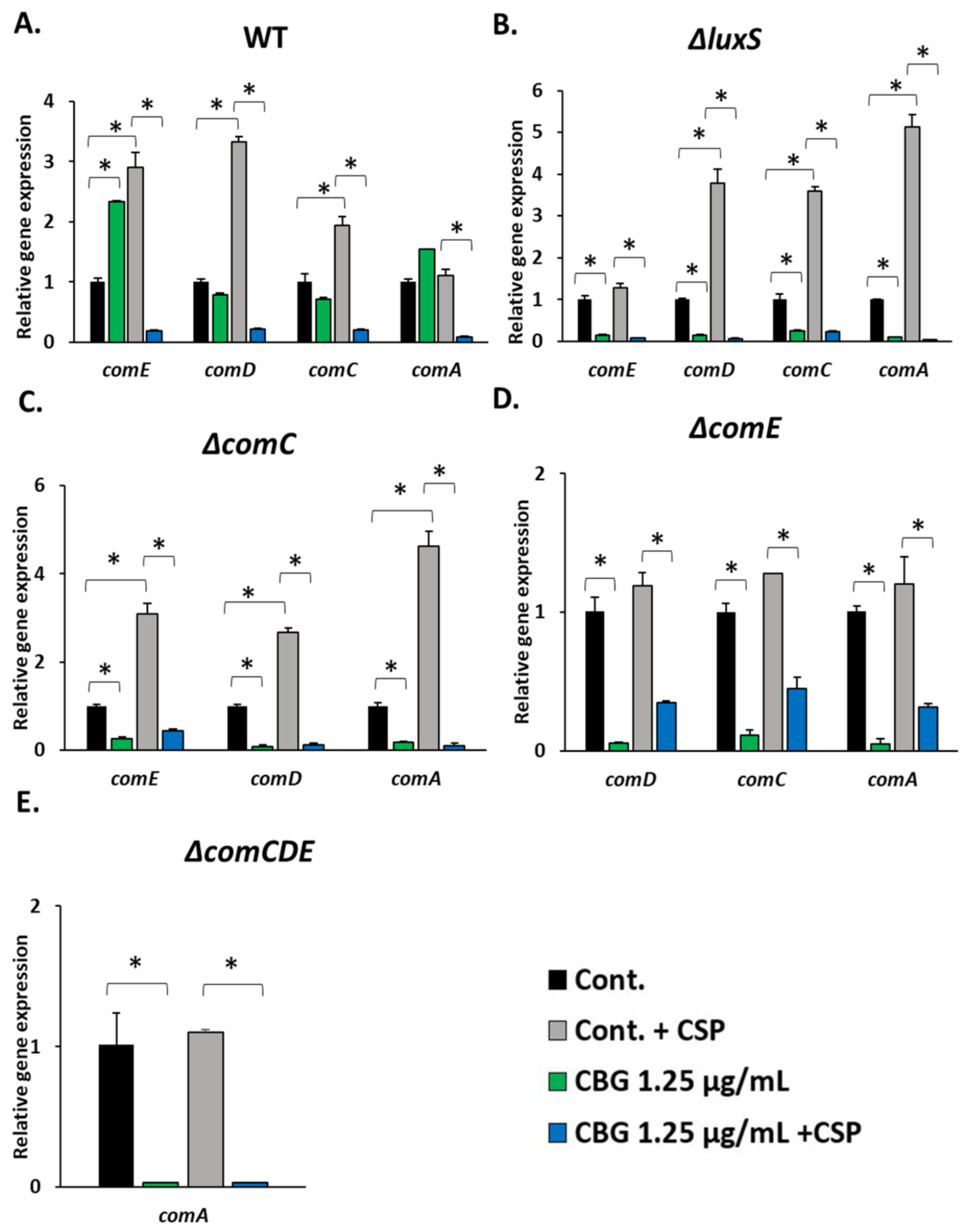
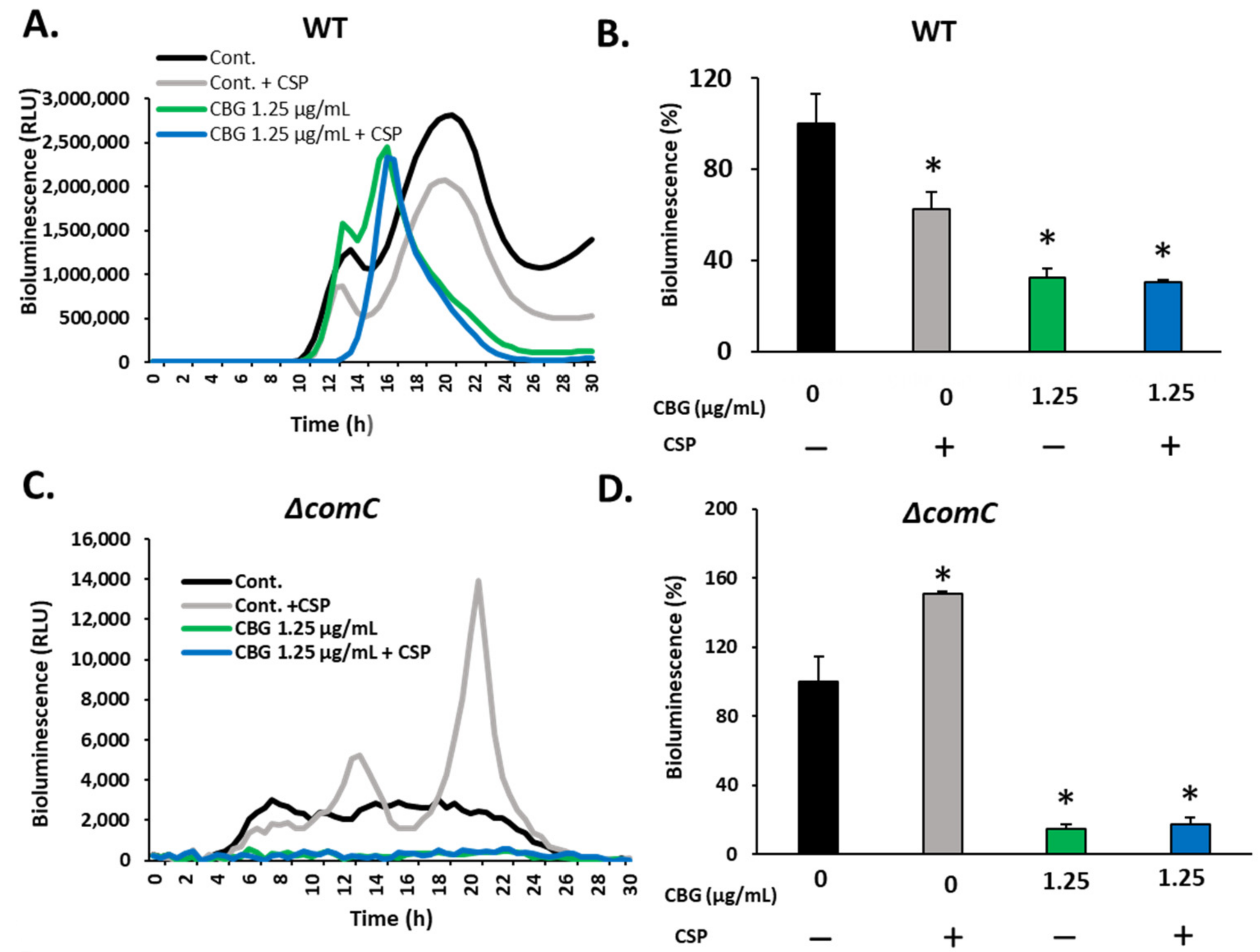
3.2. 21-CSP Prevented the Anti-Bacterial Effect of CBG on the ΔcomC and ΔcomE Strains, but Not on ΔcomCDE
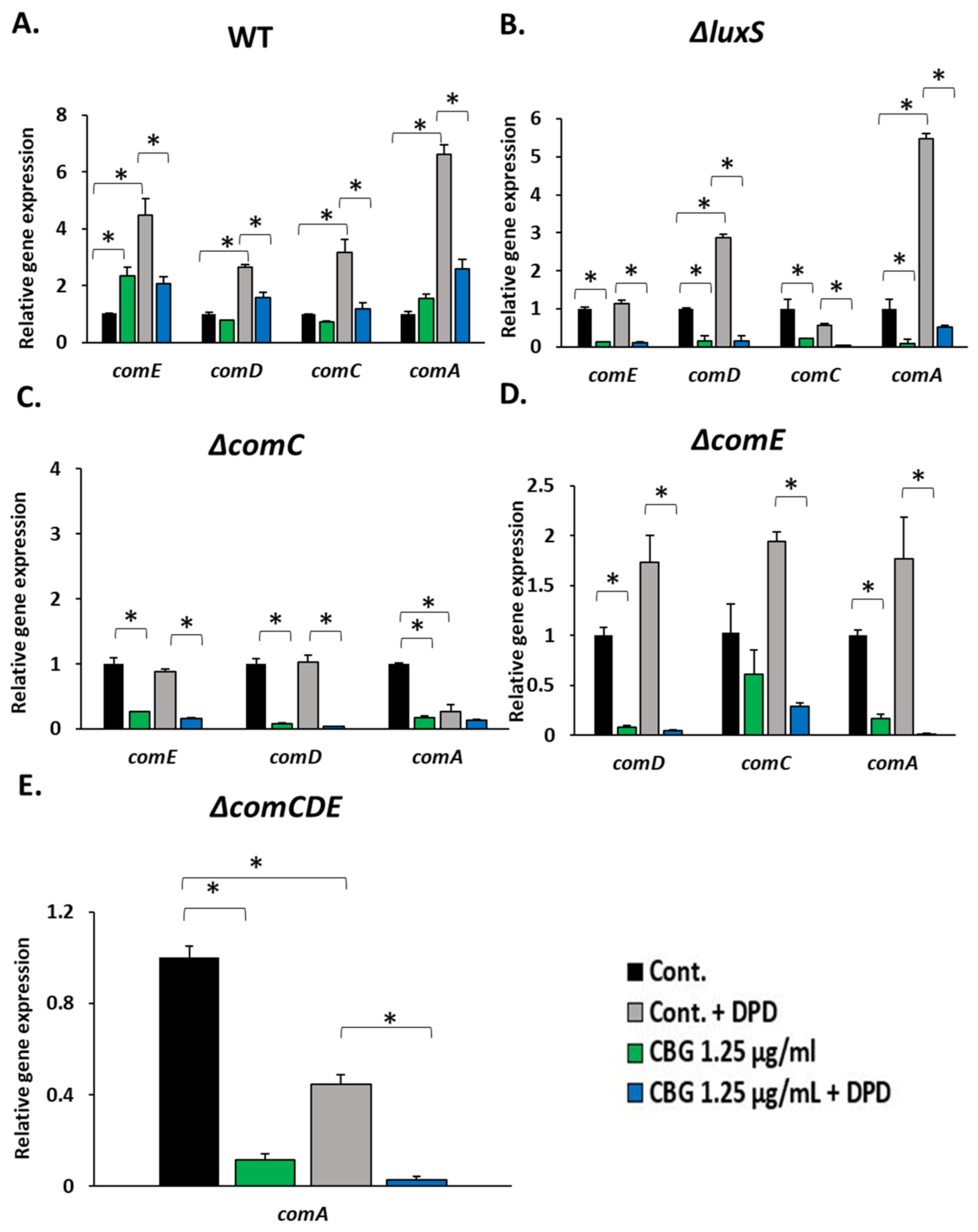
3.3. DPD Increased the Susceptibility of WT and ΔluxS, but Not of ΔcomC or ΔcomE, to CBG
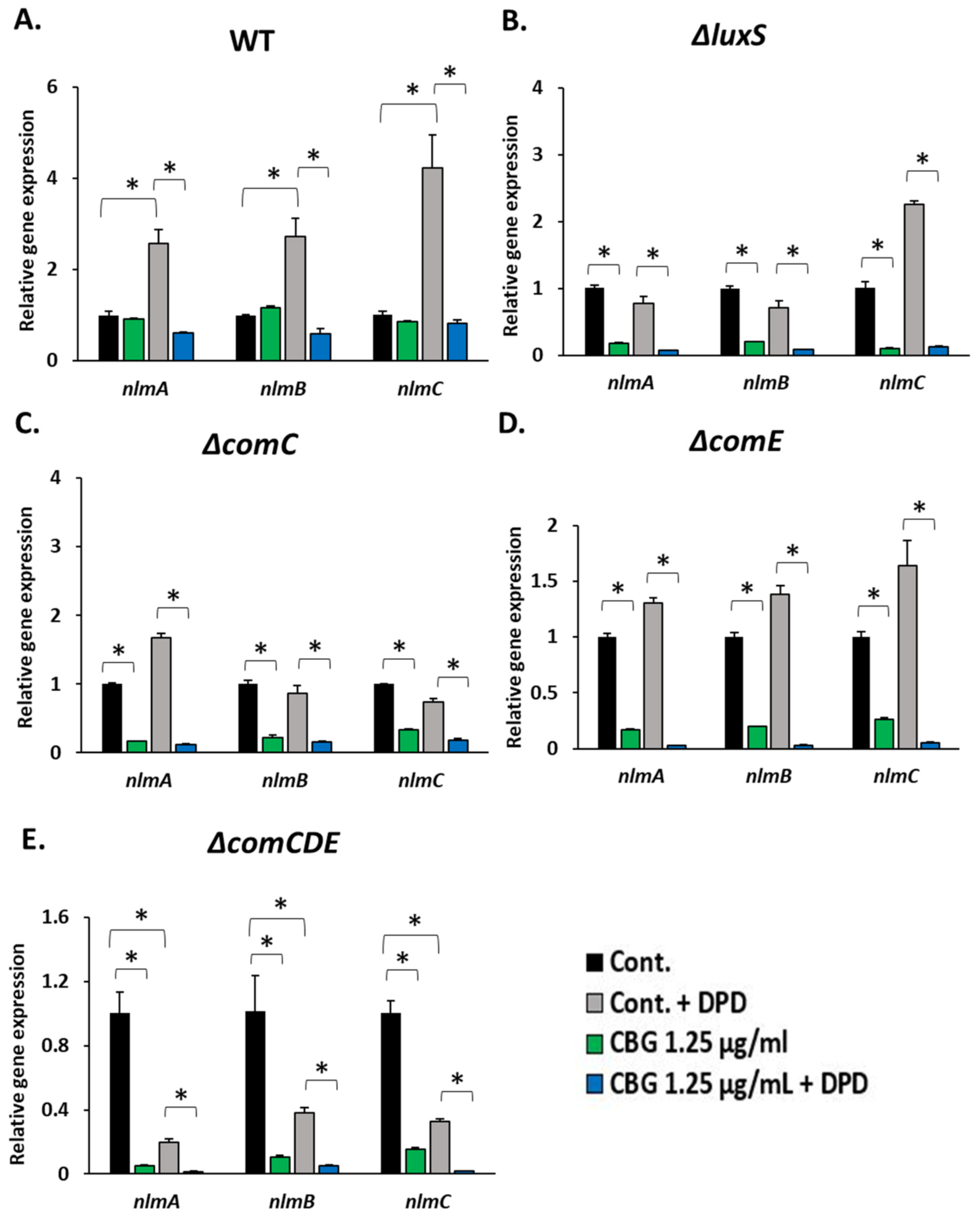
3.4. CBG Antagonizes the 21-CSP-Induced Gene Expression of nlmA, nlmB and nlmC/cipB
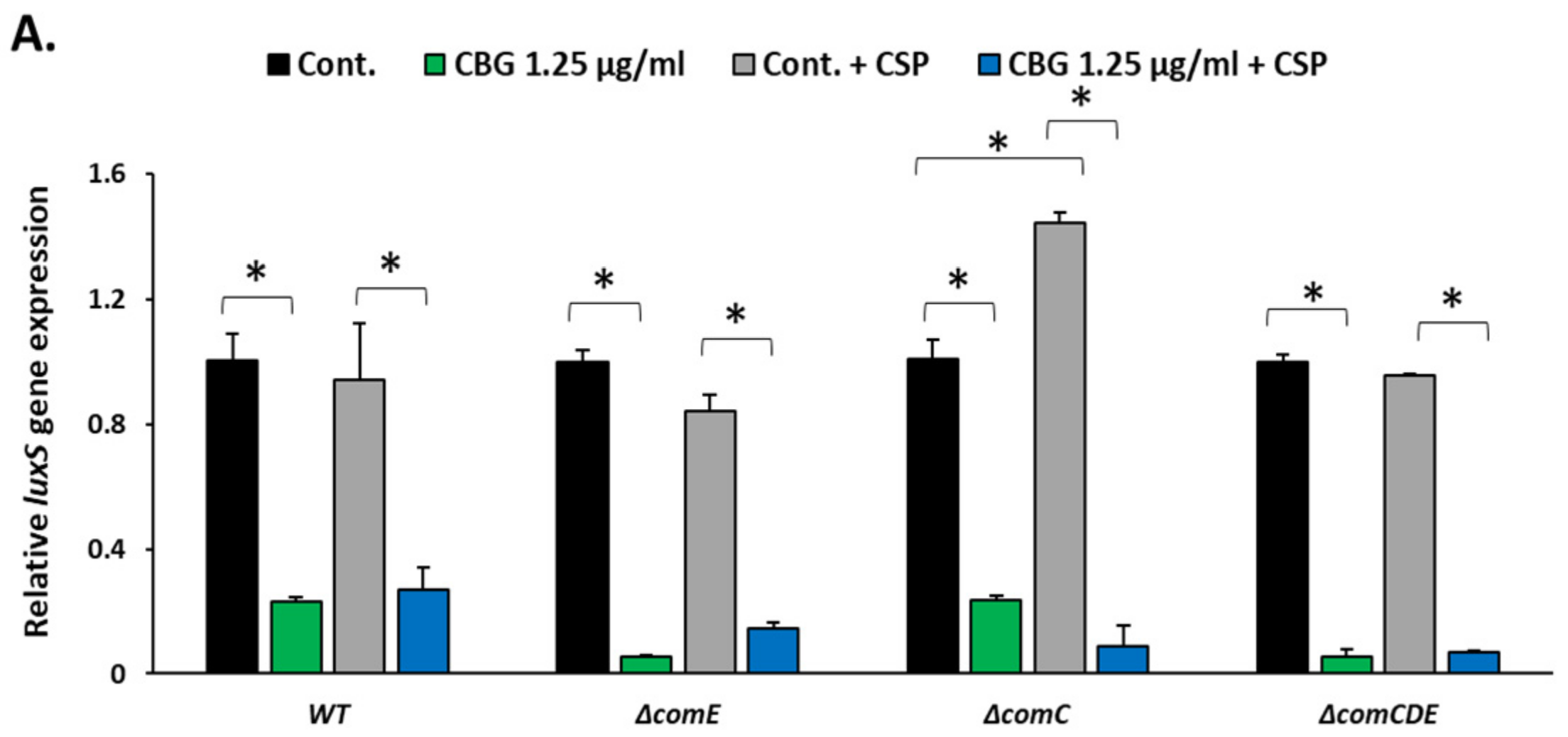
3.5. DPD Induced the Expression of comCDE and ComE-Regulated Genes, While the 21-CSP Did Not Affect the Gene Expression of luxS
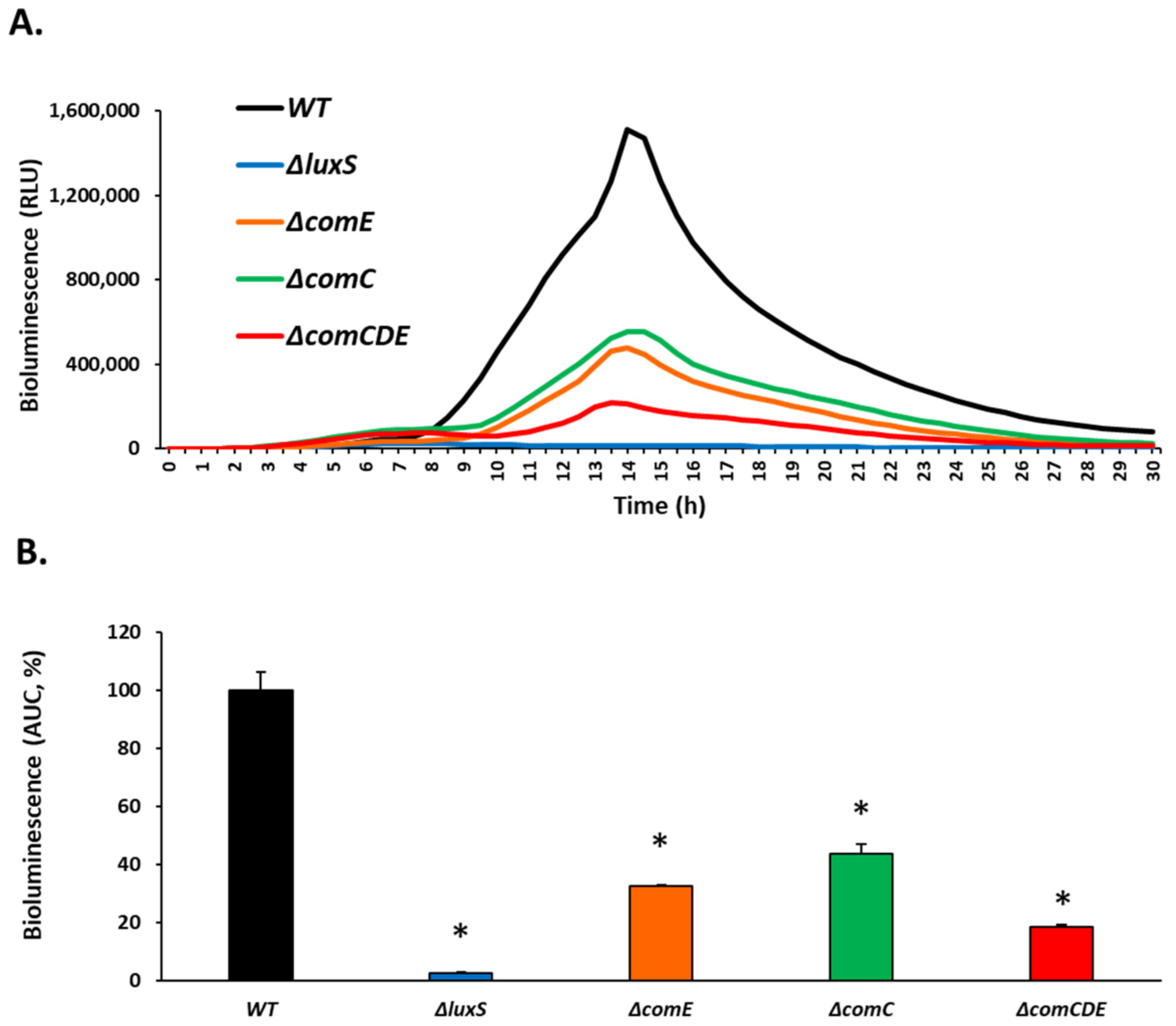
3.6. CBG Significantly Reduced the Production of AI-2
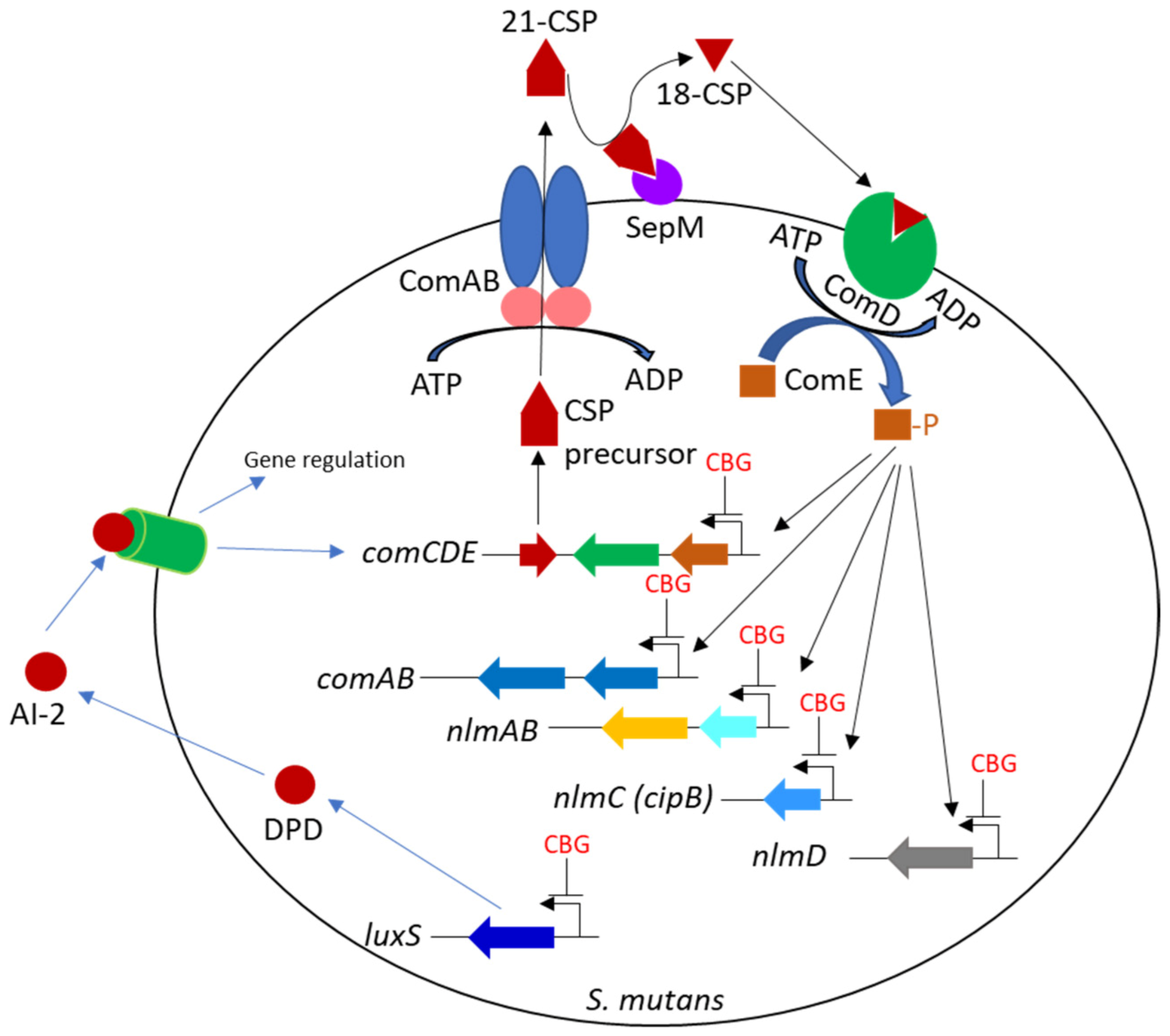
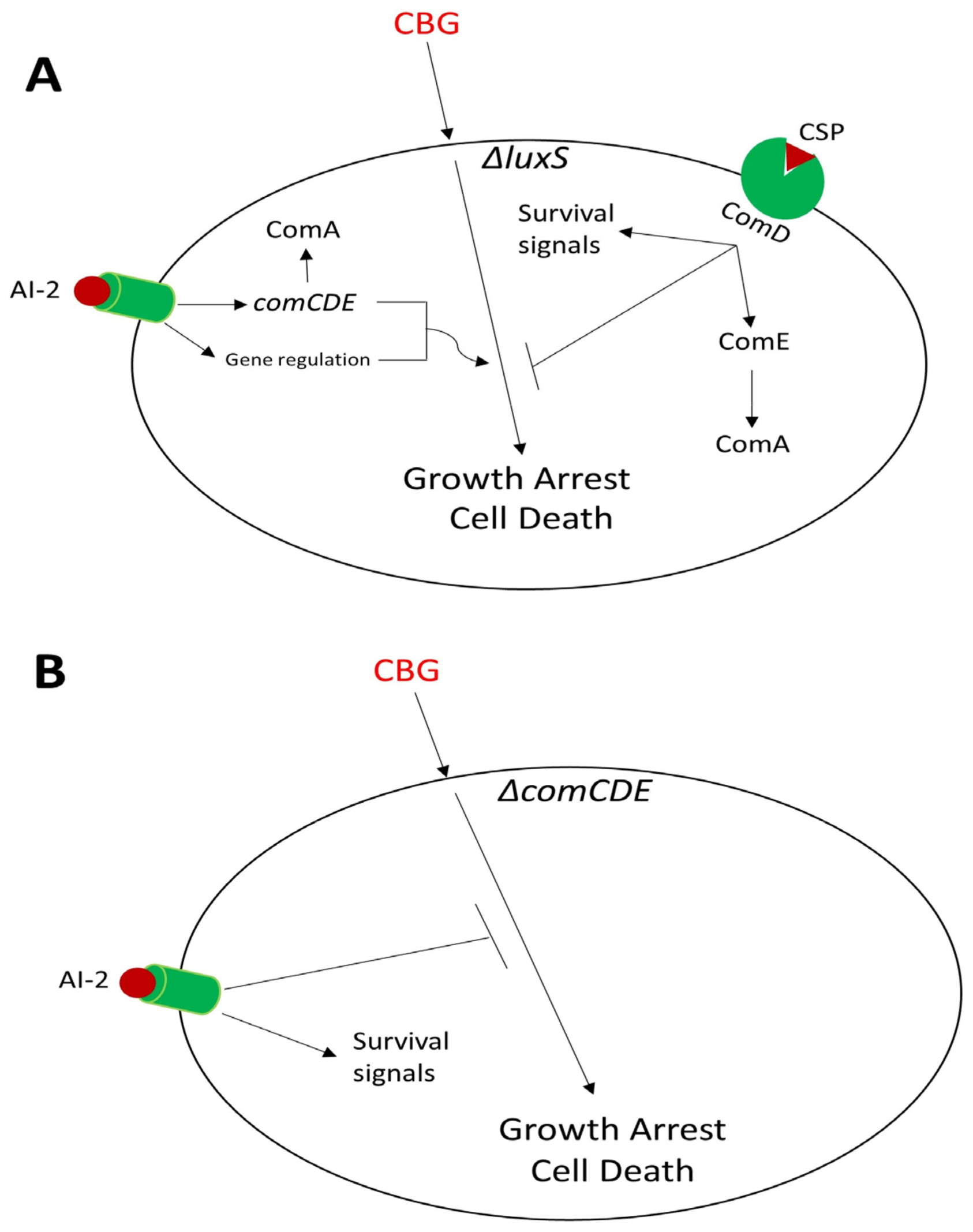
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Labbate, M.; Queck, S.Y.; Koh, K.S.; Rice, S.A.; Givskov, M.; Kjelleberg, S. Quorum sensing-controlled biofilm development in Serratia liquefaciens MG1. J. Bacteriol. 2004, 186, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Sionov, R.V.; Steinberg, D. Targeting the holy triangle of quorum sensing, biofilm formation, and antibiotic resistance in pathogenic bacteria. Microorganisms 2022, 10, 1239. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef]
- Fuqua, C.; Winans, S.C.; Greenberg, E.P. Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 1996, 50, 727–751. [Google Scholar] [CrossRef]
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 2003, 100, 14549–14554. [Google Scholar] [CrossRef]
- Sahreen, S.; Mukhtar, H.; Imre, K.; Morar, A.; Herman, V.; Sharif, S. Exploring the function of quorum sensing regulated biofilms in biological wastewater treatment: A review. Int. J. Mol. Sci. 2022, 23, 9751. [Google Scholar] [CrossRef]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef]
- Xavier, K.B.; Bassler, B.L. LuxS quorum sensing: More than just a numbers game. Curr. Opin. Microbiol. 2003, 6, 191–197. [Google Scholar] [CrossRef]
