Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities
Abstract
1. Introduction
2. Extended-Spectrum β-Lactamases (ESBL) and ESBL Producers
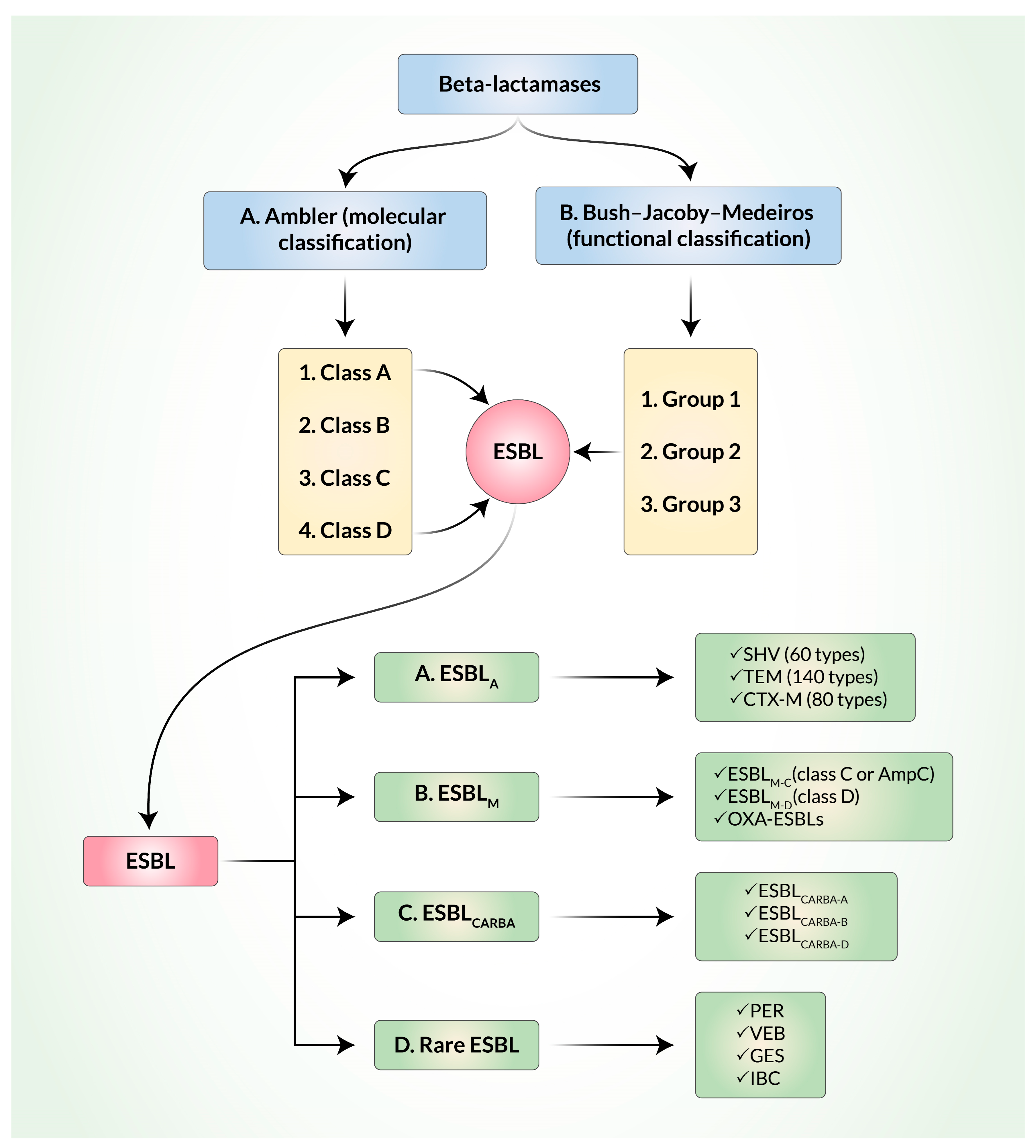
3. Classification and Evolution of ESBL
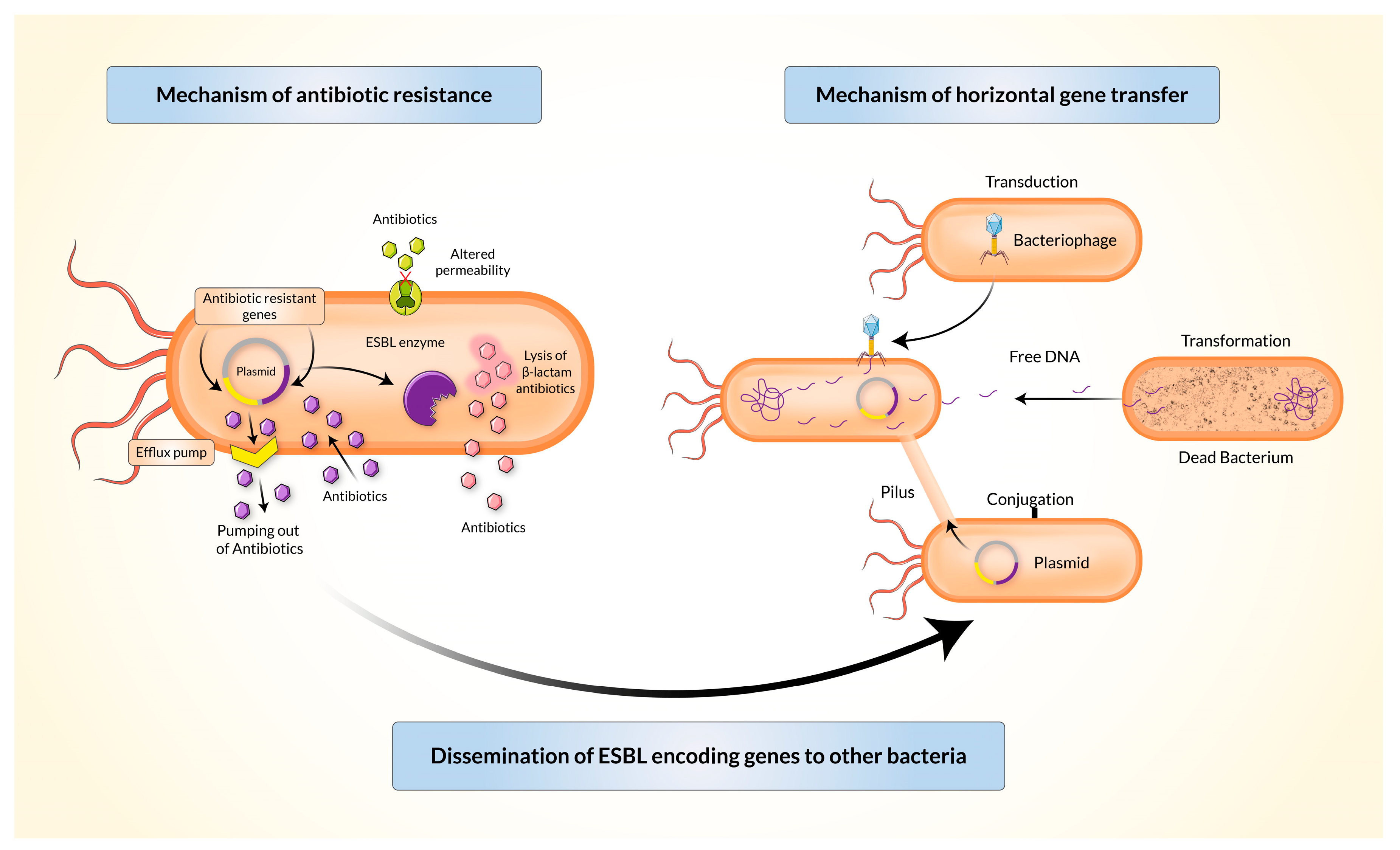
4. Mechanism of Resistance and Dissemination of Resistant Genes
5. Diagnostic Tools for Detection of ESBL
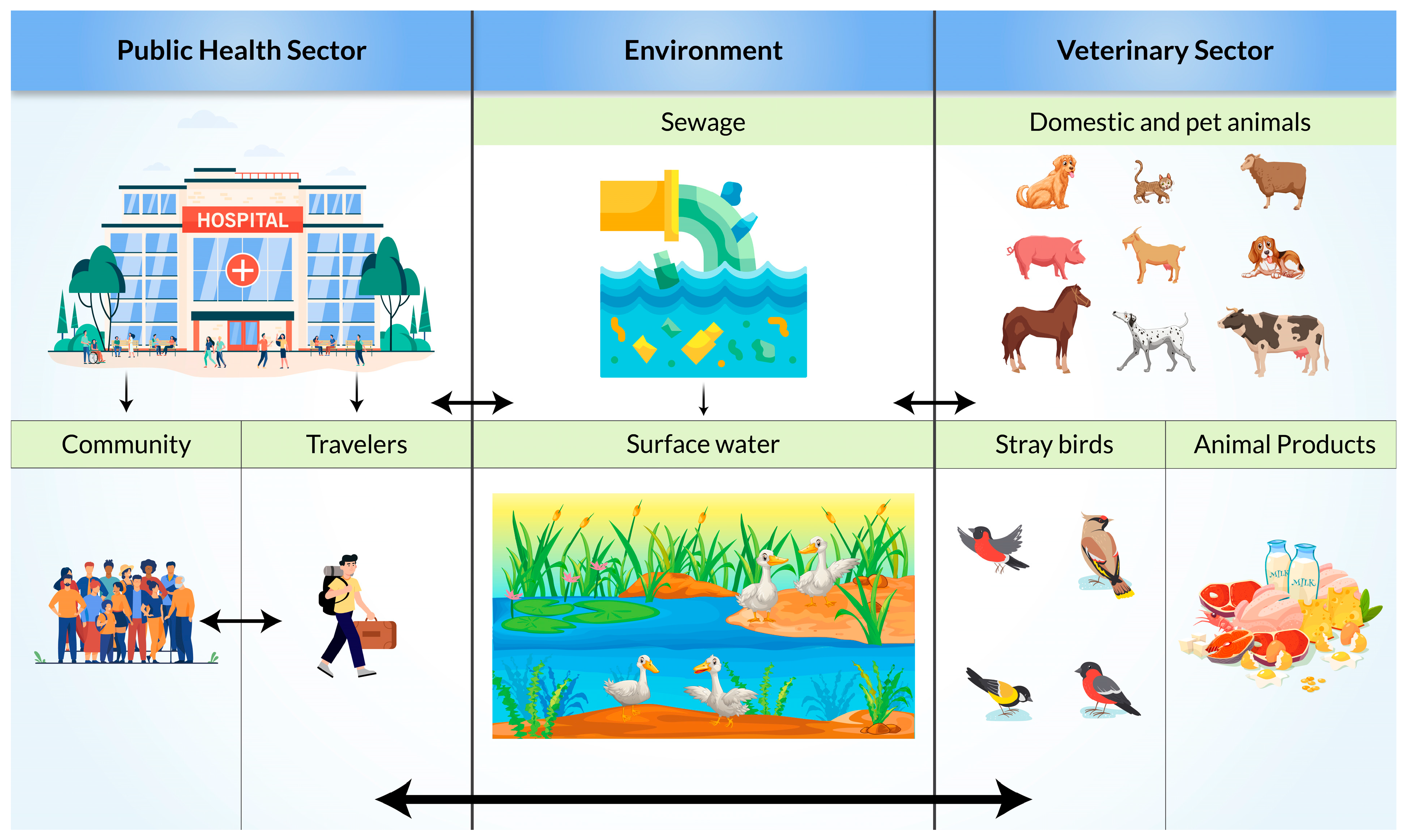
6. Risk Factors and Mode of Transmission of ESBL-Producing Bacteria
7. Possible Therapeutic Options
8. Current Status of ESBL in South Asian Developing Countries
9. Future Threats of ESBL in South Asian Developing Countries
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Forecast, M.D. Global Antibiotics Market Size, Share, Trends, COVID-19 Impact and Growth Analysis Report–Segmented by Action Mechanism, Drug Class and Region (North America, Europe, Asia pacific, Latin America, Middle East and Africa)–Industry Forecast (2022 to 2027). Antibiotics Market, 2023. Available online: https://www.marketdataforecast.com/market-reports/antibiotics-market (accessed on 10 September 2023).
