miRNA Expression Patterns in Early- and Late-Stage Prostate Cancer Patients: High-Throughput Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tumor Samples and Data Collection
2.2. miRNA Extraction, Reverse Transcription, Expression Analysis of 754 miRNAs and miRNAs Individual Assays
2.3. Bioinformatic Analysis of the TCGA Data
Statistical Analysis
3. Results
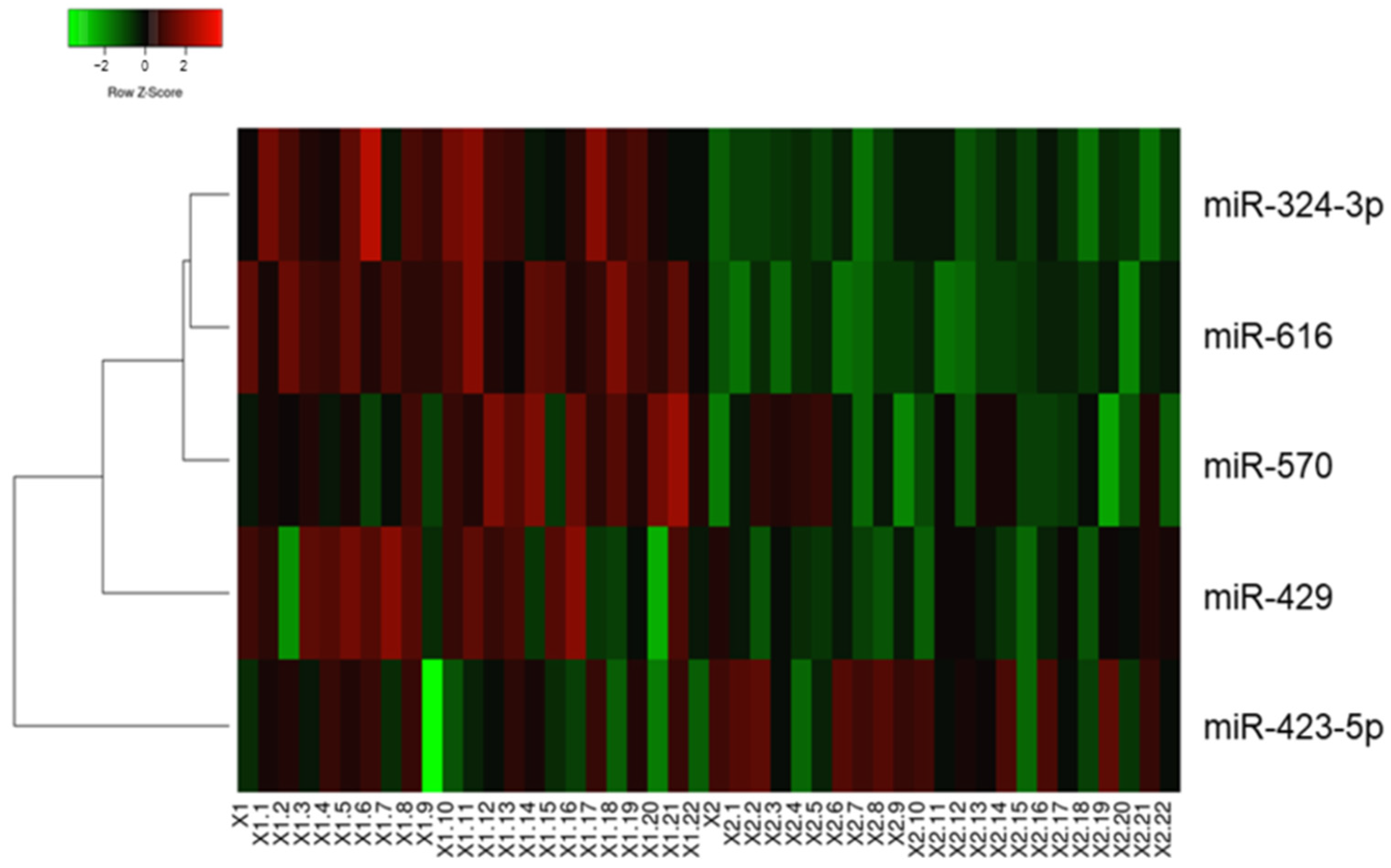
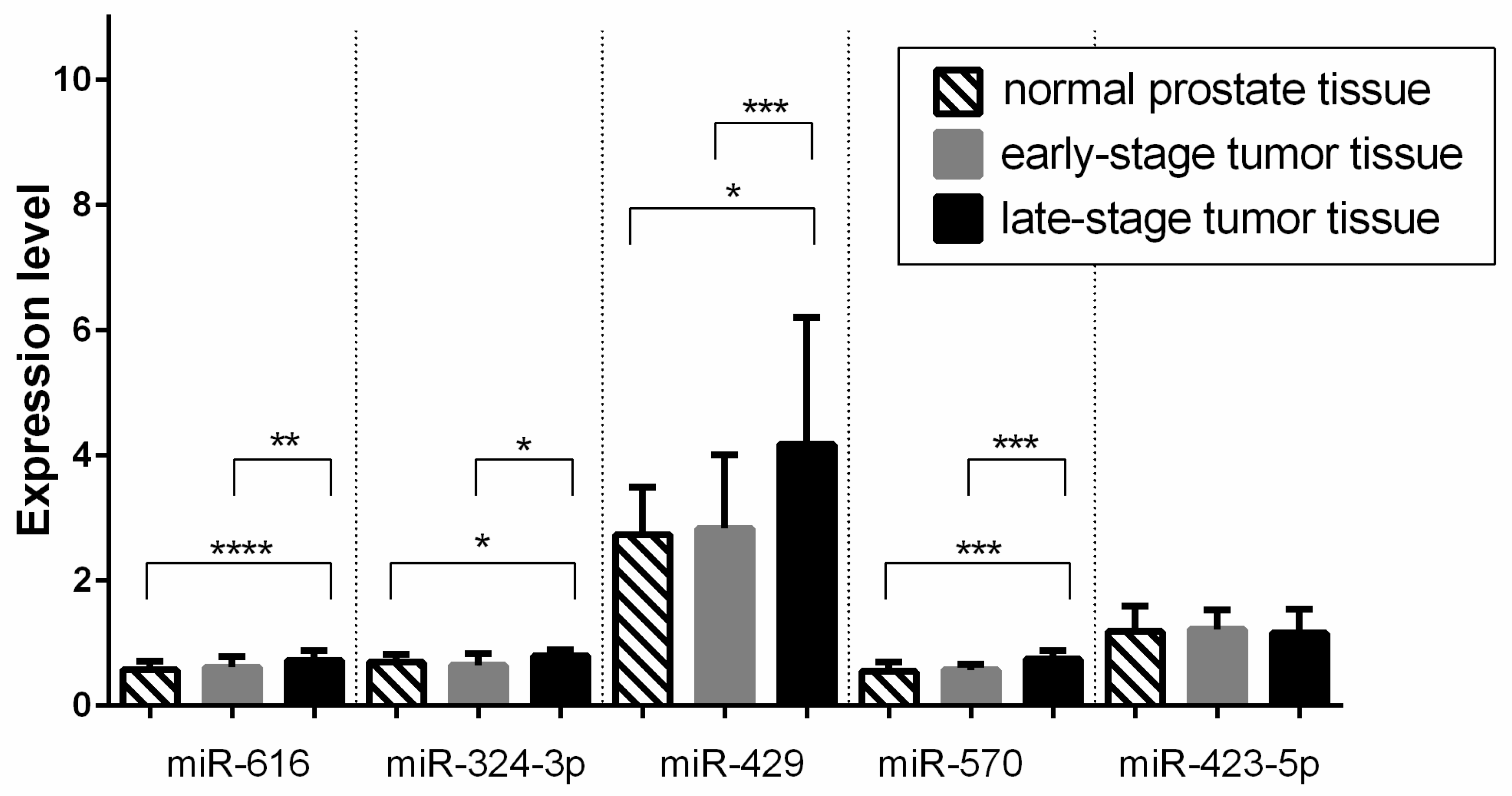
3.1. Validation Phase
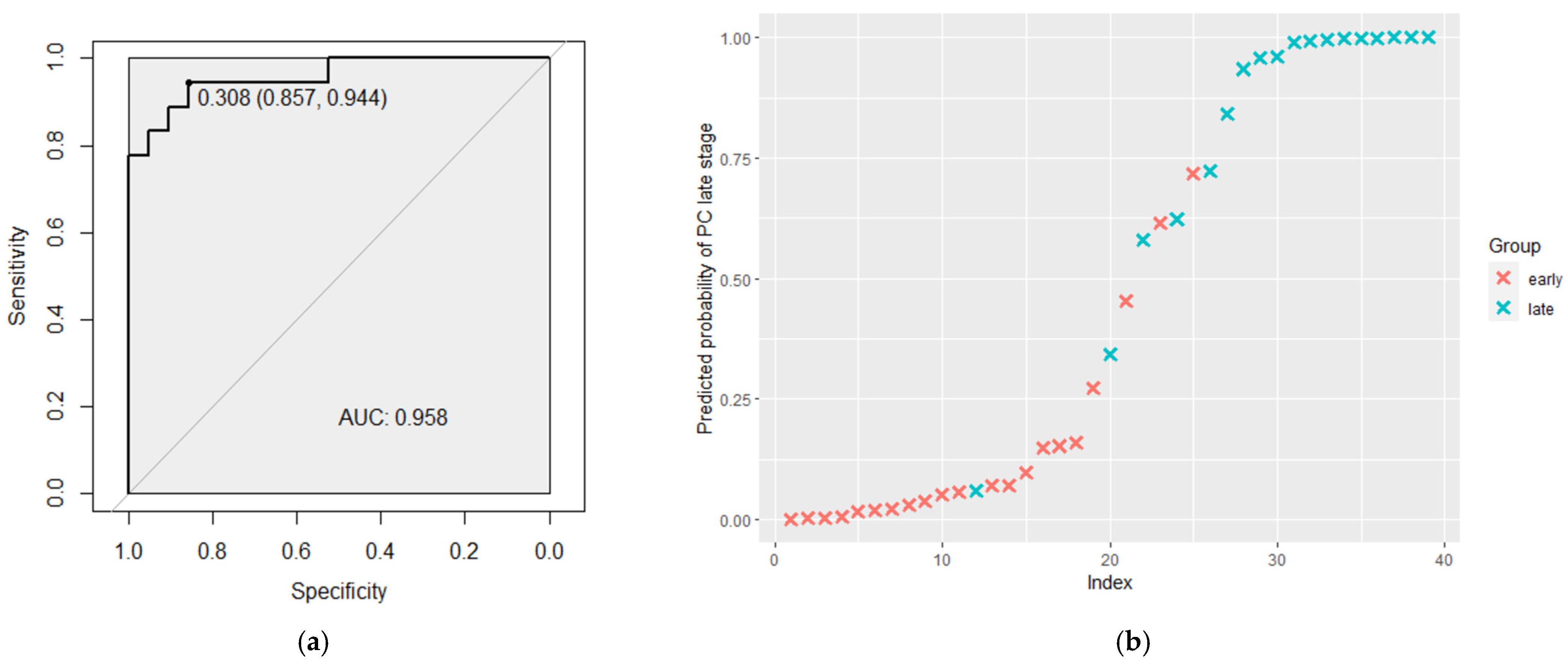
3.2. ROC Analysis
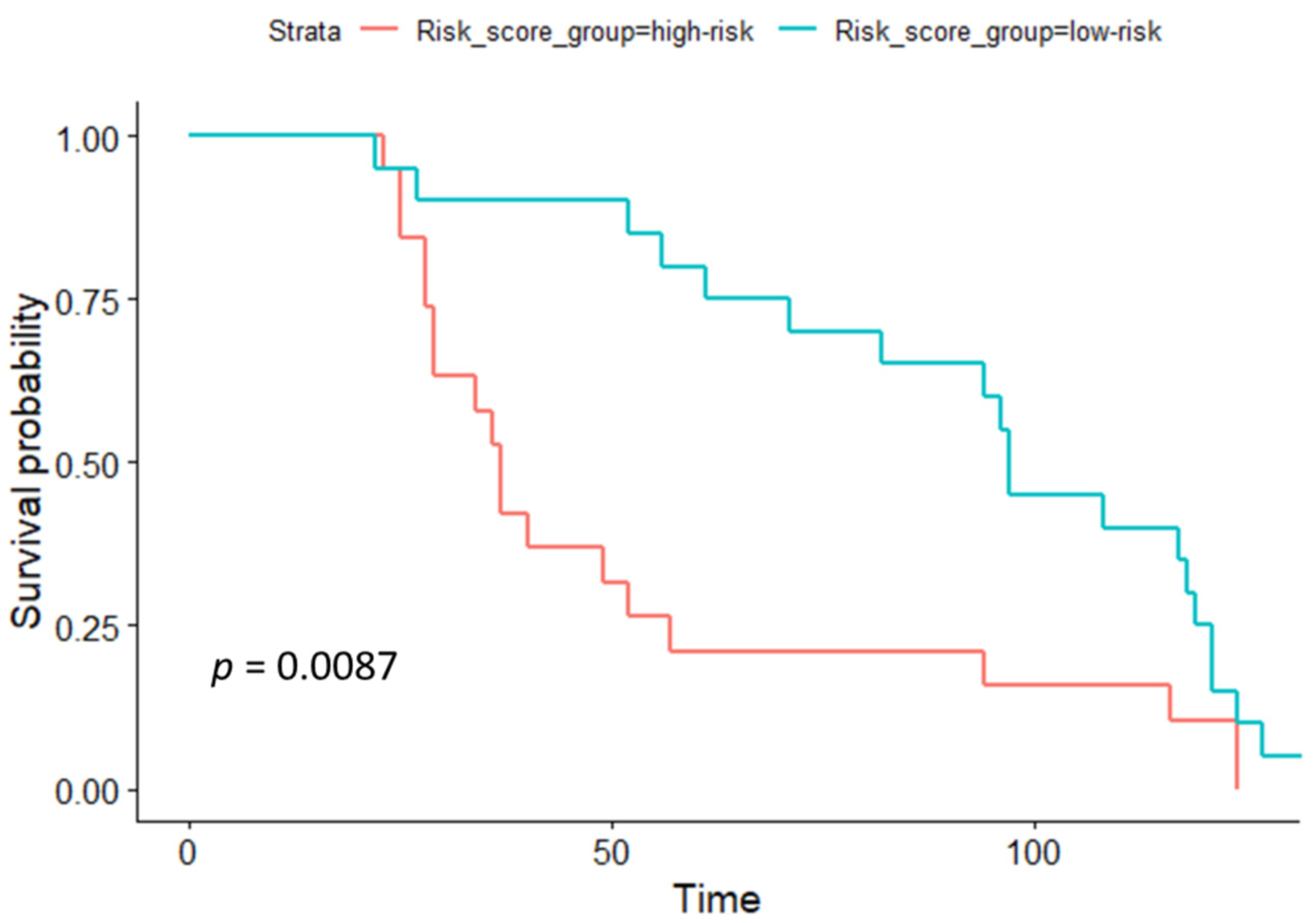
3.3. Identification of Prognosis-Related Variables
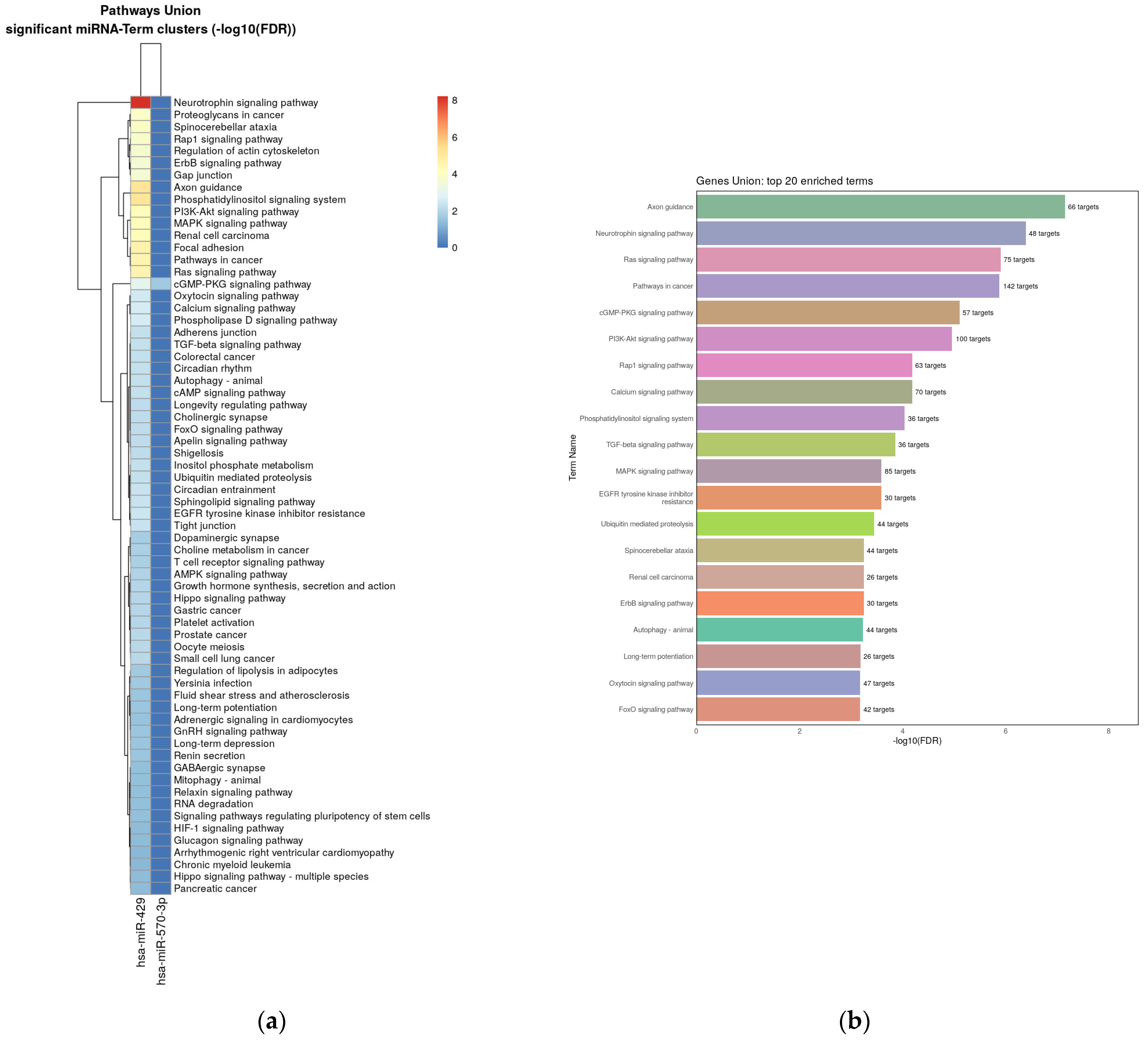
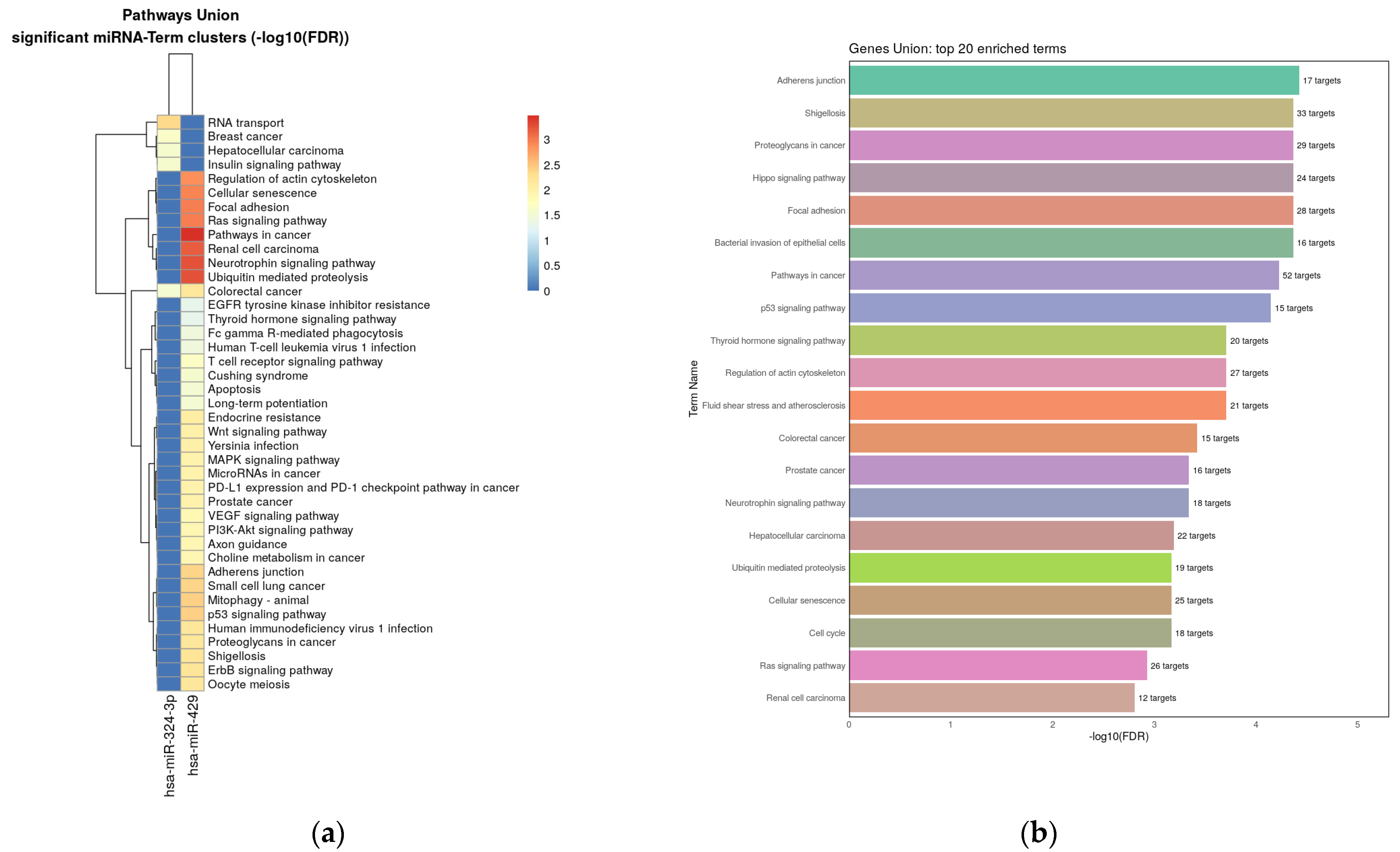
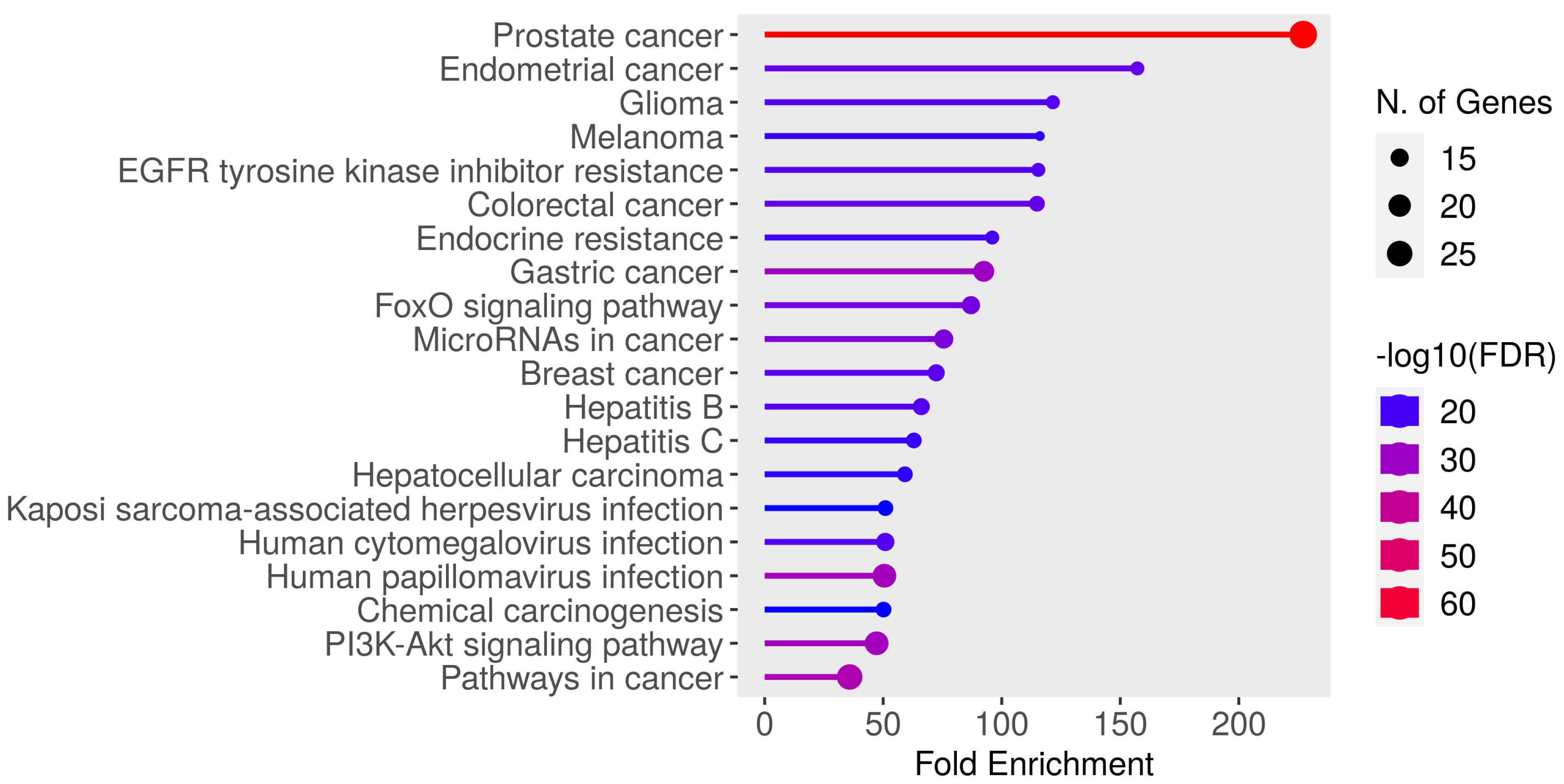
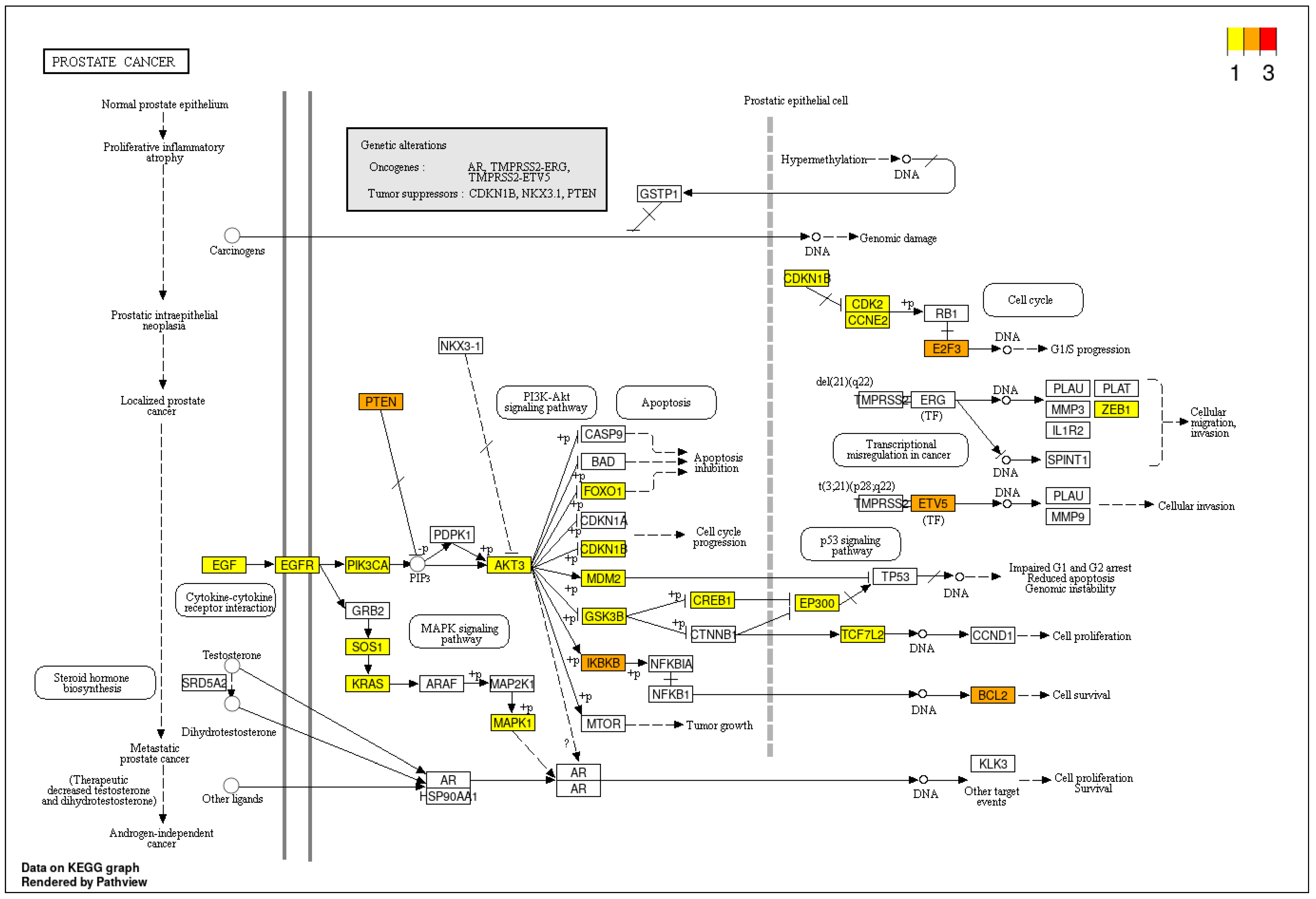
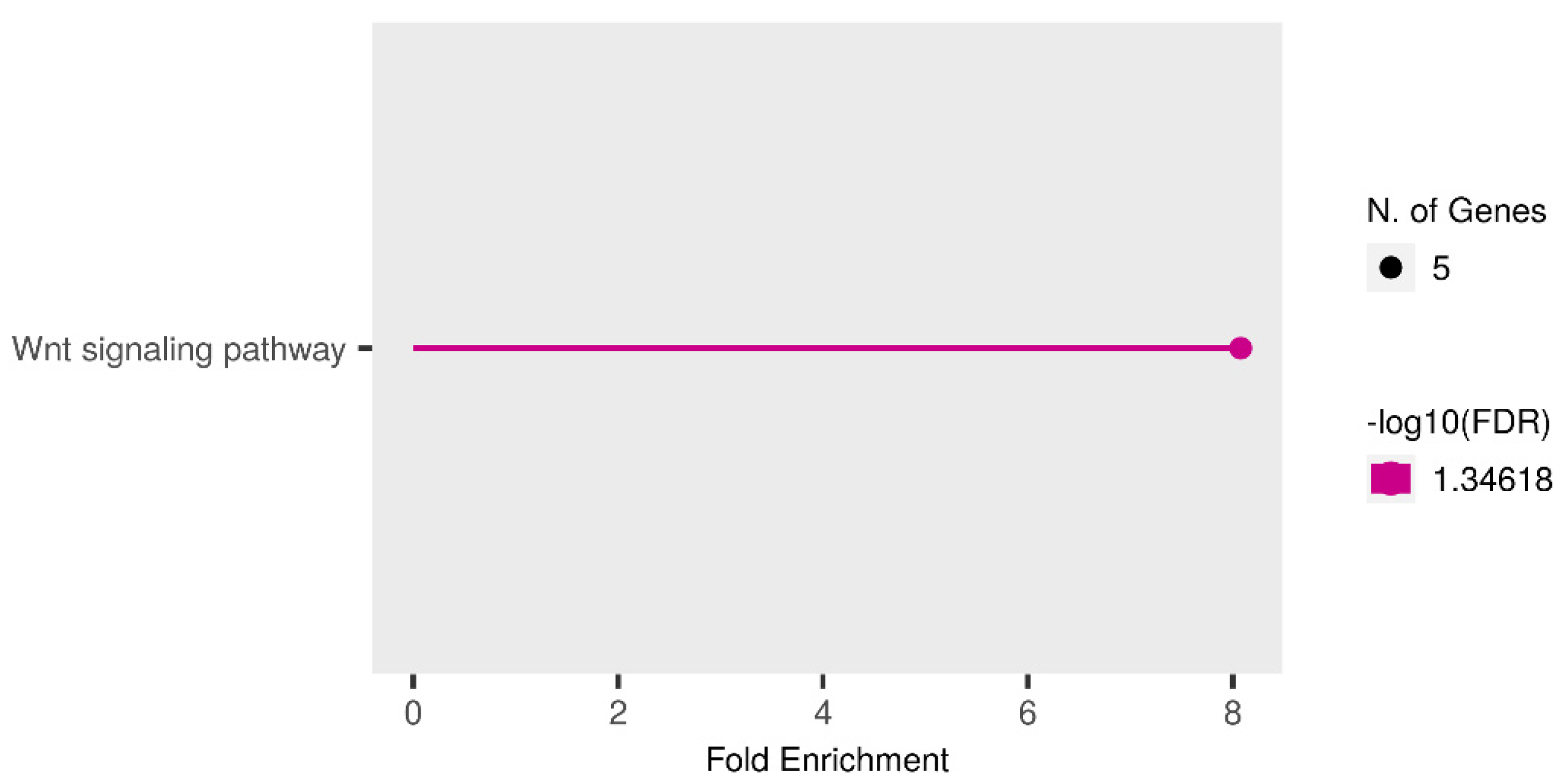
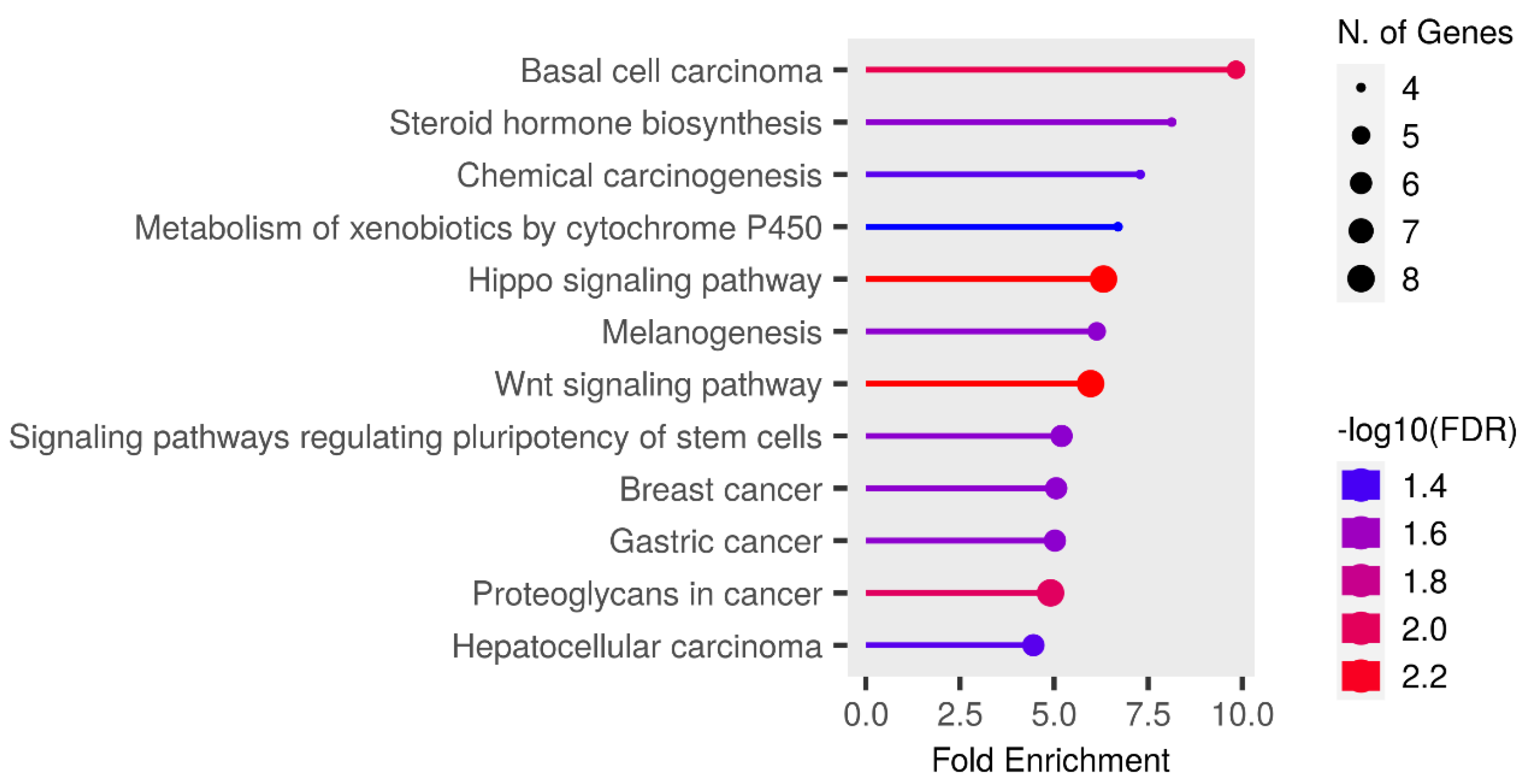
3.4. Pathway Enrichment Analysis
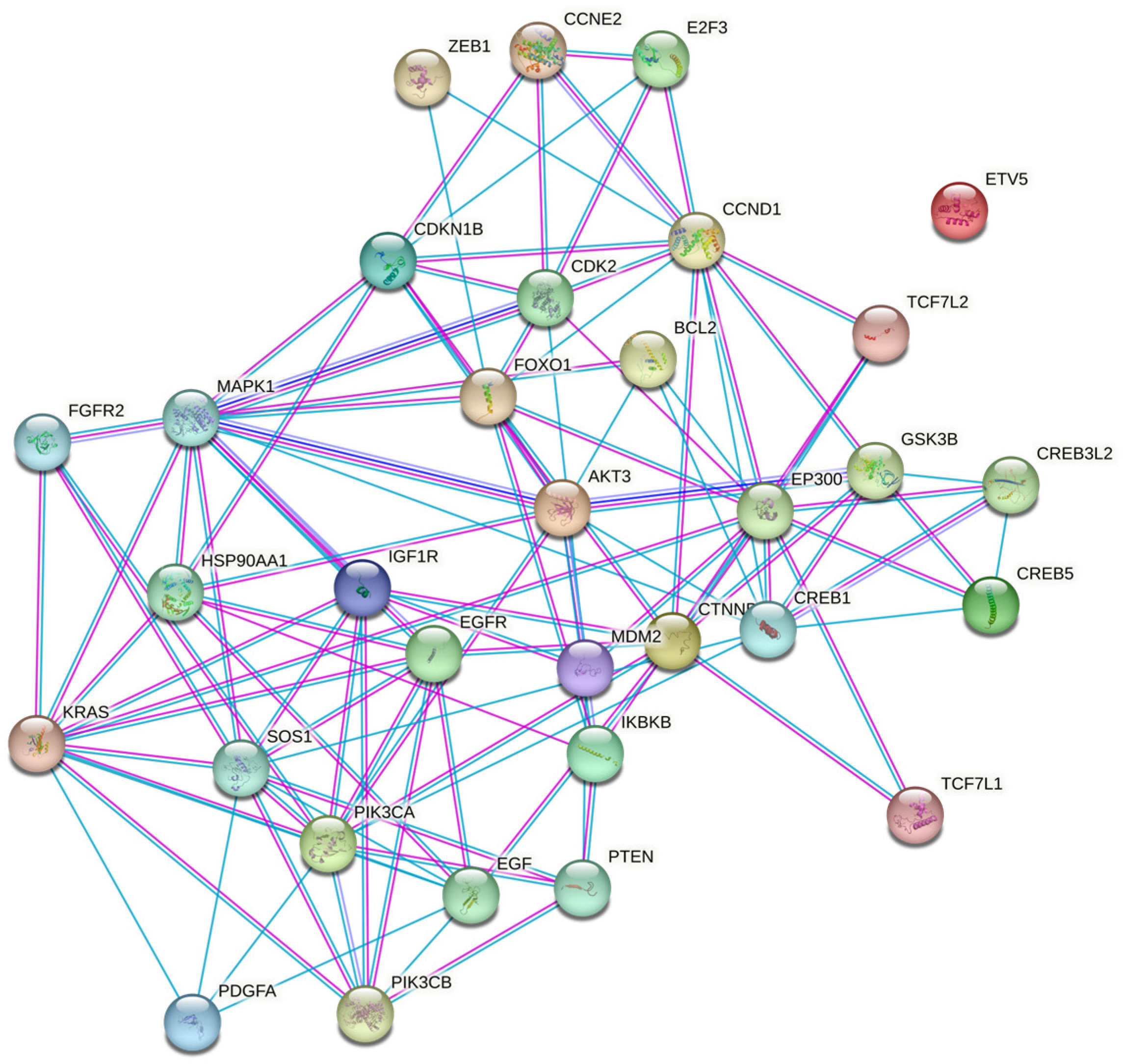
3.5. Expression Analysis of miRNA Target Genes Based on TCGA Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, J.H.; Hong, S.K. Clinical utility of current biomarkers for prostate cancer detection. Investig. Clin. Urol. 2021, 62, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Farha, M.W.; Salami, S.S. Biomarkers for prostate cancer detection and risk stratification. Ther. Adv. Urol. 2022, 14, 17562872221103988. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Becerra, M.F.; Atluri, V.S.; Bhattu, A.S.; Punnen, S. Serum and urine biomarkers for detecting clinically significant prostate cancer. Urol. Oncol. 2021, 39, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Yao, J.; Xu, L.; Xu, Z.; Ge, Y.; Zhou, L.; Zhao, F.; Jia, R. Diagnosis accuracy of PCA3 level in patients with prostate cancer: A systematic review with meta-analysis. Int. Braz. J. Urol. 2020, 46, 691–704. [Google Scholar] [CrossRef]