- Tabchoury, C.P.M.; Sousa, M.C.K.; Arthur, R.A.; Mattos-Graner, R.O.; Del Bel Cury, A.A.; Cury, J.A. Evaluation of genotypic diversity of Streptococcus mutans using distinct arbitrary primers. J. Appl. Oral Sci. 2008, 16, 403–407. [Google Scholar] [CrossRef]
- da Silva, A.C.B.; dos Santos Cruz, J.; Sampaio, F.C.; de Araújo, D.A.M. Detection of oral streptococci in dental biofilm from caries-active and caries-free children. Braz. J. Microbiol. 2008, 39, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Rajesh, S.; Princy, S.A. Plausible drug targets in the Streptococcus mutans quorum sensing pathways to combat dental biofilms and associated risks. Indian J. Microbiol. 2015, 55, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Quadri, L.E.; Kuipers, O.P.; De Vos, W.M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 1997, 24, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Aspiras, M.B.; Ellen, R.P.; Cvitkovitch, D.G. ComX activity of Streptococcus mutans growing in biofilms. FEMS Microbiol. Lett. 2004, 238, 167–174. [Google Scholar]
- Dunny, G.M.; Leonard, B.A. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 1997, 51, 527–564. [Google Scholar] [CrossRef]
- Wright, P.P.; Ramachandra, S.S. Quorum sensing and quorum quenching with a focus on cariogenic and periodontopathic oral biofilms. Microorganisms 2022, 10, 1783. [Google Scholar] [CrossRef]
- Leung, V.; Dufour, D.; Lévesque, C.M. Death and survival in Streptococcus mutans: Differing outcomes of a quorum-sensing signaling peptide. Front. Microbiol. 2015, 6, 1176. [Google Scholar] [CrossRef]
- Zu, Y.; Li, W.; Wang, Q.; Chen, J.; Guo, Q. ComDE two-component signal transduction systems in oral Streptococci: Structure and function. Curr. Issues Mol. Biol. 2019, 32, 201–258. [Google Scholar] [CrossRef]