- Maslikowska, J.A.; Walker, S.A.; Elligsen, M.; Mittmann, N.; Palmay, L.; Daneman, N.; Simor, A. Impact of infection with extended-spectrum beta-lactamase-producing Escherichia coli or Klebsiella species on outcome and hospitalization costs. J. Hosp. Infect. 2016, 92, 33–41. [Google Scholar] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC-Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [PubMed]
- Vardakas, K.Z.; Tansarli, G.S.; Rafailidis, P.I.; Falagas, M.E. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum beta-lactamases: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2012, 67, 2793–2803. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-H.; Liu, C.-W.; Liu, P.-Y. Extended-Spectrum β-Lactamases (ESBL) Producing Bacteria in Animals. Antibiotics 2023, 12, 661. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control. 2017, 6, 47. [Google Scholar] [CrossRef]
- Woerther, P.L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: Toward the globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Rabbi, B.; Sultana, S. Antibiotic resistance in Bangladesh: A systematic review. Int. J. Infect. Dis. 2019, 80, 54–61. [Google Scholar] [CrossRef]
- Rahman, S.U.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The Growing Genetic and Functional Diversity of Extended Spectrum Beta-Lactamases. BioMed Res. Int. 2018, 2018, 9519718. [Google Scholar] [CrossRef]
- Pana, Z.D.; Zaoutis, T. Treatment of extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBLs) infections: What have we learned until now? F1000Research 2018, 7, F1000. [Google Scholar]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar]
- Mammeri, H.; Van De Loo, M.; Poirel, L.; Martinez-Martinez, L.; Nordmann, P. Emergence of Plasmid-Mediated Quinolone Resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 2005, 49, 71–76. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [PubMed]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Bradford, P.A. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [PubMed]
- Rawat, D.; Nair, D. Extended-spectrum beta-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar]
- Bialvaei, A.Z.; Kafil, H.S.; Asgharzadeh, M.; Yousefi, M. CTX-M extended-spectrum β-lactamase-producing Klebsiella spp., Salmonella spp., Shigella spp. and Escherichia coli isolates in Iranian hospitals. Braz. J. Microbiol. 2016, 47, 706–711. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Vatopoulos, A.C.; Katsanis, G.; Tzelepi, E. Rare case of failure by an automated system to detect extended-spectrum beta-lactamase in a cephalosporin-resistant Klebsiella pneumoniae isolate. J. Clin. Microbiol. 1999, 37, 2388. [Google Scholar] [CrossRef]
- Madec, J.Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-spectrum beta-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: A threat for humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995, 39, 1211–1233. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar]
- Amelia, A.; Nugroho, A.; Harijanto, P.N. Diagnosis and Management of Infections Caused by Enterobacteriaceae Producing Extended-Spectrum b-Lactamase. Acta Med. Indones. 2016, 48, 156–166. [Google Scholar]
- Castanheira, M.; Mendes, R.E.; Jones, R.N.; Sader, H.S. Changes in the Frequencies of beta-Lactamase Genes among Enterobacteriaceae Isolates in U.S. Hospitals, 2012 to 2014: Activity of Ceftazidime-Avibactam Tested against beta-Lactamase-Producing Isolates. Antimicrob. Agents Chemother. 2016, 60, 4770–4777. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, A.; Schweighart, S.; Grimm, H. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 1990, 18, 294–298. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-type beta-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [PubMed]
- Poirel, L.; Lartigue, M.F.; Decousser, J.W.; Nordmann, P. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 447–450. [Google Scholar] [CrossRef] [PubMed]
- Seiffert, S.N.; Hilty, M.; Perreten, V.; Endimiani, A. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: An emerging problem for human health? Drug Resist. Update 2013, 16, 22–45. [Google Scholar]
- Zhao, W.-H.; Hu, Z.-Q. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef]
- Waldor, M.K. Mobilizable genomic islands: Going mobile with oriT mimicry. Mol. Microbiol. 2010, 78, 537–540. [Google Scholar] [CrossRef]
- Cambray, G.; Guerout, A.M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- Archer, G.L.; Polk, R.E. Treatment and prophylaxis of bacterial infections. Harrisons Princ. Intern. Med. 2005, 16, 789. [Google Scholar]
- Rahman, M.M.; Jahan, W.A. Clinical Laboratory and Molecular Detection of Extended Spectrum beta lactamases: A Review Update. Bangladesh J. Infect. Dis. 2015, 1, 12–17. [Google Scholar] [CrossRef][Green Version]
- CLSI 2012; Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012.
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008, 14, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R.; Cantón, R.; Brown, D.F.J.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef]
- Correa-Martínez, C.L.; Idelevich, E.A.; Sparbier, K.; Kostrzewa, M.; Becker, K. Rapid Detection of Extended-Spectrum β-Lactamases (ESBL) and AmpC β-Lactamases in Enterobacterales: Development of a Screening Panel Using the MALDI-TOF MS-Based Direct-on-Target Microdroplet Growth Assay. Front. Microbiol. 2019, 10, 13. [Google Scholar] [CrossRef]
- Colodner, R.; Reznik, B.; Gal, V.; Yamazaki, H.; Hanaki, H.; Kubo, R. Evaluation of a novel kit for the rapid detection of extended-spectrum beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, Y.; Kosai, K.; Yamakawa, H.; Kaku, N.; Uno, N.; Morinaga, Y.; Hasegawa, H.; Yanagihara, K. Detection of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae using the MALDI Biotyper Selective Testing of Antibiotic Resistance–β-Lactamase (MBT STAR-BL) assay. J. Microbiol. Methods 2019, 160, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Keshta, A.S.; Elamin, N.; Hasan, M.R.; Pérez-López, A.; Roscoe, D.; Tang, P.; Suleiman, M. Evaluation of Rapid Immunochromatographic Tests for the Direct Detection of Extended Spectrum Beta-Lactamases and Carbapenemases in Enterobacterales Isolated from Positive Blood Cultures. Microbiol. Spectr. 2021, 9, e0078521. [Google Scholar] [CrossRef] [PubMed]
- Zboromyrska, Y.; Rico, V.; Pitart, C.; Fernández-Pittol, M.J.; Soriano, Á.; Bosch, J. Implementation of a New Protocol for Direct Identification from Urine in the Routine Microbiological Diagnosis. Antibiotics 2022, 11, 582. [Google Scholar] [CrossRef]
- Wang, S.; Wang, S.; Tang, Y.; Peng, G.; Hao, T.; Wu, X.; Wei, J.; Qiu, X.; Zhou, D.; Zhu, S.; et al. Detection of Klebsiella pneumonia DNA and ESBL positive strains by PCR-based CRISPR-LbCas12a system. Front. Microbiol. 2023, 14, 1128261. [Google Scholar] [CrossRef]
- Carattoli, A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef]
- Karaiskos, I.; Giamarellou, H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics 2020, 9, 61. [Google Scholar] [CrossRef]