- Li, M.; Zhou, D.; Zhang, W.; Gao, S.; Zhou, X. Urine PCA3 mRNA level in diagnostic of prostate cancer. J. Cancer Res. Ther. 2018, 14, 864–866. [Google Scholar] [CrossRef]
- Cao, L.; Lee, C.H.; Ning, J.; Handy, B.C.; Wagar, E.A.; Meng, Q.H. Combination of Prostate Cancer Antigen 3 and Prostate-Specific Antigen Improves Diagnostic Accuracy in Men at Risk of Prostate Cancer. Arch. Pathol. Lab. Med. 2018, 142, 1106–1112. [Google Scholar] [CrossRef]
- Sanda, M.G.; Feng, Z.; Howard, D.H.; Tomlins, S.A.; Sokoll, L.J.; Chan, D.W.; Regan, M.M.; Groskopf, J.; Chipman, J.; Patil, D.H.; et al. Association Between Combined TMPRSS2:ERG and PCA3 RNA Urinary Testing and Detection of Aggressive Prostate Cancer. JAMA Oncol. 2017, 3, 1085–1093. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.; Li, X.; Jiang, M. Urinary Biomarker Panel to Improve Accuracy in Predicting Prostate Biopsy Result in Chinese Men with PSA 4–10 ng/mL. BioMed. Res. Int. 2017, 2017, 2512536. [Google Scholar] [CrossRef]
- Bernardeau, S.; Charles, T.; Fromont-Hankard, G.; Irani, J. The Role of a Single PCA3 Test before a First Negative Prostate Biopsy: 5-Year Follow-Up. Prog. Urol. 2017, 27, 325–330. [Google Scholar] [CrossRef]
- Wang, T.; Qu, X.; Jiang, J.; Gao, P.; Zhao, D.; Lian, X.; Li, X. Diagnostic significance of urinary long non-coding PCA3 RNA in prostate cancer. Oncotarget 2017, 8, 58577–58586. [Google Scholar] [CrossRef]
- Catalano, M.; Generali, D.; Gatti, M.; Riboli, B.; Paganini, L.; Nesi, G.; Roviello, G. DNA repair deficiency as circulating biomarker in prostate cancer. Front. Oncol. 2023, 13, 1115241. [Google Scholar] [CrossRef] [PubMed]
- de Souza Dutra, C.; da Cruz Schafhauser, D.; Hentz, M.; Mayer, N.R.; Pinheiro, R.M.; Baierle, G.; Kist, D.R.; Bullé, D.J.; Donaduzzi, R.C.; Bohmgahren, M.F.; et al. Urinary endogenous peptides as biomarkers for prostate cancer. Oncol. Lett. 2023, 25, 1–10. [Google Scholar] [CrossRef]