- Schauder, S.; Bassler, B.L. The languages of bacteria. Genes Dev. 2001, 15, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Sztajer, H.; Lemme, A.; Vilchez, R.; Schulz, S.; Geffers, R.; Yip, C.Y.Y.; Levesque, C.M.; Cvitkovitch, D.G.; Wagner-Döbler, I. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 2008, 190, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Spatafora, G. Gene regulation in S. mutans: Complex control in a complex environment. J. Dent. Res. 2012, 91, 133–141. [Google Scholar] [CrossRef]
- Martínez, V.; Iriondo De-Hond, A.; Borrelli, F.; Capasso, R.; Del Castillo, M.D.; Abalo, R. Cannabidiol and other non-psychoactive cannabinoids for prevention and treatment of gastrointestinal disorders: Useful nutraceuticals? Int.J. Mol. Sci. 2020, 21, 3067. [Google Scholar] [CrossRef]
- Nachnani, R.; Raup-Konsavage, W.M.; Vrana, K.E. The pharmacological case for cannabigerol. J. Pharmacol. Exp. Ther. 2021, 376, 204–212. [Google Scholar] [CrossRef]
- Sionov, R.V.; Steinberg, D. Anti-microbial activity of phytocannabinoids and endocannabinoids in the light of their physiological and pathophysiological roles. Biomedicines 2022, 10, 631. [Google Scholar] [CrossRef] [PubMed]
- Zagzoog, A.; Mohamed, K.A.; Kim, H.J.J.; Kim, E.D.; Frank, C.S.; Black, T.; Jadhav, P.D.; Holbrook, L.A.; Laprairie, R.B. In vitro and in vivo pharmacological activity of minor cannabinoids isolated from Cannabis sativa. Sci. Rep. 2020, 10, 20405. [Google Scholar] [CrossRef]
- Borrelli, F.; Pagano, E.; Romano, B.; Panzera, S.; Maiello, F.; Coppola, D.; De Petrocellis, L.; Buono, L.; Orlando, P.; Izzo, A.A. Colon carcinogenesis is inhibited by the TRPM8 antagonist cannabigerol, a Cannabis-derived non-psychotropic cannabinoid. Carcinogenesis 2014, 35, 2787–2797. [Google Scholar] [CrossRef]
- Pagano, E.; Iannotti, F.A.; Piscitelli, F.; Romano, B.; Lucariello, G.; Venneri, T.; Di Marzo, V.; Izzo, A.A.; Borrelli, F. Efficacy of combined therapy with fish oil and phytocannabinoids in murine intestinal inflammation. Phytother. Res. 2021, 35, 517–529. [Google Scholar] [CrossRef]
