Advances of Genome Editing with CRISPR/Cas9 in Neurodegeneration: The Right Path towards Therapy
Abstract
1. Introduction
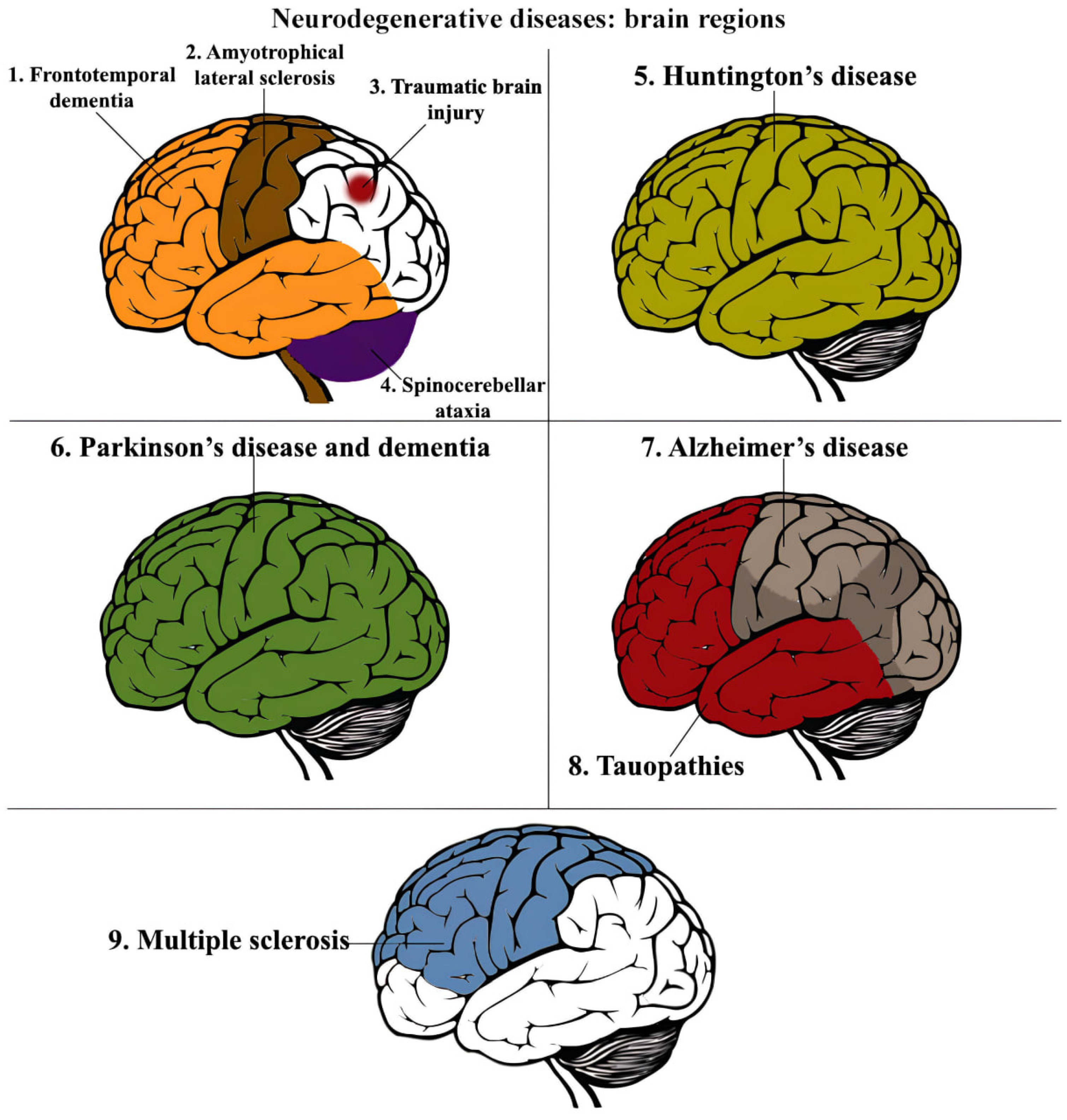
2. Hallmarks of Neurodegenerative Disorders
- Disrupted proteostasis [18];
- Metabolic changes in neuroimmune cells that result in morphological alterations in glial cells and the microenvironment of the neuroimmune system [19];
- Chronic inflammation, which was traditionally viewed as a protective function of the body but is now recognized as a hallmark of NDDs. Chronic inflammation can lead to focal cell death as a containment strategy, limiting the ability of pathogens and oncogenic cells to divide and spread. And this can manifest into NDDs [22].
3. Neuroimmune Dysfunction
- Microglia, which play an active role in the immune response of the central nervous system by producing pro-inflammatory and anti-inflammatory cytokines (M1 and M2 subtypes of microglia) [28].
- Astrocytes, which regulate the restoration of the nervous system through their control over biochemical processes in epithelial cells of the blood–brain barrier (BBB) and their activation of the repair and scarring processes following the innate immune response [29].
- Oligodendrocytes, which provide support, protection, and growth of axons [30].
3.1. Effect of Microglia in Neuroinflammation
3.2. Astroglial Scar Formation
3.3. Oligodendroglia and Myelin
| CNS disease | |||
| Acute damage | Amyotrophic lateral sclerosis | Multiple sclerosis | Alzheimer’s disease |
| Responder cells | |||
| Astrocytes, microglia, meningeal cells, fibroblasts | Astrocytes, microglia, meningeal cells, fibroblasts, oligodendrocytes | Astrocytes, microglia, meningeal cells, fibroblasts, oligodendrocytes, endothelial cells | Astrocytes, microglia, fibroblasts, smooth muscle cells |
| Cell mediators and biomarkers | |||
| Thrombin, MMP-9, ATP, PDGFRβ, TGFβ, GFAP | IL-6, CXCL1, CXCL10, CXCL12, TNFα, TGFβ, NGF, INFγ, PGD2, ADAMTS-4, CTGF, S100A4, MMP-9, GFAP | PDGFRβ, TGFβ, myelin, GFAP | PDGFRβ, TGFβ, GFAP |
| Extracellular matrix proteins | |||
| Fibronectin, laminin, collagen, CSPGs, tenasein, HSPGs | Fibronectin, collagen type IV, CSPGs, Sema3A, fibrin, vimentin, thrombin | Fibronectin, collagen, biglycan, decorin, CSPGs | Fibronectin, collagen, biglycan, decorin, CSPGs |
4. New Types of Treatment: Gene Therapy and Genome Editing Technologies
- Viral vectors can efficiently deliver therapeutic cargo to target cells and ensure its sustained presence over an extended period. This is important for diseases requiring long-term treatment, such as chronic neurological disorders;
- Viral vectors’ ability to efficiently infect postmitotic cells, including neurons in the brain, is a valuable characteristic. Many neurological disorders involve dysfunctional or damaged neurons, and viral vectors offer an effective means of delivering therapeutic cargo directly to these cells;
- Viral vectors used in gene transfer have been engineered to have low immunogenicity, meaning they are less likely to trigger an immune response. Additionally, extensive research has focused on reducing the toxicity associated with viral vectors, making them safer for use in gene therapy;
- Viral vectors’ compatibility with other forms of therapy approaches, including pharmacological treatments or surgical interventions. This compatibility allows for combination therapies that may enhance overall treatment outcomes.