- Li, Z.; Bui, D.T.; Shao, Y.; Kitova, E.N.; White, S.; Vesprini, D.; Liu, S.K.; Mahal, L.K.; Leong, H.S.; Klassen, J.S. Native Mass Spectrometry Quantitation of α2-3-Linked N-Acetylneuraminic Acid Content of Prostate-Specific Antigen: An Accurate Liquid Biopsy for Clinically Significant Prostate Cancer. Anal. Chem. 2023, 95, 10903–10912. [Google Scholar] [CrossRef]
- Diao, Y.; Zhu, B.; Ding, T.; Li, R.; Li, J.; Yang, L.; Zhou, L.; Hao, X.; Liu, J. Tumor-derived extracellular vesicle nucleic acids as promising diagnostic biomarkers for prostate cancer. Front. Oncol. 2023, 13, 1201554. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.G.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker–Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Day, J.R.; Lonigro, R.J.; Hovelson, D.H.; Siddiqui, J.; Kunju, L.P.; Dunn, R.L.; Meyer, S.; Hodge, P.; Groskopf, J.; et al. Urine TMPRSS2:ERG Plus PCA3 for Individualized Prostate Cancer Risk Assessment. Eur. Urol. 2016, 70, 45–53. [Google Scholar] [CrossRef]
- Mytsyk, Y.; Borzhiyevs’kyy, A.; Kobilnyk, Y.; Shulyak, A.; Dutka, I.; Borzhiyevs’kyy, O.; Górecki, A. Personalized management of prostate cancer: From molecular and imaging markers to radiogenomics. Pol. J. Radiol. 2022, 87, 58–62. [Google Scholar] [CrossRef]
- Gilyazova, I.; Ivanova, E.; Izmailov, A.; Sharifgaliev, I.; Karunas, A.; Pudova, E.; Kobelyatskaya, A.; Gilyazova, G.; Izmailova, A.; Pavlov, V.; et al. MicroRNA Expression Signatures in Clear Cell Renal Cell Carcinoma: High-Throughput Searching for Key miRNA Markers in Patients from the Volga-Ural Region of Eurasian Continent. Int. J. Mol. Sci. 2023, 24, 6909. [Google Scholar] [CrossRef]
- Song, C.-J.; Chen, H.; Chen, L.-Z.; Ru, G.-M.; Guo, J.-J.; Ding, Q.-N. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J. Cell. Biochem. 2018, 119, 2763–2786. [Google Scholar] [CrossRef]