- Hill, A.J.; Williams, C.M.; Whalley, B.J.; Stephens, G.J. Phytocannabinoids as novel therapeutic agents in CNS disorders. Pharmacol. Ther. 2012, 133, 79–97. [Google Scholar] [CrossRef]
- Appendino, G.; Gibbons, S.; Giana, A.; Pagani, A.; Grassi, G.; Stavri, M.; Smith, E.; Rahman, M.M. Antibacterial cannabinoids from Cannabis sativa: A structure− activity study. J. Nat. Prod. 2008, 71, 1427–1430. [Google Scholar] [CrossRef] [PubMed]
- Farha, M.A.; El-Halfawy, O.M.; Gale, R.T.; MacNair, C.R.; Carfrae, L.A.; Zhang, X.; Jentsch, N.G.; Magolan, J.; Brown, E.D. Uncovering the hidden antibiotic potential of Cannabis. ACS Infect. Dis. 2020, 6, 338–346. [Google Scholar] [CrossRef]
- Aqawi, M.; Sionov, R.V.; Gallily, R.; Friedman, M.; Steinberg, D. Anti-bacterial properties of cannabigerol toward Streptococcus mutans. Front. Microbiol. 2021, 12, 922. [Google Scholar] [CrossRef] [PubMed]
- Aqawi, M.; Sionov, R.V.; Gallily, R.; Friedman, M.; Steinberg, D. Anti-biofilm activity of cannabigerol against Streptococcus mutans. Microorganisms 2021, 9, 2031. [Google Scholar] [CrossRef] [PubMed]
- Aqawi, M.; Steinberg, D.; Feuerstein, O.; Friedman, M.; Gingichashvili, S. Cannabigerol effect on Streptococcus mutans biofilms—A computational approach to confocal image analysis. Front. Microbiol. 2022, 13, 880993. [Google Scholar] [CrossRef]
- Aqawi, M.; Gallily, R.; Sionov, R.V.; Zaks, B.; Friedman, M.; Steinberg, D. Cannabigerol prevents quorum sensing and biofilm formation of Vibrio harveyi. Front. Microbiol. 2020, 11, 858. [Google Scholar] [CrossRef] [PubMed]
- Asfour, H.Z. Anti-quorum sensing natural compounds. J. Microsc. Ultrastruct. 2018, 6, 1. [Google Scholar] [CrossRef]
- Wen, Z.T.; Burne, R.A. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 2004, 186, 2682–2691. [Google Scholar] [CrossRef]
- Yoshida, A.; Kuramitsu, H.K. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 2002, 68, 6283–6291. [Google Scholar] [CrossRef]