4.1. Lentiviral Vectors
4.2. AAV Vectors
4.3. Adenoviral Vectors
4.4. Genome Editing Technologies
5. Neurodegenerative Disorders’ Therapeutic Targets and Their Application
5.1. Sox9
5.2. RGMa
5.3. MAG
5.4. Lin28
5.5. Notch1
5.6. Msi1
5.7. Prom1
6. Overview of Clinical Trials for Treatment of NDDs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AAV | adeno-associated virus |
| AD | Alzheimer’s disease |
| ALS | amyotrophic lateral sclerosis |
| BBB | blood brain barrier |
| CSPG | chondroitin sulfate proteoglycans |
| CNS | central nervous system |
| CRISPR/Cas | clustered regularly interspaced short palindromic repeats/CRISPR-associated protein |
| crRNA | crisprRNA |
| DAM | disease-associated microglia |
| DAMP | damage-associated molecular patterns |
| DRG | dorsal root ganglion |
| DSB | double-strand break |
| IDLV | integrase-deficient lentivirus |
| ECM | extracellular matrix |
| HR | homologous recombination |
| LV | lentivirus |
| MAG | myelin-associated glycoprotein |
| MBP | myelin basic protein |
| MCT | monocarboxylate transporter |
| MSA | multiple system atrophy |
| MS | multiple sclerosis |
| NAMP | neurodegeneration-associated molecular patterns |
| NDD | neurodegenerative disorder |
| NHEJ | non-homologous end joining |
| OPC | oligodendrocyte precursor cell |
| ORF | open reading frame |
| PAM | protospacer adjacent motif |
| PAMP | pathogen-associated molecular patterns |
| PD | Parkinson’s disease |
| PNS | peripheral nervous system |
| PRR | pattern recognition receptors |
| RGMa | repulsive guidance molecule A |
| RNP | ribonucleoprotein complex |
| sgRNA | single guide RNA |
| TALEN | transcription activator-like effector nuclease |
| TLR | Toll-like receptor |
| VLP | virus-like particle |
| ZNF | zinc finger nuclease |
References
- GBD 2019 Dementia Forecasting Collaborators. Estimation of the Global Prevalence of Dementia in 2019 and Forecasted Prevalence in 2050: An Analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105–e125. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M.; Cookson, M.R.; Van Den Bosch, L.; Zetterberg, H.; Holtzman, D.M.; Dewachter, I. Hallmarks of Neurodegenerative Diseases. Cell 2023, 186, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, S.; Janelidze, S.; Quiroz, Y.T.; Zetterberg, H.; Lopera, F.; Stomrud, E.; Su, Y.; Chen, Y.; Serrano, G.E.; Leuzy, A.; et al. Discriminative Accuracy of Plasma Phospho-Tau217 for Alzheimer Disease vs Other Neurodegenerative Disorders. JAMA J. Am. Med. Assoc. 2020, 324, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Watt, A.D.; Jenkins, N.L.; McColl, G.; Collins, S.; Desmond, P.M. Ethical Issues in the Treatment of Late-Stage Alzheimer’s Disease. J. Alzheimer’s Dis. 2019, 68, 1311–1316. [Google Scholar] [CrossRef] [PubMed]
- Mattsson-Carlgren, N.; Janelidze, S.; Palmqvist, S.; Cullen, N.; Svenningsson, A.L.; Strandberg, O.; Mengel, D.; Walsh, D.M.; Stomrud, E.; Dage, J.L.; et al. Longitudinal Plasma P-Tau217 Is Increased in Early Stages of Alzheimer’s Disease. Brain 2021, 143, 3234–3241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S.; Dong, Z. Classification of Alzheimer Disease Based on Structural Magnetic Resonance Imaging by Kernel Support Vector Machine Decision Tree. Prog. Electromagn. Res. 2014, 144, 171–184. [Google Scholar] [CrossRef]
- Leuzy, A.; Chiotis, K.; Lemoine, L.; Gillberg, P.G.; Almkvist, O.; Rodriguez-Vieitez, E.; Nordberg, A. Tau PET Imaging in Neurodegenerative Tauopathies—Still a Challenge. Mol. Psychiatry 2019, 24, 1112–1134. [Google Scholar] [CrossRef]
- Foran, A.M.; Mathias, J.L.; Bowden, S.C. Effectiveness of Sorting Tests for Detecting Cognitive Decline in Older Adults with Dementia and Other Common Neurodegenerative Disorders: A Meta-Analysis. Neurosci. Biobehav. Rev. 2021, 120, 442–454. [Google Scholar] [CrossRef]
- Aung, Y.Y.M.; Wong, D.C.S.; Ting, D.S.W. The Promise of Artificial Intelligence: A Review of the Opportunities and Challenges of Artificial Intelligence in Healthcare. Br. Med. Bull. 2021, 139, 4–15. [Google Scholar] [CrossRef]
- Termine, A.; Fabrizio, C.; Strafella, C.; Caputo, V.; Petrosini, L.; Caltagirone, C.; Giardina, E.; Cascella, R. Multi-Layer Picture of Neurodegenerative Diseases: Lessons from the Use of Big Data through Artificial Intelligence. J. Pers. Med. 2021, 11, 280. [Google Scholar] [CrossRef]
- Grasso, M.; Piscopo, P.; Confaloni, A.; Denti, M.A. Circulating miRNAs as Biomarkers for Neurodegenerative Disorders. Molecules 2014, 19, 6891–6910. [Google Scholar] [CrossRef] [PubMed]
- Wolinsky, F.D.; Mahncke, H.W.; Kosinski, M.; Unverzagt, F.W.; Smith, D.M.; Jones, R.N.; Stoddard, A.; Tennstedt, S.L. The ACTIVE Cognitive Training Trial and Predicted Medical Expenditures. BMC Health Serv. Res. 2009, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Parkinson’s Disease in Adults: Diagnosis and Management; National Institute for Health and Care Excellence: Guidelines; National Institute for Health and Care Excellence (NICE): London, UK, 2017; ISBN 978-1-4731-2530-8.