- Zhou, H.; Zhu, X. MicroRNA-21 and microRNA-30c as diagnostic biomarkers for prostate cancer: A meta-analysis. Cancer Manag. Res. 2019, 11, 2039–2050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-Q.; Sun, Y.; Li, J.-Q.; Huang, L.-M.; Tan, S.-S.; Yang, F.-Y.; Li, H. The expression of microRNA-324-3p as a tumor suppressor in nasopharyngeal carcinoma and its clinical significance. OncoTargets Ther. 2017, 10, 4935–4943. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Hu, H.; Zhou, J. Knockdown of LncRNA SNHG7 inhibited epithelial-mesenchymal transition in prostate cancer though miR-324-3p/WNT2B axis in vitro. Pathol.-Res. Pract. 2019, 215, 152537. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.-L.; Li, Z.; Wang, W.-Z.; Chen, Z.; Zhang, L.; Li, Q.; Wei, S.; Li, B.-W.; Xu, J.-H.; Chen, L.; et al. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J. Gastroenterol. 2018, 53, 725–739. [Google Scholar] [CrossRef]
- Tuo, H.; Wang, Y.; Wang, L.; Yao, B.; Li, Q.; Wang, C.; Liu, Z.; Han, S.; Yin, G.; Tu, K.; et al. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/β-catenin pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 65687–65698. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Cai, S.; Yu, S.; Lin, H. Metformin induces ferroptosis by targeting miR-324-3p/GPX4 axis in breast cancer. Acta Biochim. Biophys. Sin. 2021, 53, 333–341. [Google Scholar] [CrossRef]
- Gao, X.; Wang, Y.; Zhao, H.; Wei, F.; Zhang, X.; Su, Y.; Wang, C.; Li, H.; Ren, X. Plasma miR-324-3p and miR-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma. Oncotarget 2016, 7, 59664–59675. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C. miR-616 promotes breast cancer migration and invasion by targeting TIMP2 and regulating MMP signaling. Oncol. Lett. 2019, 18, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Meng, P.; Tang, B.; Ke, Z.; Liu, L.; Chen, Y.; Zhu, F. MiR-616 promotes the progression of pancreatic carcinoma by targeting OXR1. Minerva Medica 2021, 112, 528–529. [Google Scholar] [CrossRef]