- Shemesh, M.; Tam, A.; Aharoni, R.; Steinberg, D. Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces. BMC Microbiol. 2010, 10, 51. [Google Scholar] [CrossRef]
- Mok, K.C.; Wingreen, N.S.; Bassler, B.L. Vibrio harveyi quorum sensing: A coincidence detector for two autoinducers controls gene expression. EMBO J. 2003, 22, 870–881. [Google Scholar] [CrossRef]
- Anetzberger, C.; Reiger, M.; Fekete, A.; Schell, U.; Stambrau, N.; Plener, L.; Kopka, J.; Schmitt-Kopplin, P.; Hilbi, H.; Jung, K. Autoinducers act as biological timers in Vibrio harveyi. PLoS ONE 2012, 7, e48310. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Chiou, J.; Chatterjee, P.; Otto, M. Alternative approaches to treat bacterial infections: Targeting quorum-sensing. Expert Rev. Anti-Infect. Ther. 2020, 18, 499–510. [Google Scholar] [CrossRef]
- Zhou, Z.; Wu, X.; Li, J.; Zhang, Y.; Huang, Y.; Zhang, W.; Shi, Y.; Wang, J.; Chen, S. A novel quorum quencher, Rhodococcus pyridinivorans XN-36, is a powerful agent for the biocontrol of soft rot disease in various host plants. Biol. Control 2022, 169, 104889. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, W.-J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.-H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2022, 13, 1063393. [Google Scholar] [CrossRef]
- Haque, S.; Ahmad, F.; Dar, S.A.; Jawed, A.; Mandal, R.K.; Wahid, M.; Lohani, M.; Khan, S.; Singh, V.; Akhter, N. Developments in strategies for quorum sensing virulence factor inhibition to combat bacterial drug resistance. Microb. Pathog. 2018, 121, 293–302. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef]
- Koh, C.-L.; Sam, C.-K.; Yin, W.-F.; Tan, L.Y.; Krishnan, T.; Chong, Y.M.; Chan, K.-G. Plant-derived natural products as sources of anti-quorum sensing compounds. Sensors 2013, 13, 6217–6228. [Google Scholar] [CrossRef]