- Kallio, E.L.; Öhman, H.; Kautiainen, H.; Hietanen, M.; Pitkälä, K. Cognitive Training Interventions for Patients with Alzheimer’s Disease: A Systematic Review. J. Alzheimer’s Dis. 2017, 56, 1349–1372. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, D.; Hernández-Martínez, A.; Valenzuela, P.L.; Morales, J.S.; Soriano-Maldonado, A. Effects of Physical Exercise on Plasma Brain-Derived Neurotrophic Factor in Neurodegenerative Disorders: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Neurosci. Biobehav. Rev. 2021, 128, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P.; Brown, R.H.; Cleveland, D.W. Decoding ALS: From Genes to Mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.C.; Polymenidou, M.; Cleveland, D.W. Converging Mechanisms in Als and FTD: Disrupted RNA and Protein Homeostasis. Neuron 2013, 79, 416–438. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Proteostasis and Aging. Nat. Med. 2015, 21, 1406–1415. [Google Scholar] [CrossRef]
- Ransohoff, R.M. Ransohoff2016. Science 2016, 353, 168–175. [Google Scholar]
- Barnham, K.J.; Masters, C.L.; Bush, A.I. Neurodegenerative Diseases and Oxidatives Stress. Nat. Rev. Drug Discov. 2004, 3, 205–214. [Google Scholar] [CrossRef]
- Hetz, C.; Saxena, S. ER Stress and the Unfolded Protein Response in Neurodegeneration. Nat. Rev. Neurol. 2017, 13, 477–491. [Google Scholar] [CrossRef]
- Richards, R.I.; Robertson, S.A.; Kastner, D.L. Neurodegenerative Diseases Have Genetic Hallmarks of Autoinflammatory Disease. Hum. Mol. Genet. 2018, 27, R108–R118. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.P.; Patil, C.K.; Rodier, F.; Sun, Y.; Muñoz, D.P.; Goldstein, J.; Nelson, P.S.; Desprez, P.Y.; Campisi, J. Senescence-Associated Secretory Phenotypes Reveal Cell-Nonautonomous Functions of Oncogenic RAS and the P53 Tumor Suppressor. PLoS Biol. 2008, 6, e301. [Google Scholar] [CrossRef] [PubMed]
- Bussian, T.J.; Aziz, A.; Meyer, C.F.; Swenson, B.L.; van Deursen, J.M.; Baker, D.J. Clearance of Senescent Glial Cells Prevents Tau-Dependent Pathology and Cognitive Decline. Nature 2018, 562, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; Lebrasseur, N.K.; Childs, B.G.; Van De Sluis, B.; Kirkland, J.L.; Van Deursen, J.M. Clearance of P16 Ink4a-Positive Senescent Cells Delays Ageing-Associated Disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A New Immune–Metabolic Viewpoint for Age-Related Diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Scheiblich, H.; Trombly, M.; Ramirez, A.; Heneka, M.T. Neuroimmune Connections in Aging and Neurodegenerative Diseases. Trends Immunol. 2020, 41, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Appel, S.H.; Zhao, W.; Beers, D.R.; Henkel, J.S. The Microglial-Motoneuron Dialogue in ALS. Acta Myol. 2011, 30, 4–8. [Google Scholar]
- Phatnani, H.; Maniatis, T. Astrocytes in Neurodegenerative Disease. Cold Spring Harb. Perspect. Biol. 2015, 7, a020628. [Google Scholar] [CrossRef]
- Han, S.; Gim, Y.; Jang, E.H.; Hur, E.M. Functions and Dysfunctions of Oligodendrocytes in Neurodegenerative Diseases. Front. Cell. Neurosci. 2022, 16, 1083159. [Google Scholar] [CrossRef]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System during Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef]
- Dadwal, S.; Heneka, M.T. Microglia Heterogeneity in Health and Disease. FEBS Open Bio. 2023. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.S.; Sohn, D.H. Damage-Associated Molecular Patterns in Inflammatory Diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate Immune Activation in Neurodegenerative Disease. Nat. Rev. Immunol. 2014, 14, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Poupot, R.; Bergozza, D.; Fruchon, S. Nanoparticle-Based Strategies to Treat Neuro-Inflammation. Materials 2018, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Kigerl, K.A.; de Rivero Vaccari, J.P.; Dietrich, W.D.; Popovich, P.G.; Keane, R.W. Pattern Recognition Receptors and Central Nervous System Repair. Exp. Neurol. 2014, 258, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Stuart, L.M.; Bell, S.A.; Stewart, C.R.; Silver, J.M.; Richard, J.; Goss, J.L.; Tseng, A.A.; Zhang, A.; El Khoury, J.B.; Moore, K.J. CD36 Signals to the Actin Cytoskeleton and Regulates Microglial Migration via a p130Cas Complex. J. Biol. Chem. 2007, 282, 27392–27401. [Google Scholar] [CrossRef]
- Stewart, C.R.; Stuart, L.M.; Wilkinson, K.; Van Gils, J.M.; Deng, J.; Halle, A.; Rayner, K.J.; Boyer, L.; Zhong, R.; Frazier, W.A.; et al. CD36 Ligands Promote Sterile Inflammation through Assembly of a Toll-like Receptor 4 and 6 Heterodimer. Nat. Immunol. 2010, 11, 155–161. [Google Scholar] [CrossRef]
- Coraci, I.S.; Husemann, J.; Berman, J.W.; Hulette, C.; Dufour, J.H.; Campanella, G.K.; Luster, A.D.; Silverstein, S.C.; El Khoury, J.B. CD36, a Class B Scavenger Receptor, Is Expressed on Microglia in Alzheimer’s Disease Brains and Can Mediate Production of Reactive Oxygen Species in Response to β-Amyloid Fibrils. Am. J. Pathol. 2002, 160, 101–112. [Google Scholar] [CrossRef]
- El Khoury, J.B.; Moore, K.J.; Means, T.K.; Leung, J.; Terada, K.; Toft, M.; Freeman, M.W.; Luster, A.D. CD36 Mediates the Innate Host Response to β-Amyloid. J. Exp. Med. 2003, 197, 1657–1666. [Google Scholar] [CrossRef]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef] [PubMed]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Dai, Y.J.; Chen, G.; Cui, S. Sen Dissecting the Dual Role of the Glial Scar and Scar-Forming Astrocytes in Spinal Cord Injury. Front. Cell. Neurosci. 2020, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Escartin, C.; Galea, E.; Lakatos, A.; O’Callaghan, J.P.; Petzold, G.C.; Serrano-Pozo, A.; Steinhäuser, C.; Volterra, A.; Carmignoto, G.; Agarwal, A.; et al. Reactive Astrocyte Nomenclature, Definitions, and Future Directions. Nat. Neurosci. 2021, 24, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Gallo, V. The Diversity and Disparity of the Glial Scar. Nat. Neurosci. 2018, 21, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lei, Z.; Guo, Z.; Pei, Z.; Chen, Y.; Zhang, F.; Cai, A.; Mok, G.; Lee, G.; Swaminathan, V.; et al. Development of Neuroregenerative Gene Therapy to Reverse Glial Scar Tissue Back to Neuron-Enriched Tissue. Front. Cell. Neurosci. 2020, 14, 594170. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.D. Glial Cells, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2012; ISBN 9780123750006. [Google Scholar]
- Nave, K.A.; Werner, H.B. Myelination of the Nervous System: Mechanisms and Functions. Annu. Rev. Cell Dev. Biol. 2014, 30, 503–533. [Google Scholar] [CrossRef]