- Wu, Z.-H.; Lin, C.; Liu, C.-C.; Jiang, W.-W.; Huang, M.-Z.; Liu, X.; Guo, W.-J. MiR-616-3p promotes angiogenesis and EMT in gastric cancer via the PTEN/AKT/mTOR pathway. Biochem. Biophys. Res. Commun. 2018, 501, 1068–1073. [Google Scholar] [CrossRef]
- Sa, R.; Song, H.; Wei, M.; Su, H.; Hong, Y.; Zhou, L.; Bai, Y. MiR-616 plays oncogenic role in hepatocellular carcinoma progression through suppressing PTEN expression and activating PI3K/AKT pathway. Artif. Cells Nanomed. Biotechnol. 2020, 48, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Hu, J.; Li, H.; Chen, J.; Ding, J.; Zu, X.; Fan, B. miR-616-5p Promotes Invasion and Migration of Bladder Cancer via Downregulating NR2C2 Expression. Front. Oncol. 2021, 11, 762946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, X.; Chen, G.; Zheng, X.; Zhu, X.; Shan, L. Hsa_circ_0007494 suppresses prostate cancer progression via miR-616/PTEN axis. Exp. Cell Res. 2020, 395, 112233. [Google Scholar] [CrossRef]
- Yang, Y.; Ban, D.; Zhang, C.; Shen, L. Downregulation of circ_0000673 Promotes Cell Proliferation and Migration in Endometriosis via the Mir-616-3p/PTEN Axis. Int. J. Med. Sci. 2021, 18, 3506–3515. [Google Scholar] [CrossRef] [PubMed]
- Arabkari, V.; Sultana, A.; Barua, D.; Webber, M.; Smith, T.; Gupta, A.; Gupta, S. UPR-Induced miR-616 Inhibits Human Breast Cancer Cell Growth and Migration by Targeting c-MYC. Int. J. Mol. Sci. 2023, 24, 13034. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Zhang, R.; Yang, X.; Zhang, W.; Hou, Y. Positive correlation between miR-570 and prognosis of colon cancer: Inhibition of cell proliferation and invasion. Clin. Exp. Med. 2022, 22, 193–200. [Google Scholar] [CrossRef]