- Pilmis, B.; Zahar, J.-R. Ventilator-associated pneumonia related to ESBL-producing gram negative bacilli. Ann. Transl. Med. 2018, 6, 424. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.I.; Wi, Y.M.; Lee, M.Y.; Ko, K.S.; Chung, D.R.; Peck, K.R.; Lee, N.Y.; Song, J.H. Epidemiology and risk factors of community onset infections caused by extended-spectrum beta-lactamase-producing Escherichia coli strains. J. Clin. Microbiol. 2012, 50, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.; Furuya-Kanamori, L.; Ezure, Y.; Harris, P.N.A.; Paterson, D.L. Adverse clinical outcomes associated with infections by Enterobacterales producing ESBL (ESBL-E): A systematic review and meta-analysis. JAC-Antimicrob. Resist. 2021, 3, dlab068. [Google Scholar] [CrossRef]
- Chong, Y.; Shimoda, S.; Shimono, N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2018, 61, 185–188. [Google Scholar] [CrossRef]
- Batchelor, M.; Threlfall, E.J.; Liebana, E. Cephalosporin resistance among animal-associated Enterobacteria: A current perspective. Expert Rev. Anti-Infect. Ther. 2005, 3, 403–417. [Google Scholar] [CrossRef]
- Kruse, H.; Sørum, H. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 1994, 60, 4015–4021. [Google Scholar] [CrossRef]
- Brinas, L.; Moreno, M.A.; Teshager, T.; Zarazaga, M.; Saenz, Y.; Porrero, C.; Dominguez, L.; Torres, C. Beta-lactamase characterization in Escherichia coli isolates with diminished susceptibility or resistance to extended-spectrum cephalosporins recovered from sick animals in Spain. Microb. Drug Resist. 2003, 9, 201–209. [Google Scholar] [CrossRef]
- Donati, V.; Feltrin, F.; Hendriksen, R.S.; Svendsen, C.A.; Cordaro, G.; García-Fernández, A.; Lorenzetti, S.; Lorenzetti, R.; Battisti, A.; Franco, A. Extended-Spectrum-Beta-Lactamases, AmpC Beta-Lactamases and Plasmid Mediated Quinolone Resistance in Klebsiella spp. from Companion Animals in Italy. PLoS ONE 2014, 9, e90564. [Google Scholar] [CrossRef]
- Day, M.J.; Rodriguez, I.; van Essen-Zandbergen, A.; Dierikx, C.; Kadlec, K.; Schink, A.K.; Wu, G.; Chattaway, M.A.; DoNascimento, V.; Wain, J.; et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antimicrob. Chemother. 2016, 71, 1178–1182. [Google Scholar] [CrossRef]
- Uivaraseanu, B.; Bungau, S.; Tit, D.M.; Fratila, O.; Rus, M.; Maghiar, T.A.; Maghiar, O.; Pantis, C.; Vesa, C.M.; Zaha, D.C. Clinical, Pathological and Microbiological Evaluation of Diabetic Foot Syndrome. Medicina 2020, 56, 380. [Google Scholar] [CrossRef] [PubMed]
- Jolivet, S.; Lescure, F.-X.; Armand-Lefevre, L.; Raffoul, R.; Dilly, M.-P.; Ghodbane, W.; Nataf, P.; Lucet, J.-C. Surgical site infection with extended-spectrum β-lactamase-producing Enterobacteriaceae after cardiac surgery: Incidence and risk factors. Clin. Microbiol. Infect. 2017, 24, 283–288. [Google Scholar] [CrossRef]
- Lazarus, B.; Paterson, D.L.; Mollinger, J.L.; Rogers, B.A. Do Human Extraintestinal Escherichia coli Infections Resistant to Expanded-Spectrum Cephalosporins Originate from Food-Producing Animals? A Systematic Review. Clin. Infect. Dis. 2014, 60, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Huijbers, P.M.; van Hoek, A.H.; Graat, E.A.; Haenen, A.P.; Florijn, A.; Hengeveld, P.D.; van Duijkeren, E. Methicillin-resistant Staphylococcus aureus and extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in broilers and in people living and/or working on organic broiler farms. Vet. Microbiol. 2015, 176, 120–125. [Google Scholar] [PubMed]
- Dohmen, W.; Bonten, M.J.; Bos, M.E.; van Marm, S.; Scharringa, J.; Wagenaar, J.A.; Heederik, D.J. Carriage of extended-spectrum beta-lactamases in pig farmers is associated with occurrence in pigs. Clin. Microbiol. Infect. 2015, 21, 917–923. [Google Scholar] [CrossRef]
- Brechet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum beta-lactamase-producing Escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [PubMed]
- Hernandez, J.; Johansson, A.; Stedt, J.; Bengtsson, S.; Porczak, A.; Granholm, S.; Gonzalez-Acuna, D.; Olsen, B.; Bonnedahl, J.; Drobni, M. Characterization and comparison of extended-spectrum beta-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin’s gulls (Leucophaeus pipixcan) and humans in Chile. PLoS ONE 2013, 8, e76150. [Google Scholar]
- Hasan, B.; Melhus, Å.; Sandegren, L.; Alam, M.; Olsen, B. The Gull (Chroicocephalus brunnicephalus) as an Environmental Bioindicator and Reservoir for Antibiotic Resistance on the Coastlines of the Bay of Bengal. Microb. Drug Resist. 2014, 20, 466–471. [Google Scholar] [CrossRef]
- Mohsin, M.; Raza, S.; Schaufler, K.; Roschanski, N.; Sarwar, F.; Semmler, T.; Schierack, P.; Guenther, S. High Prevalence of CTX-M-15-Type ESBL-Producing E. coli from Migratory Avian Species in Pakistan. Front. Microbiol. 2017, 8, 2476. [Google Scholar] [CrossRef]
- Islam, M.A.; Amin, M.B.; Roy, S.; Asaduzzaman, M.; Islam, R.; Navab-Daneshmand, T.; Mattioli, M.C.; Kile, M.L.; Levy, K.; Julian, T.R. Fecal Colonization with Multidrug-Resistant E. coli Among Healthy Infants in Rural Bangladesh. Front. Microbiol. 2019, 10, 640. [Google Scholar] [CrossRef]
- Tamma, P.D.; Rodriguez-Bano, J. The Use of Noncarbapenem beta-Lactams for the Treatment of Extended-Spectrum beta-Lactamase Infections. Clin. Infect. Dis. 2017, 64, 972–980. [Google Scholar] [CrossRef]
- Maseda, E.; Suárez de la Rica, A. Controversies in the management of ESBL-producing Enterabacterales. Clinical Implications. Rev. Esp. Quim. 2022, 35 (Suppl. 3), 41–45. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, D.R.; Bassetti, M.; De Rosa, F.G.; Del Bono, V.; Grossi, P.A.; Menichetti, F.; Pea, F.; Rossolini, G.M.; Tumbarello, M.; Viale, P.; et al. Ceftolozane/tazobactam: Place in therapy. Expert Rev. Anti-Infect. Ther. 2018, 16, 307–320. [Google Scholar] [CrossRef]
- Karaiskos, I.; Giamarellou, H. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: Current and emerging therapeutic approaches. Expert Opin. Pharmacother. 2014, 15, 1351–1370. [Google Scholar] [CrossRef] [PubMed]
- Chastain, D.B.; White, B.P.; Cretella, D.A.; Bland, C.M. Is It Time to Rethink the Notion of Carbapenem-Sparing Therapy Against Extended-Spectrum beta-Lactamase-Producing Enterobacteriaceae Bloodstream Infections? A Critical Review. Ann. Pharmacother. 2018, 52, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Endimiani, A.; Perez, F.; Bonomo, R.A. Cefepime: A reappraisal in an era of increasing antimicrobial resistance. Expert Rev. Anti-Infect. Ther. 2008, 6, 805–824. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martinez-Martinez, L.; Pascual, A. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Updates 2016, 29, 13–29. [Google Scholar] [CrossRef]
- Fernandez-Martinez, M.; Ruiz Del Castillo, B.; Lecea-Cuello, M.J.; Rodriguez-Bano, J.; Pascual, A.; Martinez-Martinez, L. Prevalence of Aminoglycoside-Modifying Enzymes in Escherichia coli and Klebsiella pneumoniae Producing Extended Spectrum beta-Lactamases Collected in Two Multicenter Studies in Spain. Microbial. Drug Resist. 2018, 24, 367–376. [Google Scholar] [CrossRef]
- FDA. FDA Updates Warnings for Fluoroquinolone Antibiotics on Risks of Mental Health and Low Blood Sugar Adverse Reactions; U.S. Food and Drug Administration (FDA). Available online: https://www.fda.gov/news-events/press-announcements/fda-updates-warnings-fluoroquinolone-antibiotics-risks-mental-health-and-low-blood-sugar-adverse (accessed on 10 September 2023).