- Miller, D.J.; Duka, T.; Stimpson, C.D.; Schapiro, S.J.; Baze, W.B.; McArthur, M.J.; Fobbs, A.J.; Sousa, A.M.M.; Šestan, N.; Wildman, D.E.; et al. Prolonged Myelination in Human Neocortical Evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 16480–16485. [Google Scholar] [CrossRef]
- Duncan, G.J.; Simkins, T.J.; Emery, B. Neuron-Oligodendrocyte Interactions in the Structure and Integrity of Axons. Front. Cell Dev. Biol. 2021, 9, 653101. [Google Scholar] [CrossRef] [PubMed]
- Mot, A.I.; Depp, C.; Nave, K.-A. An Emerging Role of Dysfunctional Axon-Oligodendrocyte Coupling in Neurodegenerative Diseases. Dialogues Clin. Neurosci. 2018, 20, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Morrison, B.M.; Li, Y.; Lengacher, S.; Farah, M.H.; Hoffman, P.N.; Liu, Y.; Tsingalia, A.; Jin, L.; Zhang, P.-W.; et al. Oligodendroglia Metabolically Support Axons and Contribute to Neurodegeneration. Nature 2012, 487, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, V.; Kursula, P. Multiple Sclerosis and Myelin Basic Protein: Insights into Protein Disorder and Disease. Amino Acids 2022, 54, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Psenicka, M.W.; Smith, B.C.; Tinkey, R.A.; Williams, J.L. Connecting Neuroinflammation and Neurodegeneration in Multiple Sclerosis: Are Oligodendrocyte Precursor Cells a Nexus of Disease? Front. Cell. Neurosci. 2021, 15, 654284. [Google Scholar] [CrossRef]
- Kjell, J.; Götz, M. Filling the Gaps—A Call for Comprehensive Analysis of Extracellular Matrix of the Glial Scar in Region- and Injury-Specific Contexts. Front. Cell. Neurosci. 2020, 14, 32. [Google Scholar] [CrossRef]
- Rittiner, J.E.; Moncalvo, M.; Chiba-Falek, O.; Kantor, B. Gene-Editing Technologies Paired With Viral Vectors for Translational Research Into Neurodegenerative Diseases. Front. Mol. Neurosci. 2020, 13, 148. [Google Scholar] [CrossRef]
- Blömer, U.; Naldini, L.; Kafri, T.; Trono, D.; Verma, I.M.; Gage, F.H. Highly Efficient and Sustained Gene Transfer in Adult Neurons with a Lentivirus Vector. J. Virol. 1997, 71, 6641–6649. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, J.; Ericson, C.; Jansson, M.; Björk, E.; Lundberg, C. Targeted Transgene Expression in Rat Brain Using Lentiviral Vectors. J. Neurosci. Res. 2003, 73, 876–885. [Google Scholar] [CrossRef]
- Azzouz, M.; Martin-Rendon, E.; Barber, R.D.; Mitrophanous, K.A.; Carter, E.E.; Rohll, J.B.; Kingsman, S.M.; Kingsman, A.J.; Mazarakis, N.D. Multicistronic Lentiviral Vector-Mediated Striatal Gene Transfer of Aromatic L-Amino Acid Decarboxylase, Tyrosine Hydroxylase, and GTP Cyclohydrolase I Induces Sustained Transgene Expression, Dopamine Production, and Functional Improvement in a Rat Model. J. Neurosci. Off. J. Soc. Neurosci. 2002, 22, 10302–10312. [Google Scholar] [CrossRef]
- Consiglio, A.; Quattrini, A.; Martino, S.; Bensadoun, J.C.; Dolcetta, D.; Trojani, A.; Benaglia, G.; Marchesini, S.; Cestari, V.; Oliverio, A.; et al. In Vivo Gene Therapy of Metachromatic Leukodystrophy by Lentiviral Vectors: Correction of Neuropathology and Protection against Learning Impairments in Affected Mice. Nat. Med. 2001, 7, 310–316. [Google Scholar] [CrossRef]
- Kantor, B.; Ma, H.; Webster-Cyriaque, J.; Monahan, P.E.; Kafri, T. Epigenetic Activation of Unintegrated HIV-1 Genomes by Gut-Associated Short Chain Fatty Acids and Its Implications for HIV Infection. Proc. Natl. Acad. Sci. USA 2009, 106, 18786–18791. [Google Scholar] [CrossRef] [PubMed]
- Bayer, M.; Kantor, B.; Cockrell, A.; Ma, H.; Zeithaml, B.; Li, X.; McCown, T.; Kafri, T. A Large U3 Deletion Causes Increased in Vivo Expression from a Nonintegrating Lentiviral Vector. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 1968–1976. [Google Scholar] [CrossRef] [PubMed]
- Milone, M.C.; O’Doherty, U. Clinical Use of Lentiviral Vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Dautzenberg, I.J.C.; Rabelink, M.J.W.E.; Hoeben, R.C. The Stability of Envelope-Pseudotyped Lentiviral Vectors. Gene Ther. 2021, 28, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Kantor, B.; Bailey, R.M.; Wimberly, K.; Kalburgi, S.N.; Gray, S.J. Methods for Gene Transfer to the Central Nervous System. Adv. Genet. 2014, 87, 125–197. [Google Scholar] [CrossRef] [PubMed]
- Cronin, J.; Zhang, X.-Y.; Reiser, J. Altering the Tropism of Lentiviral Vectors through Pseudotyping. Curr. Gene Ther. 2005, 5, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Ranzani, M.; Annunziato, S.; Calabria, A.; Brasca, S.; Benedicenti, F.; Gallina, P.; Naldini, L.; Montini, E. Lentiviral Vector-Based Insertional Mutagenesis Identifies Genes Involved in the Resistance to Targeted Anticancer Therapies. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 2056–2068. [Google Scholar] [CrossRef]
- Ortinski, P.I.; O’Donovan, B.; Dong, X.; Kantor, B. Integrase-Deficient Lentiviral Vector as an All-in-One Platform for Highly Efficient CRISPR/Cas9-Mediated Gene Editing. Mol. Ther. Methods Clin. Dev. 2017, 5, 153–164. [Google Scholar] [CrossRef]
- Wanisch, K.; Yáñez-Muñoz, R.J. Integration-Deficient Lentiviral Vectors: A Slow Coming of Age. Mol. Ther. J. Am. Soc. Gene Ther. 2009, 17, 1316–1332. [Google Scholar] [CrossRef]
- Li, M.; Snider, B.J. Chapter 1—Gene Therapy Methods and Their Applications in Neurological Disorders; Li, M., Snider, B.J., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 3–39. ISBN 978-0-12-809813-4. [Google Scholar]
- Gene Therapy: The Age of AAV. Available online: https://wyss.harvard.edu/news/gene-therapy-the-age-of-aav/ (accessed on 1 November 2023).
- Maurer, A.C.; Weitzman, M.D. Adeno-Associated Virus Genome Interactions Important for Vector Production and Transduction. Hum. Gene Ther. 2020, 31, 499–511. [Google Scholar] [CrossRef]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., 3rd; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs Clin. Immunother. Biopharm. Gene Ther. 2017, 31, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Daya, S.; Berns, K.I. Gene Therapy Using Adeno-Associated Virus Vectors. Clin. Microbiol. Rev. 2008, 21, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Maurya, S.; Sarangi, P.; Jayandharan, G.R. Safety of Adeno-Associated Virus-Based Vector-Mediated Gene Therapy—Impact of Vector Dose. Cancer Gene Ther. 2022, 29, 1305–1306. [Google Scholar] [CrossRef] [PubMed]
- Large, E.E.; Silveria, M.A.; Zane, G.M.; Weerakoon, O.; Chapman, M.S. Adeno-Associated Virus (AAV) Gene Delivery: Dissecting Molecular Interactions upon Cell Entry. Viruses 2021, 13, 1336. [Google Scholar] [CrossRef] [PubMed]