- Wang, L.L.; Huang, W.W.; Huang, J.; Huang, R.F.; Li, N.N.; Hong, Y.; Chen, M.L.; Wu, F.; Liu, J. Protective effect of hsa-miR-570-3p targeting CD274 on triple negative breast cancer by blocking PI3K/AKT/mTOR signaling pathway. Kaohsiung J. Med. Sci. 2020, 36, 581–591. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, S.; Su, L.; Su, Q.; Lin, J.; Huang, X.; Wang, C. miR-570 Inhibits Proliferation, Angiogenesis, and Immune Escape of Hepatocellular Carcinoma. Cancer Biother. Radiopharm. 2018, 33, 252–257. [Google Scholar] [CrossRef]
- Wu, X.; Liu, D.; Wang, S.; Liu, J. Circ_0007444 Inhibits the Progression of Ovarian Cancer via Mediating the miR-570-3p/PTEN Axis. OncoTargets Ther. 2021, 14, 97–110. [Google Scholar] [CrossRef]
- Ouyang, Y.; Gao, P.; Zhu, B.; Chen, X.; Lin, F.; Wang, X.; Wei, J.; Zhang, H. Downregulation of microRNA-429 inhibits cell proliferation by targeting p27Kip1 in human prostate cancer cells. Mol. Med. Rep. 2015, 11, 1435–1441. [Google Scholar] [CrossRef]
- Cavallari, I.; Ciccarese, F.; Sharova, E.; Urso, L.; Raimondi, V.; Silic-Benussi, M.; D’agostino, D.M.; Ciminale, V. The miR-200 Family of microRNAs: Fine Tuners of Epithelial-Mesenchymal Transition and Circulating Cancer Biomarkers. Cancers 2021, 13, 5874. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Lin, L.; Liu, Q.; Gao, W.; Chen, L.; Sha, C.; Chen, Q.; Xu, W.; Li, Y.; Zhu, X. Exosomal transfer of miR-429 confers chemoresistance in epithelial ovarian cancer. Am. J. Cancer Res. 2021, 11, 2124. [Google Scholar] [PubMed]
- Lin, H.-M.; Mahon, K.L.; Spielman, C.; Gurney, H.; Mallesara, G.; Stockler, M.R.; Bastick, P.; Briscoe, K.; Marx, G.; Swarbrick, A.; et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br. J. Cancer 2017, 116, 1002–1011. [Google Scholar] [CrossRef]