- Yu, H.; Chen, W.; Bhatt, K.; Zhou, Z.; Zhu, X.; Liu, S.; He, J.; Zhang, L.; Chen, S.; Wang, H. A novel bacterial strain Burkholderia sp. F25 capable of degrading diffusible signal factor signal shows strong biocontrol potential. Front. Plant Sci. 2022, 13, 1071693. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, N.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Community structure and functional gene profile of bacteria on healthy and diseased thalli of the red seaweed Delisea pulchra. PLoS ONE 2012, 7, e50854. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.; Shaaban, M.I.; Abdel Bar, F.M.; El-Mahdy, A.M.; Shokralla, S. Quorum sensing inhibiting activity of Streptomyces coelicoflavus isolated from soil. Front. Microbiol. 2016, 7, 659. [Google Scholar] [CrossRef]
- Bouyahya, A.; Chamkhi, I.; Balahbib, A.; Rebezov, M.; Shariati, M.A.; Wilairatana, P.; Mubarak, M.S.; Benali, T.; El Omari, N. Mechanisms, anti-quorum-sensing actions, and clinical trials of medicinal plant bioactive compounds against bacteria: A comprehensive review. Molecules 2022, 27, 1484. [Google Scholar] [CrossRef] [PubMed]
- Vargas, E.L.G.; de Almeida, F.A.; de Freitas, L.L.; Pinto, U.M.; Vanetti, M.C.D. Plant compounds and nonsteroidal anti-inflammatory drugs interfere with quorum sensing in Chromobacterium violaceum. Arch. Microbiol. 2021, 203, 5491–5507. [Google Scholar] [CrossRef] [PubMed]
- Topa, S.H.; Palombo, E.A.; Kingshott, P.; Blackall, L.L. Activity of cinnamaldehyde on quorum sensing and biofilm susceptibility to antibiotics in Pseudomonas aeruginosa. Microorganisms 2020, 8, 455. [Google Scholar] [CrossRef]
- Swem, L.R.; Swem, D.L.; O’Loughlin, C.T.; Gatmaitan, R.; Zhao, B.; Ulrich, S.M.; Bassler, B.L. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol. Cell 2009, 35, 143–153. [Google Scholar] [CrossRef]
- Vetrivel, A.; Natchimuthu, S.; Subramanian, V.; Murugesan, R. High-throughput virtual screening for a new class of antagonist targeting LasR of Pseudomonas aeruginosa. ACS Omega 2021, 6, 18314–18324. [Google Scholar] [CrossRef]