- Henckaerts, E.; Linden, R.M. Adeno-Associated Virus: A Key to the Human Genome? Future Virol. 2010, 5, 555–574. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Jang, M.J.; Yoo, B.B.; Greenbaum, A.; Ravi, N.; Wu, W.-L.; Sánchez-Guardado, L.; Lois, C.; Mazmanian, S.K.; Deverman, B.E.; et al. Engineered AAVs for Efficient Noninvasive Gene Delivery to the Central and Peripheral Nervous Systems. Nat. Neurosci. 2017, 20, 1172–1179. [Google Scholar] [CrossRef] [PubMed]
- Mandel, R.J.; Burger, C. Clinical Trials in Neurological Disorders Using AAV Vectors: Promises and Challenges. Curr. Opin. Mol. Ther. 2004, 6, 482–490. [Google Scholar]
- Cearley, C.N.; Wolfe, J.H. Transduction Characteristics of Adeno-Associated Virus Vectors Expressing Cap Serotypes 7, 8, 9, and Rh10 in the Mouse Brain. Mol. Ther. J. Am. Soc. Gene Ther. 2006, 13, 528–537. [Google Scholar] [CrossRef]
- Duque, S.; Joussemet, B.; Riviere, C.; Marais, T.; Dubreil, L.; Douar, A.-M.; Fyfe, J.; Moullier, P.; Colle, M.-A.; Barkats, M. Intravenous Administration of Self-Complementary AAV9 Enables Transgene Delivery to Adult Motor Neurons. Mol. Ther. J. Am. Soc. Gene Ther. 2009, 17, 1187–1196. [Google Scholar] [CrossRef]
- Hester, M.E.; Foust, K.D.; Kaspar, R.W.; Kaspar, B.K. AAV as a Gene Transfer Vector for the Treatment of Neurological Disorders: Novel Treatment Thoughts for ALS. Curr. Gene Ther. 2009, 9, 428–433. [Google Scholar] [CrossRef]
- Gray, S.J.; Nagabhushan Kalburgi, S.; McCown, T.J.; Jude Samulski, R. Global CNS Gene Delivery and Evasion of Anti-AAV-Neutralizing Antibodies by Intrathecal AAV Administration in Non-Human Primates. Gene Ther. 2013, 20, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. FDA Advisers Back Gene Therapy for Rare Form of Blindness. Nature 2017, 550, 314. [Google Scholar] [CrossRef] [PubMed]
- Foust, K.D.; Wang, X.; McGovern, V.L.; Braun, L.; Bevan, A.K.; Haidet, A.M.; Le, T.T.; Morales, P.R.; Rich, M.M.; Burghes, A.H.M.; et al. RETRACTED ARTICLE: Rescue of the Spinal Muscular Atrophy Phenotype in a Mouse Model by Early Postnatal Delivery of SMN. Nat. Biotechnol. 2010, 28, 271–274. [Google Scholar] [CrossRef]
- Kang, L.; Jin, S.; Wang, J.; Lv, Z.; Xin, C.; Tan, C.; Zhao, M.; Wang, L.; Liu, J. AAV Vectors Applied to the Treatment of CNS Disorders: Clinical Status and Challenges. J. Control. Release 2023, 355, 458–473. [Google Scholar] [CrossRef] [PubMed]
- McCarty, D.M.; Young, S.M.J.; Samulski, R.J. Integration of Adeno-Associated Virus (AAV) and Recombinant AAV Vectors. Annu. Rev. Genet. 2004, 38, 819–845. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, K.S.; Kleinstiver, B.P.; Garcia, S.P.; Zaborowski, M.P.; Volak, A.; Spirig, S.E.; Muller, A.; Sousa, A.A.; Tsai, S.Q.; Bengtsson, N.E.; et al. High Levels of AAV Vector Integration into CRISPR-Induced DNA Breaks. Nat. Commun. 2019, 10, 4439. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A. The Promise and Challenge of Therapeutic Genome Editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM Recognition by Miniature CRISPR-Cas12f Nucleases Triggers Programmable Double-Stranded DNA Target Cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA Destruction by Miniature CRISPR-Cas14 Enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef]
- Kim, D.Y.; Chung, Y.; Lee, Y.; Jeong, D.; Park, K.-H.; Chin, H.J.; Lee, J.M.; Park, S.; Ko, S.; Ko, J.-H.; et al. Author Correction: Hypercompact Adenine Base Editors Based on a Cas12f Variant Guided by Engineered RNA. Nat. Chem. Biol. 2023, 19, 389. [Google Scholar] [CrossRef]
- Kim, D.Y.; Lee, J.M.; Moon, S.B.; Chin, H.J.; Park, S.; Lim, Y.; Kim, D.; Koo, T.; Ko, J.-H.; Kim, Y.-S. Efficient CRISPR Editing with a Hypercompact Cas12f1 and Engineered Guide RNAs Delivered by Adeno-Associated Virus. Nat. Biotechnol. 2022, 40, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Barkats, M.; Bilang-Bleuel, A.; Buc-Caron, M.H.; Castel-Barthe, M.N.; Corti, O.; Finiels, F.; Horellou, P.; Revah, F.; Sabate, O.; Mallet, J. Adenovirus in the Brain: Recent Advances of Gene Therapy for Neurodegenerative Diseases. Prog. Neurobiol. 1998, 55, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Federici, T.; Riley, J.; Boulis, N. Gene-Based Neuromodulation; Elsevier Ltd.: Amsterdam, The Netherlands, 2009; Volume 1, ISBN 9780123742483. [Google Scholar]
- Mackey, J.K.; Wold, W.S.; Rigden, P.; Green, M. Transforming Region of Group A, B, and C Adenoviruses: DNA Homology Studies with Twenty-Nine Human Adenovirus Serotypes. J. Virol. 1979, 29, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Kaspar, B.K. Gene Therapy: Direct Viral Delivery. In Encyclopedia of Neuroscience; Elsevier: Amsterdam, The Netherlands, 2009; pp. 633–639. [Google Scholar] [CrossRef]
- Vetrini, F.; Ng, P. Gene Therapy with Helper-Dependent Adenoviral Vectors: Current Advances and Future Perspectives. Viruses 2010, 2, 1886–1917. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Shayakhmetov, D.M. Cytokine Responses to Adenovirus and Adenovirus Vectors. Viruses 2022, 14, 888. [Google Scholar] [CrossRef] [PubMed]
- Lowenstein, P.R.; Castro, M.G. Inflammation and Adaptive Immune Responses to Adenoviral Vectors Injected into the Brain: Peculiarities, Mechanisms, and Consequences. Gene Ther. 2003, 10, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hu, Y.; Ju, D. Gene Therapy for Neurodegenerative Disorders: Advances, Insights and Prospects. Acta Pharm Sin B 2020, 10, 1347–1359. [Google Scholar] [CrossRef]
- Bibikova, M.; Golic, M.; Golic, K.G.; Carroll, D. Targeted Chromosomal Cleavage and Mutagenesis in Drosophila Using Zinc-Finger Nucleases. Genetics 2002, 161, 1169–1175. [Google Scholar] [CrossRef]
- Meyer, M.; de Angelis, M.H.; Wurst, W.; Kühn, R. Gene Targeting by Homologous Recombination in Mouse Zygotes Mediated by Zinc-Finger Nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 15022–15026. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE Nuclease Architecture for Efficient Genome Editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.; Boch, J. TALE and TALEN Genome Editing Technologies. Gene Genome Ed. 2021, 2, 100007. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR-Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Gerbi, S.A. Making Ends Meet: Targeted Integration of DNA Fragments by Genome Editing. Chromosoma 2018, 127, 405–420. [Google Scholar] [CrossRef]