- Guo, C.; Zhao, D.; Zhang, Q.; Liu, S.; Sun, M.-Z. miR-429 suppresses tumor migration and invasion by targeting CRKL in hepatocellular carcinoma via inhibiting Raf/MEK/ERK pathway and epithelial-mesenchymal transition. Sci. Rep. 2018, 8, 2375. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, L.; Luan, S.; Jiang, Y.; Wang, Q. miR-429 inhibits osteosarcoma progression by targeting HOXA9 through suppressing Wnt/β-catenin signaling pathway. Oncol. Lett. 2020, 20, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Q.; Mu, Q.; Zhou, D.; Li, H.; Zhang, B.; Yin, C. MiR-429 suppresses proliferation and invasion of breast cancer via inhibiting the Wnt/β-catenin signaling pathway. Thorac. Cancer 2020, 11, 3126–3138. [Google Scholar] [CrossRef]
- Nilsen, A.; Hillestad, T.; Skingen, V.E.; Aarnes, E.; Fjeldbo, C.S.; Hompland, T.; Evensen, T.S.; Stokke, T.; Kristensen, G.B.; Grallert, B.; et al. miR-200a/b/-429 downregulation is a candidate biomarker of tumor radioresistance and independent of hypoxia in locally advanced cervical cancer. Mol. Oncol. 2022, 16, 1402. [Google Scholar] [CrossRef]

| Discovery Phase (n = 46) | Validation Phase (n = 39) | |
|---|---|---|
| Age, mean (range) | 65 (56–75) | 63 (52–70) |
| Fuhrman grade | ||
| 2 | 20 | 19 |
| 3–4 | 26 | 20 |
| TNM | ||
| I–II | 23 | 21 |
| III–IV | 23 | 18 |
| Metastasis | 10 | 8 |
| PSA, mean (range) | 16.02 (0.27–75.04) | 14.3 (2.04–56.11) |
| Gleason Score | ||
| <8 | 28 | 23 |
| ≥8 | 18 | 16 |
| miRNAs | Fold Change | PFDR-Value |
|---|---|---|
| miRNA-324-3p | 0.67 | <0.0001 |
| miRNA-616 | 0.48 | <0.0001 |
| miRNA-570 | 0.82 | 0.0006 |
| miRNA-429 | 0.39 | 0.0021 |
| miRNA-423-5p | 1.80 | 0.0145 |
| Estimate | Std. Error | z Value | Pr (>|z|) | |
|---|---|---|---|---|
| (Intercept) | −35.419466 | 12.487824 | −2.836 | 0.00456 |
| miR.616 | 0.009438 | 0.042614 | 0.221 | 0.82473 |
| miR.324 | 3.047531 | 1.291730 | 2.359 | 0.01831 |
| miR.570 | 5.923752 | 3.640018 | 1.627 | 0.10365 |
| PSA | 6.618535 | 4.263967 | 1.552 | 0.12061 |
| Gleason_score | 8.181656 | 3.991785 | 2.050 | 0.04040 |
| Early Stage | Not Early (Late) Stage | Row Total | Predictive Values | |