- Tapia-Rodriguez, M.R.; Hernandez-Mendoza, A.; Gonzalez-Aguilar, G.A.; Martinez-Tellez, M.A.; Martins, C.M.; Ayala-Zavala, J.F. Carvacrol as potential quorum sensing inhibitor of Pseudomonas aeruginosa and biofilm production on stainless steel surfaces. Food Control 2017, 75, 255–261. [Google Scholar] [CrossRef]
- Ryu, D.-H.; Lee, S.-W.; Mikolaityte, V.; Kim, Y.-W.; Jeong, H.Y.; Lee, S.J.; Lee, C.-H.; Lee, J.-K. Identification of a second type of AHL-lactonase from Rhodococcus sp. BH4, belonging to the α/β hydrolase superfamily. J. Microbiol. Biotechnol. 2020, 30, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Bouyahya, A.; Dakka, N.; Et-Touys, A.; Abrini, J.; Bakri, Y. Medicinal plant products targeting quorum sensing for combating bacterial infections. Asian Pac. J. Trop. Med. 2017, 10, 729–743. [Google Scholar] [CrossRef]
- Lei, L.; Long, L.; Yang, X.; Qiu, Y.; Zeng, Y.; Hu, T.; Wang, S.; Li, Y. The VicRK two-component system regulates Streptococcus mutans virulence. Curr. Issues Mol. Biol. 2019, 32, 167–200. [Google Scholar] [CrossRef]
- Biswas, S.; Cao, L.; Kim, A.; Biswas, I. SepM, a streptococcal protease involved in quorum sensing, displays strict substrate specificity. J. Bacteriol. 2016, 198, 436–447. [Google Scholar] [CrossRef]
- Vendeville, A.; Winzer, K.; Heurlier, K.; Tang, C.M.; Hardie, K.R. Making’sense’of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 2005, 3, 383–396. [Google Scholar] [CrossRef]
- Wang, Y.; Bian, Z.; Wang, Y. Biofilm formation and inhibition mediated by bacterial quorum sensing. Appl. Microbiol. Biotechnol. 2022, 106, 6365–6381. [Google Scholar] [CrossRef]
- Niazy, A.A. LuxS quorum sensing system and biofilm formation of oral microflora: A short review article. Saudi Dent. J. 2021, 33, 116–123. [Google Scholar] [CrossRef]