- Nambiar, T.S.; Baudrier, L.; Billon, P.; Ciccia, A. CRISPR-Based Genome Editing through the Lens of DNA Repair. Mol. Cell 2022, 82, 348–388. [Google Scholar] [CrossRef]
- György, B.; Lööv, C.; Zaborowski, M.P.; Takeda, S.; Kleinstiver, B.P.; Commins, C.; Kastanenka, K.; Mu, D.; Volak, A.; Giedraitis, V.; et al. CRISPR/Cas9 Mediated Disruption of the Swedish APP Allele as a Therapeutic Approach for Early-Onset Alzheimer’s Disease. Mol. Ther. Nucleic Acids 2018, 11, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Barman, N.C.; Khan, N.M.; Islam, M.; Nain, Z.; Roy, R.K.; Haque, A.; Barman, S.K. CRISPR-Cas9: A Promising Genome Editing Therapeutic Tool for Alzheimer’s Disease-A Narrative Review. Neurol. Ther. 2020, 9, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC Locus with CRISPR/Cas9 Enhances Tumour Rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef]
- Ustiugova, A.S.; Ekaterina, D.M.; Nataliya, M.V.; Alexey, D.A.; Dmitry, K.V.; Marina, A.A. CRISPR/Cas9 Genome Editing Demonstrates Functionality of the Autoimmunity-Associated SNP Rs12946510. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166599. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome Editing with CRISPR-Cas Nucleases, Base Editors, Transposases and Prime Editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A Review of the Challenges and Approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, C.A.; Brandes, N.; Bueno, R.; Trinidad, M.; Mazumder, T.; Yu, B.; Hwang, B.; Chang, C.; Liu, J.; Sun, Y.; et al. Mitigation of Chromosome Loss in Clinical CRISPR-Cas9-Engineered T Cells. Cell 2023, 186, 4567–4582.e20. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.P.K.; Liu, M.-S.; Hibshman, G.N.; Dangerfield, T.L.; Jung, K.; McCool, R.S.; Johnson, K.A.; Taylor, D.W. Structural Basis for Mismatch Surveillance by CRISPR-Cas9. Nature 2022, 603, 343–347. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.-H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed Evolution of CRISPR-Cas9 to Increase Its Specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A High-Fidelity Cas9 Mutant Delivered as a Ribonucleoprotein Complex Enables Efficient Gene Editing in Human Hematopoietic Stem and Progenitor Cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Jo, A.; Denduluri, S.; Zhang, B.; Wang, Z.; Yin, L.; Yan, Z.; Kang, R.; Shi, L.L.; Mok, J.; Lee, M.J.; et al. The Versatile Functions of Sox9 in Development, Stem Cells, and Human Diseases. Genes. Dis. 2014, 1, 149–161. [Google Scholar] [CrossRef]
- Tedeschi, A.; Dupraz, S.; Laskowski, C.J.; Xue, J.; Ulas, T.; Beyer, M.; Schultze, J.L.; Bradke, F. The Calcium Channel Subunit Alpha2delta2 Suppresses Axon Regeneration in the Adult CNS. Neuron 2016, 92, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Cornwell, A.; Li, J.; Peng, S.; Osorio, M.J.; Aalling, N.; Wang, S.; Benraiss, A.; Lou, N.; Goldman, S.A.; et al. SOX9 Is an Astrocyte-Specific Nuclear Marker in the Adult Brain Outside the Neurogenic Regions. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 4493–4507. [Google Scholar] [CrossRef] [PubMed]
- McKillop, W.M.; Dragan, M.; Schedl, A.; Brown, A. Conditional Sox9 Ablation Reduces Chondroitin Sulfate Proteoglycan Levels and Improves Motor Function Following Spinal Cord Injury. Glia 2013, 61, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Cehajic-Kapetanovic, J.; Birtel, J.; McClements, M.E.; Shanks, M.E.; Clouston, P.; Downes, S.M.; Charbel Issa, P.; MacLaren, R.E. Clinical and Molecular Characterization of PROM1-Related Retinal Degeneration. JAMA Netw. Open 2019, 2, e195752. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Shin, J.E.; Lee, B.; Kim, H.; Jeon, Y.; Ahn, S.H.; Chi, S.W.; Cho, Y. The Stem Cell Marker Prom1 Promotes Axon Regeneration by down-Regulating Cholesterol Synthesis via Smad Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 15955–15966. [Google Scholar] [CrossRef] [PubMed]
- Victor, M.B.; Richner, M.; Hermanstyne, T.O.; Ransdell, J.L.; Sobieski, C.; Deng, P.-Y.; Klyachko, V.A.; Nerbonne, J.M.; Yoo, A.S. Generation of Human Striatal Neurons by microRNA-Dependent Direct Conversion of Fibroblasts. Neuron 2014, 84, 311–323. [Google Scholar] [CrossRef]
- Hou, S.; Lu, P. Direct Reprogramming of Somatic Cells into Neural Stem Cells or Neurons for Neurological Disorders. Neural Regen. Res. 2016, 11, 28–31. [Google Scholar] [CrossRef]
- Müller, M.; Bhattacharya, S.S.; Moore, T.; Prescott, Q.; Wedig, T.; Herrmann, H.; Magin, T.M. Dominant Cataract Formation in Association with a Vimentin Assembly Disrupting Mutation. Hum. Mol. Genet. 2009, 18, 1052–1057. [Google Scholar] [CrossRef]
- Sottile, V.; Li, M.; Scotting, P.J. Stem Cell Marker Expression in the Bergmann Glia Population of the Adult Mouse Brain. Brain Res. 2006, 1099, 8–17. [Google Scholar] [CrossRef]
- Mukhopadhyay, G.; Doherty, P.; Walsh, F.S.; Crocker, P.R.; Filbin, M.T. A Novel Role for Myelin-Associated Glycoprotein as an Inhibitor of Axonal Regeneration. Neuron 1994, 13, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Quarles, R.H. Myelin-Associated Glycoprotein (MAG): Past, Present and Beyond. J. Neurochem. 2007, 100, 1431–1448. [Google Scholar] [CrossRef] [PubMed]
- Cafferty, W.B.J.; Duffy, P.; Huebner, E.; Strittmatter, S.M. MAG and OMgp Synergize with Nogo-A to Restrict Axonal Growth and Neurological Recovery after Spinal Cord Trauma. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 6825–6837. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Geoffroy, C.G.; Chan, A.F.; Tolentino, K.E.; Crawford, M.J.; Leal, M.A.; Kang, B.; Zheng, B. Assessing Spinal Axon Regeneration and Sprouting in Nogo-, MAG-, and OMgp-Deficient Mice. Neuron 2010, 66, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, U.; Bandtlow, C.E.; Schnell, L.; Bartsch, S.; Spillmann, A.A.; Rubin, B.P.; Hillenbrand, R.; Montag, D.; Schwab, M.E.; Schachner, M. Lack of Evidence That Myelin-Associated Glycoprotein Is a Major Inhibitor of Axonal Regeneration in the CNS. Neuron 1995, 15, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Kinter, J.; Lazzati, T.; Schmid, D.; Zeis, T.; Erne, B.; Lützelschwab, R.; Steck, A.J.; Pareyson, D.; Peles, E.; Schaeren-Wiemers, N. An Essential Role of MAG in Mediating Axon-Myelin Attachment in Charcot-Marie-Tooth 1A Disease. Neurobiol. Dis. 2013, 49, 221–231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nguyen, T.; Mehta, N.R.; Conant, K.; Kim, K.-J.; Jones, M.; Calabresi, P.A.; Melli, G.; Hoke, A.; Schnaar, R.L.; Ming, G.-L.; et al. Axonal Protective Effects of the Myelin-Associated Glycoprotein. J. Neurosci. Off. J. Soc. Neurosci. 2009, 29, 630–637. [Google Scholar] [CrossRef]