|---|---|---|---|---|
| Test Positive | 18 | 1 | 19 | PPV = 18/19 (94%) |
| Test Negative | 3 | 17 | 20 | NPV = 17/20 (85%) |
| Variables | Coef | HR | HR.95 L | HR.95 H | p |
|---|---|---|---|---|---|
| PSA | 0.01355 | 1.01364 | 0.9915 | 1.036 | 0.22904 |
| Gleason_score | 1.04361 | 2.83945 | 1.4593 | 5.525 | 0.00212 |
| miR.616 | 2.34229 | 10.405 | 1.6563 | 65.365 | 0.01249 |
| miR.324 | 2.52039 | 12.43345 | 1.6794 | 92.052 | 0.01361 |
| miR.570 | 0.594 | 1.81122 | 0.3964 | 8.275 | 0.44349 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilyazova, I.; Ivanova, E.; Gupta, H.; Mustafin, A.; Ishemgulov, R.; Izmailov, A.; Gilyazova, G.; Pudova, E.; Pavlov, V.; Khusnutdinova, E. miRNA Expression Patterns in Early- and Late-Stage Prostate Cancer Patients: High-Throughput Analysis. Biomedicines 2023, 11, 3073. https://doi.org/10.3390/biomedicines11113073
Gilyazova I, Ivanova E, Gupta H, Mustafin A, Ishemgulov R, Izmailov A, Gilyazova G, Pudova E, Pavlov V, Khusnutdinova E. miRNA Expression Patterns in Early- and Late-Stage Prostate Cancer Patients: High-Throughput Analysis. Biomedicines. 2023; 11(11):3073. https://doi.org/10.3390/biomedicines11113073
Chicago/Turabian StyleGilyazova, Irina, Elizaveta Ivanova, Himanshu Gupta, Artur Mustafin, Ruslan Ishemgulov, Adel Izmailov, Gulshat Gilyazova, Elena Pudova, Valentin Pavlov, and Elza Khusnutdinova. 2023. "miRNA Expression Patterns in Early- and Late-Stage Prostate Cancer Patients: High-Throughput Analysis" Biomedicines 11, no. 11: 3073. https://doi.org/10.3390/biomedicines11113073
APA StyleGilyazova, I., Ivanova, E., Gupta, H., Mustafin, A., Ishemgulov, R., Izmailov, A., Gilyazova, G., Pudova, E., Pavlov, V., & Khusnutdinova, E. (2023). miRNA Expression Patterns in Early- and Late-Stage Prostate Cancer Patients: High-Throughput Analysis. Biomedicines, 11(11), 3073. https://doi.org/10.3390/biomedicines11113073