- Bouxom, H.; Fournier, D.; Bouiller, K.; Hocquet, D.; Bertrand, X. Which non-carbapenem antibiotics are active against extended-spectrum beta-lactamase-producing Enterobacteriaceae? Int. J. Antimicrob. Agents 2018, 52, 100–103. [Google Scholar] [CrossRef]
- Karaiskos, I.; Souli, M.; Giamarellou, H. Plazomicin: An investigational therapy for the treatment of urinary tract infections. Expert Opin. Investig. Drugs 2015, 24, 1501–1511. [Google Scholar] [CrossRef]
- Karaiskos, I.; Lagou, S.; Pontikis, K.; Rapti, V.; Poulakou, G. The "Old" and the "New" Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front. Public Health 2019, 7, 151. [Google Scholar] [PubMed]
- Morrissey, I.; Olesky, M.; Hawser, S.; Lob, S.H.; Karlowsky, J.A.; Corey, G.R.; Bassetti, M.; Fyfe, C. In Vitro Activity of Eravacycline against Gram-Negative Bacilli Isolated in Clinical Laboratories Worldwide from 2013 to 2017. Antimicrob. Agents Chemother. 2020, 64, e01699-19. [Google Scholar] [CrossRef]
- Huband, M.D.; Pfaller, M.A.; Shortridge, D.; Flamm, R.K. Surveillance of omadacycline activity tested against clinical isolates from the United States and Europe: Results from the SENTRY Antimicrobial Surveillance Programme, 2017. J. Glob. Antimicrob. Resist. 2019, 19, 56–63. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Walkty, A.J.; Karlowsky, J.A. Fosfomycin: A First-Line Oral Therapy for Acute Uncomplicated Cystitis. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 2082693. [Google Scholar] [CrossRef]
- Pardo, J.R.P.; Villar, S.S.; Ramos, J.C.R.; Pintado, V. Infections caused by carbapenemase-producing Enterobacteriaceae: Risk factors, clinical features and prognosis. Enferm. Infecc. Microbiol. Clin. 2014, 32 (Suppl. 4), 41–48. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.; Roy, D.N.; Tajmim, A.; Rajib, S.S.; Hossain, M.; Farzana, F.; Yasmen, N. Prescription antibiotics for outpatients in Bangladesh: A cross-sectional health survey conducted in three cities. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 15. [Google Scholar] [CrossRef]
- Mustufa, A.; Ahmed, I.; Fareed, M.; Anwar, T. Factors Leading to Acquired Bacterial Resistance Due to Antibiotics in Pakistan. Curr. Trends Biotechnol. Microbiol. 2018, 1, 1–7. [Google Scholar]
- Mitu, F.S.; Al Maruf, M.A.; Mahanty, A.; Huda, A.N.; Khan, S.A.; Rahman, M.M. Prevalence of extended spectrum beta-lactamase (ESBL) and AmpC beta-lactamase producing bacteria in urinary tract infection patients in Bangladesh. Malays. J. Microbiol. 2019, 15, 204–212. [Google Scholar]
- Hawkey, P.M. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin. Microbiol. Infect. 2008, 14, 159–165. [Google Scholar] [CrossRef]
- Begum, N.; Shamsuzzaman, S.M. Emergence of CTX-M-15 producing E. coli O25b-ST131 clone in a tertiary care hospital of Bangladesh. Malays. J. Pathol. 2016, 38, 241–249. [Google Scholar]
- Ranjan, A.; Shaik, S.; Nandanwar, N.; Hussain, A.; Tiwari, S.K.; Semmler, T.; Jadhav, S.; Wieler, L.H.; Alam, M.; Colwell, R.R.; et al. Comparative Genomics of Escherichia coli Isolated from Skin and Soft Tissue and Other Extraintestinal Infections. mBio 2017, 8, e01070-17. [Google Scholar] [CrossRef] [PubMed]
- Suresh, A.; Ranjan, A.; Jadhav, S.; Hussain, A.; Shaik, S.; Alam, M.; Baddam, R.; Wieler, L.H.; Ahmed, N. Molecular Genetic and Functional Analysis of pks-Harboring, Extra-Intestinal Pathogenic Escherichia coli from India. Front. Microbiol. 2018, 9, 2631. [Google Scholar] [CrossRef]
- Parvez, A.K.; Marzan, M.; Liza, S.M.; Mou, T.J.; Azmi, I.J.; Mahmud, S.R.A.Z.H. Prevalence of Inhibitor Resistant Beta Lactamase Producing E. coli in Human and Poultry Origin of Bangladesh. J. Bacteriol. Parasitol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Souverein, D.; Euser, S.M.; van der Reijden, W.A.; Herpers, B.L.; Kluytmans, J.; Rossen, J.W.A.; Boer, J.W.D. Clinical sensitivity and specificity of the Check-Points Check-Direct ESBL Screen for BD MAX, a real-time PCR for direct ESBL detection from rectal swabs. J. Antimicrob. Chemother. 2017, 72, 2512–2518. [Google Scholar] [CrossRef][Green Version]
- Khan, E.R.; Aung, M.S.; Paul, S.K.; Ahmed, S.; Haque, N.; Ahamed, F.; Sarkar, S.R.; Roy, S.; Rahman, M.M.; Mahmud, M.C.; et al. Prevalence and Molecular Epidemiology of Clinical Isolates of Escherichia coli and Klebsiella pneumoniae Harboring Extended-Spectrum Beta-Lactamase and Carbapenemase Genes in Bangladesh. Microbial. Drug Resist. 2018, 24, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, S.; Islam, R. Beta-lactamase-producing Escherichia coli in Bangladesh: Their phenotypic and molecular characteristics. Dhaka Univ. J. Biol. Sci. 2019, 28, 71–81. [Google Scholar] [CrossRef]
- Chakraborty, S.; Mohsina, K.; Sarker, P.K.; Alam, Z.; Karim, M.I.A.; Abu Sayem, S.M. Prevalence, antibiotic susceptibility profiles and ESBL production in Klebsiella pneumoniae and Klebsiella oxytoca among hospitalized patients. Period. Biol. 2016, 118, 8486742. [Google Scholar] [CrossRef]
- Yasmin, T.; Hossain, A.; Paul, S.K.; Mowla, G.; Sultana, S. Prevalence of CTX-M? lactamases among Gram negative bacteria in a tertiary care hospital in Bangladesh. Ibrahim Med. Coll. J. 2016, 9, 26–30. [Google Scholar] [CrossRef][Green Version]
- Gajamer, V.R.; Bhattacharjee, A.; Paul, D.; Ingti, B.; Sarkar, A.; Kapil, J.; Singh, A.K.; Pradhan, N.; Tiwari, H.K. High prevalence of carbapenemase, AmpC β-lactamase and aminoglycoside resistance genes in extended-spectrum β-lactamase-positive uropathogens from Northern India. J. Glob. Antimicrob. Resist. 2020, 20, 197–203. [Google Scholar] [PubMed]