- Jones, M.V.; Nguyen, T.T.; Ewaleifoh, O.; Lebson, L.; Whartenby, K.A.; Griffin, J.W.; Calabresi, P.A. Accelerated Axon Loss in MOG35-55 Experimental Autoimmune Encephalomyelitis (EAE) in Myelin-Associated Glycoprotein-Deficient (MAGKO) Mice. J. Neuroimmunol. 2013, 262, 53–61. [Google Scholar] [CrossRef]
- Geoffroy, C.G.; Zheng, B. Myelin-Associated Inhibitors in Axonal Growth after CNS Injury. Curr. Opin. Neurobiol. 2014, 27, 31–38. [Google Scholar] [CrossRef]
- Thornton, J.E.; Gregory, R.I. How Does Lin28 Let-7 Control Development and Disease? Trends Cell Biol. 2012, 22, 474–482. [Google Scholar] [CrossRef]
- Yue, Y.; Zhang, D.; Jiang, S.; Li, A.; Guo, A.; Wu, X.; Xia, X.; Cheng, H.; Tao, T.; Gu, X. LIN28 Expression in Rat Spinal Cord after Injury. Neurochem. Res. 2014, 39, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Nathan, F.M.; Ohtake, Y.; Wang, S.; Jiang, X.; Sami, A.; Guo, H.; Zhou, F.-Q.; Li, S. Upregulating Lin28a Promotes Axon Regeneration in Adult Mice with Optic Nerve and Spinal Cord Injury. Mol. Ther. J. Am. Soc. Gene Ther. 2020, 28, 1902–1917. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Li, Q.; Liu, C.-M.; Hall, P.A.; Jiang, J.-J.; Katchis, C.D.; Kang, S.; Dong, B.C.; Li, S.; Zhou, F.-Q. Lin28 Signaling Supports Mammalian PNS and CNS Axon Regeneration. Cell Rep. 2018, 24, 2540–2552.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, Q.; Deng, K.; Guo, X.; Xia, J. Lin28: An Emerging Important Oncogene Connecting Several Aspects of Cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 2841–2848. [Google Scholar] [CrossRef] [PubMed]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and Regulation in Development and Beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef] [PubMed]
- Stier, S.; Cheng, T.; Dombkowski, D.; Carlesso, N.; Scadden, D.T. Notch1 Activation Increases Hematopoietic Stem Cell Self-Renewal in Vivo and Favors Lymphoid over Myeloid Lineage Outcome. Blood 2002, 99, 2369–2378. [Google Scholar] [CrossRef]
- Kumano, K.; Chiba, S.; Shimizu, K.; Yamagata, T.; Hosoya, N.; Saito, T.; Takahashi, T.; Hamada, Y.; Hirai, H. Notch1 Inhibits Differentiation of Hematopoietic Cells by Sustaining GATA-2 Expression. Blood 2001, 98, 3283–3289. [Google Scholar] [CrossRef]
- Yu, X.; Zou, J.; Ye, Z.; Hammond, H.; Chen, G.; Tokunaga, A.; Mali, P.; Li, Y.-M.; Civin, C.; Gaiano, N.; et al. Notch Signaling Activation in Human Embryonic Stem Cells Is Required for Embryonic, but Not Trophoblastic, Lineage Commitment. Cell Stem Cell 2008, 2, 461–471. [Google Scholar] [CrossRef]
- Umemoto, T.; Yamato, M.; Nishida, K.; Kohno, C.; Yang, J.; Tano, Y.; Okano, T. Rat Limbal Epithelial Side Population Cells Exhibit a Distinct Expression of Stem Cell Markers That Are Lacking in Side Population Cells from the Central Cornea. FEBS Lett. 2005, 579, 6569–6574. [Google Scholar] [CrossRef]
- Pierfelice, T.; Alberi, L.; Gaiano, N. Notch in the Vertebrate Nervous System: An Old Dog with New Tricks. Neuron 2011, 69, 840–855. [Google Scholar] [CrossRef]
- Alberi, L.; Liu, S.; Wang, Y.; Badie, R.; Smith-Hicks, C.; Wu, J.; Pierfelice, T.J.; Abazyan, B.; Mattson, M.P.; Kuhl, D.; et al. Activity-Induced Notch Signaling in Neurons Requires Arc/Arg3.1 and Is Essential for Synaptic Plasticity in Hippocampal Networks. Neuron 2011, 69, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Lütolf, S.; Radtke, F.; Aguet, M.; Suter, U.; Taylor, V. Notch1 Is Required for Neuronal and Glial Differentiation in the Cerebellum. Development 2002, 129, 373–385. [Google Scholar] [CrossRef] [PubMed]
- El Bejjani, R.; Hammarlund, M. Notch Signaling Inhibits Axon Regeneration. Neuron 2012, 73, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.N.R.; Pearse, D.D. Regulating Axonal Responses to Injury: The Intersection between Signaling Pathways Involved in Axon Myelination and The Inhibition of Axon Regeneration. Front. Mol. Neurosci. 2016, 9, 33. [Google Scholar] [CrossRef]
- Chen, J.; Leong, S.-Y.; Schachner, M. Differential Expression of Cell Fate Determinants in Neurons and Glial Cells of Adult Mouse Spinal Cord after Compression Injury. Eur. J. Neurosci. 2005, 22, 1895–1906. [Google Scholar] [CrossRef]
- Yamamoto, S.; Nagao, M.; Sugimori, M.; Kosako, H.; Nakatomi, H.; Yamamoto, N.; Takebayashi, H.; Nabeshima, Y.; Kitamura, T.; Weinmaster, G.; et al. Transcription Factor Expression and Notch-Dependent Regulation of Neural Progenitors in the Adult Rat Spinal Cord. J. Neurosci. Off. J. Soc. Neurosci. 2001, 21, 9814–9823. [Google Scholar] [CrossRef]
- Yamamoto, N.; Yamamoto, S.; Inagaki, F.; Kawaichi, M.; Fukamizu, A.; Kishi, N.; Matsuno, K.; Nakamura, K.; Weinmaster, G.; Okano, H.; et al. Role of Deltex-1 as a Transcriptional Regulator Downstream of the Notch Receptor. J. Biol. Chem. 2001, 276, 45031–45040. [Google Scholar] [CrossRef]
- Sakakibara, S.; Okano, H. Expression of Neural RNA-Binding Proteins in the Postnatal CNS: Implications of Their Roles in Neuronal and Glial Cell Development. J. Neurosci. Off. J. Soc. Neurosci. 1997, 17, 8300–8312. [Google Scholar] [CrossRef]
- Sakakibara, S.; Imai, T.; Hamaguchi, K.; Okabe, M.; Aruga, J.; Nakajima, K.; Yasutomi, D.; Nagata, T.; Kurihara, Y.; Uesugi, S.; et al. Mouse-Musashi-1, a Neural RNA-Binding Protein Highly Enriched in the Mammalian CNS Stem Cell. Dev. Biol. 1996, 176, 230–242. [Google Scholar] [CrossRef]
- Kaneko, Y.; Sakakibara, S.; Imai, T.; Suzuki, A.; Nakamura, Y.; Sawamoto, K.; Ogawa, Y.; Toyama, Y.; Miyata, T.; Okano, H. Musashi1: An Evolutionally Conserved Marker for CNS Progenitor Cells Including Neural Stem Cells. Dev. Neurosci. 2000, 22, 139–153. [Google Scholar] [CrossRef]
- Sakakibara, S.; Nakamura, Y.; Yoshida, T.; Shibata, S.; Koike, M.; Takano, H.; Ueda, S.; Uchiyama, Y.; Noda, T.; Okano, H. RNA-Binding Protein Musashi Family: Roles for CNS Stem Cells and a Subpopulation of Ependymal Cells Revealed by Targeted Disruption and Antisense Ablation. Proc. Natl. Acad. Sci. USA 2002, 99, 15194–15199. [Google Scholar] [CrossRef] [PubMed]