- Senadheera, D.B.; Cordova, M.; Ayala, E.A.; Chávez de Paz, L.E.; Singh, K.; Downey, J.S.; Svensäter, G.; Goodman, S.D.; Cvitkovitch, D. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans. J. Bacteriol. 2012, 194, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Prudhomme, M.; Alloing, G.; Granadel, C.; Claverys, J.P. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 2000, 38, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.A.; Petersen, F.C.; Scheie, A.A. AI-2/LuxS is involved in increased biofilm formation by Streptococcus intermedius in the presence of antibiotics. Antimicrob. Agents Chemother. 2009, 53, 4258–4263. [Google Scholar] [CrossRef]
- Cao, M.; Feng, Y.; Wang, C.; Zheng, F.; Li, M.; Liao, H.; Mao, Y.; Pan, X.; Wang, J.; Hu, D. Functional definition of LuxS, an autoinducer-2 (AI-2) synthase and its role in full virulence of Streptococcus suis serotype 2. J. Microbiol. 2011, 49, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, J.R.; Walker, A.R. Expanding the vocabulary of peptide signals in Streptococcus mutans. Front. Cell. Infect. Microbiol. 2019, 9, 194. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aqawi, M.; Sionov, R.V.; Friedman, M.; Steinberg, D. The Antibacterial Effect of Cannabigerol toward Streptococcus mutans Is Influenced by the Autoinducers 21-CSP and AI-2. Biomedicines 2023, 11, 668. https://doi.org/10.3390/biomedicines11030668
Aqawi M, Sionov RV, Friedman M, Steinberg D. The Antibacterial Effect of Cannabigerol toward Streptococcus mutans Is Influenced by the Autoinducers 21-CSP and AI-2. Biomedicines. 2023; 11(3):668. https://doi.org/10.3390/biomedicines11030668
Chicago/Turabian StyleAqawi, Muna, Ronit Vogt Sionov, Michael Friedman, and Doron Steinberg. 2023. "The Antibacterial Effect of Cannabigerol toward Streptococcus mutans Is Influenced by the Autoinducers 21-CSP and AI-2" Biomedicines 11, no. 3: 668. https://doi.org/10.3390/biomedicines11030668
APA StyleAqawi, M., Sionov, R. V., Friedman, M., & Steinberg, D. (2023). The Antibacterial Effect of Cannabigerol toward Streptococcus mutans Is Influenced by the Autoinducers 21-CSP and AI-2. Biomedicines, 11(3), 668. https://doi.org/10.3390/biomedicines11030668