- Singh, N.; Pattnaik, D.; Neogi, D.K.; Jena, J.; Mallick, B. Prevalence of ESBL in Escherichia coli Isolates Among ICU Patients in a Tertiary Care Hospital. J. Clin. Diagn. Res. 2016, 10, 1. [Google Scholar] [CrossRef]
- Ravikant, K.P.; Ranotkar, S.; Zutshi, S.; Lahkar, M.; Phukan, C.; Saikia, K.K. Prevalence and identification of extended spectrum β-lactamases (ESBL) in Escherichia coli isolated from a tertiary care hospital in North-East India. Indian J. Exp. Biol. 2016, 54, 108–114. [Google Scholar]
- Karunasagar, I.; Rohit, A.; Deekshit, V.K.; Balaraj, M.; Alandur, V.S.; Abraham, G.; Karunasagar, I. CTX-M type extended-spectrum β-lactamase in Escherichia coli isolated from extra-intestinal infections in a tertiary care hospital in south India. Indian J. Med. Res. 2019, 149, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Sengupta, A.; Kumar, A.; Singh, U.K.; Jaiswal, A.K.; Das, P.; Das, S. Molecular Epidemiology of Extended-Spectrum β-Lactamase-Producing Escherichia coli Pathotypes in Diarrheal Children from Low Socioeconomic Status Communities in Bihar, India: Emergence of the CTX-M Type. Infect. Dis. 2017, 10, 1178633617739018. [Google Scholar]
- Mathur, P.; Bajpai, V.; Govindaswamy, A.; Khurana, S.; Batra, P.; Aravinda, A.; Katoch, O.; Hasan, F.; Malhotra, R. Phenotypic & genotypic profile of antimicrobial resistance in Pseudomonas species in hospitalized patients. Indian J. Med. Res. 2019, 149, 216–221. [Google Scholar] [CrossRef]
- Umair, M.; Mohsin, M.; Ali, Q.; Qamar, M.U.; Raza, S.; Ali, A.; Guenther, S.; Schierack, P. Prevalence and Genetic Relatedness of Extended Spectrum-β-Lactamase-Producing Escherichia coli Among Humans, Cattle, and Poultry in Pakistan. Microb. Drug Resist. 2019, 25, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Ullah, F.; Malik, S.A.; Ahmed, J. Antimicrobial susceptibility and ESBL prevalence in Pseudomonas aeruginosa isolated from burn patients in the North West of Pakistan. Burns 2009, 35, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Abrar, S.; Ain, N.U.; Liaqat, H.; Hussain, S.; Rasheed, F.; Riaz, S. Distribution of bla (CTX - M), bla (TEM), bla (SHV) and bla (OXA) genes in Extended-spectrum-β-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrob. Resist. Infect. Control 2019, 8, 80. [Google Scholar] [CrossRef]
- Abbas, G.; Khan, I.; Mohsin, M.; Sajjad-Ur-Rahman, S.-U.; Younas, T.; Ali, S. High rates of CTX-M group-1 extended-spectrum β-lactamases producing Escherichia coli from pets and their owners in Faisalabad, Pakistan. Infect. Drug Resist. 2019, 12, 571–578. [Google Scholar] [CrossRef]
- Chaudhry, T.H.; Aslam, B.; Arshad, M.; Nawaz, Z.; Waseem, M. Occurrence of ESBL-producing Klebsiella pneumoniae in hospital settings and waste. Pak. J. Pharm. Sci. 2019, 32, 773–778. [Google Scholar]
- Abrar, S.; Vajeeha, A.; Ul-Ain, N.; Riaz, S. Distribution of CTX-M group I and group III β-lactamases produced by Escherichia coli and klebsiella pneumoniae in Lahore, Pakistan. Microb. Pathog. 2017, 103, 8–12. [Google Scholar] [CrossRef]
- Ullah, W.; Qasim, M.; Rahman, H.; Khan, S.; Rehman, Z.U.; Ali, N.; Muhammad, N. CTX-M-15 and OXA-10 beta lactamases in multi drug resistant Pseudomonas aeruginosa: First report from Pakistan. Microb. Pathog. 2017, 105, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Saleem, R.; Ejaz, H.; Zafar, A.; Younas, S.; Rathore, A.W. Phenotypic characterization of extended-spectrum-beta-lactamase producing E. coli from healthy individuals, patients, sewage sludge, cattle, chickens and raw meat. Pak. J. Med. Sci. 2017, 33, 886–890. [Google Scholar] [CrossRef] [PubMed]
- Ehsan, B.; Haque, A.; Qasim, M.; Ali, A.; Sarwar, Y. High prevalence of extensively drug resistant and extended spectrum beta lactamases (ESBLs) producing uropathogenic Escherichia coli isolated from Faisalabad, Pakistan. World J. Microbiol. Biotechnol. 2023, 39, 132. [Google Scholar] [CrossRef]
- Hussain, A.; Shaik, S.; Ranjan, A.; Suresh, A.; Sarker, N.; Semmler, T.; Wieler, L.H.; Alam, M.; Watanabe, H.; Chakravortty, D.; et al. Genomic and Functional Characterization of Poultry Escherichia coli from India Revealed Diverse Extended-Spectrum β-Lactamase-Producing Lineages with Shared Virulence Profiles. Front. Microbiol. 2019, 10, 2766. [Google Scholar] [CrossRef]
- Parvin, M.; Talukder, S.; Ali, M.; Chowdhury, E.H.; Rahman, M.; Islam, M. Antimicrobial Resistance Pattern of Escherichia coli Isolated from Frozen Chicken Meat in Bangladesh. Pathogens 2020, 9, 420. [Google Scholar]
- Mamun, M.-M.; Hassan, J.; Nazir, K.H.M.N.H.; Islam, M.-A.; Zesmin, K.; Rahman, M.-B. Prevalence and Molecular Detection of Quinolone-Resistant E. coli in Rectal Swab of Apparently Healthy Cattle in Bangladesh. Int. J. Trop. Dis. Health 2017, 24, 1–7. [Google Scholar] [CrossRef]
- Nirupama, K.R.; Kumar, O.R.V.; Pruthvishree, B.S.; Sinha, D.K.; Murugan, M.S.; Krishnaswamy, N.; Singh, B.R. Molecular characterisation of bla(OXA-48) carbapenemase-, extended-spectrum β-lactamase- and Shiga toxin-producing Escherichia coli isolated from farm piglets in India. J. Glob. Antimicrob. Resist. 2018, 13, 201–205. [Google Scholar]
- Mahanti, A.; Ghosh, P.; Samanta, I.; Joardar, S.N.; Bandyopadhyay, S.; Bhattacharyya, D.; Banerjee, J.; Batabyal, S.; Sar, T.K.; Dutta, T.K. Prevalence of CTX-M-Producing Klebsiella spp. in Broiler, Kuroiler, and Indigenous Poultry in West Bengal State, India. Microb. Drug Resist 2018, 24, 299–306. [Google Scholar]