- Kuwako, K.; Kakumoto, K.; Imai, T.; Igarashi, M.; Hamakubo, T.; Sakakibara, S.; Tessier-Lavigne, M.; Okano, H.J.; Okano, H. Neural RNA-Binding Protein Musashi1 Controls Midline Crossing of Precerebellar Neurons through Posttranscriptional Regulation of Robo3/Rig-1 Expression. Neuron 2010, 67, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, E.A.G.; Sato, K.; Salfity, H.V.N.; Sarria, D.A.; Belecky-Adams, T. Musashi and Plasticity of Xenopus and Axolotl Spinal Cord Ependymal Cells. Front. Cell. Neurosci. 2018, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Z.; Ghosh-Roy, A.; Hubert, T.; Yan, D.; O’Rourke, S.; Bowerman, B.; Wu, Z.; Jin, Y.; Chisholm, A.D. Axon Regeneration Pathways Identified by Systematic Genetic Screening in C. elegans. Neuron 2011, 71, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Miraglia, S.; Godfrey, W.; Yin, A.H.; Atkins, K.; Warnke, R.; Holden, J.T.; Bray, R.A.; Waller, E.K.; Buck, D.W. A Novel Five-Transmembrane Hematopoietic Stem Cell Antigen: Isolation, Characterization, and Molecular Cloning. Blood 1997, 90, 5013–5021. [Google Scholar] [CrossRef] [PubMed]
- Corbeil, D.; Röper, K.; Hellwig, A.; Tavian, M.; Miraglia, S.; Watt, S.M.; Simmons, P.J.; Peault, B.; Buck, D.W.; Huttner, W.B. The Human AC133 Hematopoietic Stem Cell Antigen Is Also Expressed in Epithelial Cells and Targeted to Plasma Membrane Protrusions. J. Biol. Chem. 2000, 275, 5512–5520. [Google Scholar] [CrossRef]
- Yin, A.H.; Miraglia, S.; Zanjani, E.D.; Almeida-Porada, G.; Ogawa, M.; Leary, A.G.; Olweus, J.; Kearney, J.; Buck, D.W. AC133, a Novel Marker for Human Hematopoietic Stem and Progenitor Cells. Blood 1997, 90, 5002–5012. [Google Scholar] [CrossRef]
- Reichert, D.; Scheinpflug, J.; Karbanová, J.; Freund, D.; Bornhäuser, M.; Corbeil, D. Tunneling Nanotubes Mediate the Transfer of Stem Cell Marker CD133 between Hematopoietic Progenitor Cells. Exp. Hematol. 2016, 44, 1092–1112.e2. [Google Scholar] [CrossRef]
- Zacchigna, S.; Oh, H.; Wilsch-Bräuninger, M.; Missol-Kolka, E.; Jászai, J.; Jansen, S.; Tanimoto, N.; Tonagel, F.; Seeliger, M.; Huttner, W.B.; et al. Loss of the Cholesterol-Binding Protein Prominin-1/CD133 Causes Disk Dysmorphogenesis and Photoreceptor Degeneration. J. Neurosci. Off. J. Soc. Neurosci. 2009, 29, 2297–2308. [Google Scholar] [CrossRef]
- Walker, T.L.; Wierick, A.; Sykes, A.M.; Waldau, B.; Corbeil, D.; Carmeliet, P.; Kempermann, G. Prominin-1 Allows Prospective Isolation of Neural Stem Cells from the Adult Murine Hippocampus. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 3010–3024. [Google Scholar] [CrossRef]
- Permanyer, J.; Navarro, R.; Friedman, J.; Pomares, E.; Castro-Navarro, J.; Marfany, G.; Swaroop, A.; Gonzàlez-Duarte, R. Autosomal Recessive Retinitis Pigmentosa with Early Macular Affectation Caused by Premature Truncation in PROM1. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2656–2663. [Google Scholar] [CrossRef] [PubMed]
- Li, Z. CD133: A Stem Cell Biomarker and Beyond. Exp. Hematol. Oncol. 2013, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Wu, Y.-Z.; Wang, Y.; Wu, F.-R.; Zang, C.-B.; Tang, C.; Cao, S.; Li, S.-L. CD133 Silencing Inhibits Stemness Properties and Enhances Chemoradiosensitivity in CD133-Positive Liver Cancer Stem Cells. Int. J. Mol. Med. 2013, 31, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, L.; Selzer, M.E.; Hu, Y. Neuronal Endoplasmic Reticulum Stress in Axon Injury and Neurodegeneration. Ann. Neurol. 2013, 74, 768–777. [Google Scholar] [CrossRef] [PubMed]
- Kamei, N.; Kwon, S.-M.; Alev, C.; Nakanishi, K.; Yamada, K.; Masuda, H.; Ishikawa, M.; Kawamoto, A.; Ochi, M.; Asahara, T. Ex-Vivo Expanded Human Blood-Derived CD133+ Cells Promote Repair of Injured Spinal Cord. J. Neurol. Sci. 2013, 328, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; Xu, X.-M.; Zhang, C.-L. Regeneration Through in Vivo Cell Fate Reprogramming for Neural Repair. Front. Cell. Neurosci. 2020, 14, 107. [Google Scholar] [CrossRef]
- Zhao, N.; Francis, N.L.; Calvelli, H.R.; Moghe, P.V. Microglia-Targeting Nanotherapeutics for Neurodegenerative Diseases. APL Bioeng. 2020, 4, 30902. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Liu, T.; Mao, Y.; Peng, B. Novel Microglia-Based Therapeutic Approaches to Neurodegenerative Disorders. Neurosci. Bull. 2023, 39, 491–502. [Google Scholar] [CrossRef]
- Rahim, A.A.; Buckley, S.M.K.; Chan, J.K.Y.; Peebles, D.M.; Waddington, S.N. Perinatal Gene Delivery to the CNS. Ther. Deliv. 2011, 2, 483–491. [Google Scholar] [CrossRef]
- Puhl, D.L.; D’Amato, A.R.; Gilbert, R.J. Challenges of Gene Delivery to the Central Nervous System and the Growing Use of Biomaterial Vectors. Brain Res. Bull. 2019, 150, 216–230. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Mao, D.; Wang, Y.; Zhang, H.; Pan, Y.; Wang, Y.; Teng, S.; Huang, P. Current Trends of Clinical Trials Involving CRISPR/Cas Systems. Front. Med. 2023, 10, 1292452. [Google Scholar] [CrossRef] [PubMed]
- Ashmore-Harris, C.; Fruhwirth, G.O. The Clinical Potential of Gene Editing as a Tool to Engineer Cell-Based Therapeutics. Clin. Transl. Med. 2020, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.; Kesari, K.K.; Rachamalla, M.; Mani, S.; Ashraf, G.M.; Jha, S.K.; Kumar, P.; Ambasta, R.K.; Dureja, H.; Devkota, H.P.; et al. CRISPR/Cas9 Gene Editing: New Hope for Alzheimer’s Disease Therapeutics. J. Adv. Res. 2022, 40, 207–221. [Google Scholar] [CrossRef] [PubMed]
- The World’s First CRISPR Therapy Is Approved: Who Will Receive It? Available online: https://www.nature.com/articles/d41587-023-00016-6 (accessed on 5 December 2023).
- Mijanović, O.; Branković, A.; Borovjagin, A.V.; Butnaru, D.V.; Bezrukov, E.A.; Sukhanov, R.B.; Shpichka, A.; Timashev, P.; Ulasov, I. Battling Neurodegenerative Diseases with Adeno-Associated Virus-Based Approaches. Viruses 2020, 12, 460. [Google Scholar] [CrossRef]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klinkovskij, A.; Shepelev, M.; Isaakyan, Y.; Aniskin, D.; Ulasov, I. Advances of Genome Editing with CRISPR/Cas9 in Neurodegeneration: The Right Path towards Therapy. Biomedicines 2023, 11, 3333. https://doi.org/10.3390/biomedicines11123333
Klinkovskij A, Shepelev M, Isaakyan Y, Aniskin D, Ulasov I. Advances of Genome Editing with CRISPR/Cas9 in Neurodegeneration: The Right Path towards Therapy. Biomedicines. 2023; 11(12):3333. https://doi.org/10.3390/biomedicines11123333
Chicago/Turabian StyleKlinkovskij, Aleksandr, Mikhail Shepelev, Yuri Isaakyan, Denis Aniskin, and Ilya Ulasov. 2023. "Advances of Genome Editing with CRISPR/Cas9 in Neurodegeneration: The Right Path towards Therapy" Biomedicines 11, no. 12: 3333. https://doi.org/10.3390/biomedicines11123333
APA StyleKlinkovskij, A., Shepelev, M., Isaakyan, Y., Aniskin, D., & Ulasov, I. (2023). Advances of Genome Editing with CRISPR/Cas9 in Neurodegeneration: The Right Path towards Therapy. Biomedicines, 11(12), 3333. https://doi.org/10.3390/biomedicines11123333