- Batabyal, K.; Banerjee, A.; Pal, S.; Dey, S.; Joardar, S.N.; Samanta, I.; Isore, D.P.; Singh, A.D. Detection, characterization, and antibiogram of extended-spectrum beta-lactamase Escherichia coli isolated from bovine milk samples in West Bengal, India. Veter. World 2018, 11, 1423. [Google Scholar] [CrossRef]
- Lalruatdiki, A.; Dutta, T.K.; Roychoudhury, P.; Subudhi, P.K. Extended-spectrum β-lactamases producing multidrug resistance Escherichia coli, Salmonella and Klebsiella pneumoniae in pig population of Assam and Meghalaya, India. Vet. World 2018, 11, 868–873. [Google Scholar]
- Wajid, M.; Saleemi, M.K.; Sarwar, Y.; Ali, A. Detection and characterization of multidrug-resistant Salmonella enterica serovar Infantis as an emerging threat in poultry farms of Faisalabad, Pakistan. J. Appl. Microbiol. 2019, 127, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Rahman, S.U.; Zhang, L.; Shahid, M.; Han, D.; Gao, J.; Zhang, S.; Ruegg, P.L.; Saddique, U.; Han, B. Characteristics and genetic diversity of multi-drug resistant extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from bovine mastitis. Oncotarget 2017, 8, 90144–90163. [Google Scholar] [CrossRef]
- Saeed, M.A.; Saqlain, M.; Waheed, U.; Ehtisham-Ul-Haque, S.; Khan, A.U.; Rehman, A.U.; Sajid, M.; Atif, F.A.; Neubauer, H.; El-Adawy, H. Cross-Sectional Study for Detection and Risk Factor Analysis of ESBL-Producing Avian Pathogenic Escherichia coli Associated with Backyard Chickens in Pakistan. Antibiotics 2023, 12, 934. [Google Scholar] [CrossRef]
- Rahman, M.; Husna, A.; Elshabrawy, H.A.; Alam, J.; Runa, N.Y.; Badruzzaman, A.T.M.; Banu, N.A.; Al Mamun, M.; Paul, B.; Das, S.; et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci. Rep. 2020, 10, 21999. [Google Scholar] [CrossRef] [PubMed]
- Jobayer, M.; Afroz, Z.; Nahar, S.S.; Begum, A.; Begum, S.A.; Shamsuzzaman, S. Antimicrobial susceptibility pattern of extended-spectrum beta-lactamases producing organisms isolated in a Tertiary Care Hospital, Bangladesh. Int. J. Appl. Basic Med. Res. 2017, 7, 189–192. [Google Scholar] [CrossRef]
- Lina, T.T.; Khajanchi, B.K.; Azmi, I.J.; Islam, M.A.; Mahmood, B.; Akter, M.; Banik, A.; Alim, R.; Navarro, A.; Perez, G.; et al. Phenotypic and Molecular Characterization of Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Bangladesh. PLoS ONE 2014, 9, e108735. [Google Scholar] [CrossRef]
- Hossain, S.; Ali, S.; Hossain, M.; Uddin, S.Z.; Moniruzzaman, M.; Islam, M.R.; Shohael, A.M.; Islam, S.; Ananya, T.H.; Rahman, M.; et al. ESBL Producing Escherichia coli in Faecal Sludge Treatment Plants: An Invisible Threat to Public Health in Rohingya Camps, Cox’s Bazar, Bangladesh. Front. Public Health 2021, 9, 783019. [Google Scholar] [CrossRef]
- Rogawski, E.T.; Platts-Mills, J.A.; Seidman, J.C.; John, S.; Mahfuz, M.; Ulak, M.; Shrestha, S.K.; Soofi, S.B.; Yori, P.P.; Mduma, E.; et al. Use of antibiotics in children younger than two years in eight countries: A prospective cohort study. Bull. World Health Organ. 2016, 95, 49–61. [Google Scholar] [CrossRef]
- Haque, A.; Yoshizumi, A.; Saga, T.; Ishii, Y.; Tateda, K. ESBL-producing Enterobacteriaceae in environmental water in Dhaka, Bangladesh. J. Infect. Chemother. 2014, 20, 735–737. [Google Scholar] [CrossRef]
- Hasan, B.; Sandegren, L.; Melhus, A.; Drobni, M.; Hernandez, J.; Waldenstrom, J.; Alam, M.; Olsen, B. Antimicrobial drug-resistant Escherichia coli in wild birds and free-range poultry, Bangladesh. Emerg. Infect. Dis. 2012, 18, 2055–2058. [Google Scholar]
- Hasan, B.; Olsen, B.; Alam, A.; Akter, L.; Melhus, A. Dissemination of the multidrug-resistant extended-spectrum beta-lactamase-producing Escherichia coli O25b-ST131 clone and the role of house crow (Corvus splendens) foraging on hospital waste in Bangladesh. Clin. Microbiol. Infect. 2015, 21, 1000.e1–1000.e4. [Google Scholar]
- Hasan, B.; Islam, K.; Ahsan, M.; Hossain, Z.; Rashid, M.; Talukder, B.; Ahmed, K.U.; Olsen, B.; Abul Kashem, M. Fecal carriage of multi-drug resistant and extended spectrum β-lactamases producing E. coli in household pigeons, Bangladesh. Vet. Microbiol. 2014, 168, 221–224. [Google Scholar] [PubMed]
- Islam, M.S.; Rahman, A.T.; Hassan, J.; Rahman, M.T. Extended-spectrum beta-lactamase in Escherichia coli isolated from humans, animals, and environments in Bangladesh: A One Health perspective systematic review and meta-analysis. One Health 2023, 16, 100526. [Google Scholar] [PubMed]
- Al Azad, M.A.R.; Rahman, M.; Amin, R.; Begum, M.I.A.; Fries, R.; Husna, A.; Khairalla, A.S.; Badruzzaman, A.; El Zowalaty, M.E.; Na Lampang, K.; et al. Susceptibility and Multidrug Resistance Patterns of Escherichia coli Isolated from Cloacal Swabs of Live Broiler Chickens in Bangladesh. Pathogens 2019, 8, 118. [Google Scholar] [CrossRef]
- Saifullah, K.; Mamun, M.; Rubayet, R.; Nazir, K.; Zesmin, K.; Rahman, T. Molecular detection of Salmonella spp. isolated from apparently healthy pigeon in Mymensingh, Bangladesh and their antibiotic resistance pattern. J. Adv. Veter.-Anim. Res. 2016, 3, 51. [Google Scholar] [CrossRef]
- Akond, M.A.; Shirin, M.; Alam, S.; Hassan, S.; Rahman, M.; Hoq, M. Frequency of drug resistant Salmonella spp. isolated from poultry samples in Bangladesh. Stamford J. Microbiol. 2013, 2, 15–19. [Google Scholar] [CrossRef]
- Sultana, F.; Kamrunnahar; Afroz, H.; Jahan, A.; Fakruddin; Datta, S. Multi–antibiotic resistant bacteria in frozen food (ready to cook food) of animal origin sold in Dhaka, Bangladesh. Asian Pac. J. Trop. Biomed. 2014, 4, S268–S271. [Google Scholar] [CrossRef]
- Hossain, G.; Saha, S.; Rahman, M.; Singha, J.; Mamun, A. Isolation, Identification and Antibiogram Study of Pseudomonas Aeruginosa from Cattle in Bangladesh. J. Veter.-Adv. 2013, 3, 180–185. [Google Scholar] [CrossRef]
- Sobur, M.A.; Sabuj, A.A.M.; Sarker, R.; Rahman, A.M.M.T.; Kabir, S.M.L.; Rahman, M.T. Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Vet. World 2019, 12, 984–993. [Google Scholar] [CrossRef]
- Tanzin, T.; Nazir, K.; Zahan, M.; Parvej, S.; Zesmin, K.; Rahman, T. Antibiotic resistance profile of bacteria isolated from raw milk samples of cattle and buffaloes. J. Adv. Veter.-Anim. Res. 2016, 3, 62. [Google Scholar] [CrossRef]
- Karim, A.; Poirel, L.; Nagarajan, S.; Nordmann, P. Plasmid-mediated extended-spectrum β-lactamase (CTX-M-3 like) from India and gene association with insertion sequence IS Ecp1. FEMS Microbiol. Lett. 2001, 201, 237–241. [Google Scholar] [PubMed]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Kuralayanapalya, S.P.; Patil, S.S.; Hamsapriya, S.; Shinduja, R.; Roy, P.; Amachawadi, R.G. Prevalence of extended-spectrum beta-lactamase producing bacteria from animal origin: A systematic review and meta-analysis report from India. PLoS ONE 2019, 14, e0221771. [Google Scholar] [CrossRef] [PubMed]
- Peter, E. Deadly superbugs invade U.S. health care facilities. USA Today, 29 November 2012. [Google Scholar]
- Miriagou, V.; Cornaglia, G.; Edelstein, M.; Galani, I.; Giske, C.; Gniadkowski, M.; Malamou-Lada, E.; Martinez-Martinez, L.; Navarro, F.; Nordmann, P.; et al. Acquired carbapenemases in Gram-negative bacterial pathogens: Detection and surveillance issues. Clin. Microbiol. Infect. 2010, 16, 112–122. [Google Scholar] [CrossRef]
- Yang, H.; Aitha, M.; Hetrick, A.M.; Richmond, T.K.; Tierney, D.L.; Crowder, M.W. Mechanistic and spectroscopic studies of metallo-beta-lactamase NDM-1. Biochemistry 2012, 51, 3839–3847. [Google Scholar]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Wei, W.-J.; Yang, H.-F.; Ye, Y.; Li, J.-B. New Delhi Metallo-β-Lactamase-Mediated Carbapenem Resistance: Origin, Diagnosis, Treatment and Public Health Concern. Chin. Med. J. 2015, 128, 1969–1976. [Google Scholar]
- Liu, Z.; Li, J.; Wang, X.; Liu, D.; Ke, Y.; Wang, Y.; Shen, J. Novel Variant of New Delhi Metallo-β-lactamase, NDM-20, in Escherichia coli. Front. Microbiol. 2018, 9, 248. [Google Scholar] [CrossRef]
- Cannatelli, A.; Giani, T.; Aiezza, N.; Di Pilato, V.; Principe, L.; Luzzaro, F.; Galeotti, C.L.; Rossolini, G.M. An allelic variant of the PmrB sensor kinase responsible for colistin resistance in an Escherichia coli strain of clinical origin. Sci. Rep. 2017, 7, 5071. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Plasmid-mediated colistin resistance: An additional antibiotic resistance menace. Clin. Microbiol. Infect. 2016, 22, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Letellier, A. Resistance to colistin: What is the fate for this antibiotic in pig production? Int. J. Antimicrob. Agents 2016, 48, 119–126. [Google Scholar] [CrossRef]
- Rakhi, N.N.; Alam, A.R.U.; Sultana, M.; Rahaman, M.; Hossain, M.A. Diversity of carbapenemases in clinical isolates: The emergence of blaVIM-5 in Bangladesh. J. Infect. Chemother. 2019, 25, 444–451. [Google Scholar] [CrossRef]
- Farzana, R.; Jones, L.S.; Barratt, A.; Rahman, M.A.; Sands, K.; Portal, E.; Boostrom, I.; Espina, L.; Pervin, M.; Uddin, A.K.M.N.; et al. Emergence of Mobile Colistin Resistance (mcr-8) in a Highly Successful Klebsiella pneumoniae Sequence Type 15 Clone from Clinical Infections in Bangladesh. mSphere 2020, 5, 1110–1128. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Steinig, E.J.; Duchene, S.; Robinson, D.A.; Monecke, S.; Yokoyama, M.; Laabei, M.; Slickers, P.; Andersson, P.; Williamson, D.; Kearns, A.; et al. Evolution and Global Transmission of a Multidrug-Resistant, Community-Associated Methicillin-Resistant Staphylococcus aureus Lineage from the Indian Subcontinent. mBio 2019, 10, e01105-19. [Google Scholar] [CrossRef]
- Sachan, D. Poor antibiotic stewardship blamed as India found to be superbug’s birthplace. Chemistry World, 23 December 2019. [Google Scholar]
- Akram, J.; Khan, A.S.; Khan, H.A.; Gilani, S.A.; Akram, S.J.; Ahmad, F.J.; Mehboob, R. Extensively Drug-Resistant (XDR) Typhoid: Evolution, Prevention, and Its Management. BioMed Res. Int. 2020, 2020, 6432580. [Google Scholar] [CrossRef]
- Abubakar, U.; Al-Anazi, M.; Alanazi, Z.; Rodríguez-Baño, J. Impact of COVID-19 pandemic on multidrug resistant gram positive and gram negative pathogens: A systematic review. J. Infect. Public Health 2023, 16, 320–331. [Google Scholar] [CrossRef]
| Screening Tests | Confirmatory Tests | Rapid Kit Test | |||||
|---|---|---|---|---|---|---|---|
| Phenotypic Methods | Genotypic Methods | ||||||
| Test Name | Antibiotic | Sensitivity | Test Name | Antibiotic | Sensitivity | ||
| Kirby-Bauer disks | Cefotaxime, ceftriaxone, ceftazidime, or aztreonam | 92–93% | Double-disk synergy test (DDST) | Cefotaxime, ceftriaxone, ceftazidime, or aztreonam | 70–80% | PCR | Cica Beta Test 1/HMRZ-86/Chromogenic cephalosporin |
| Vitek | Combination disk method, | Cefotaxime and cefepime | 100% | Nucleotide sequencing | |||
| E-test ESBL strip | Cefotaxime and ceftazidime | 71–73% | Isoelectric point determination | ||||
| Cefepime | 90% | DNA probes | |||||
| Oligotyping method | |||||||
| PCR-RFLP | |||||||
| PCR-SSCP | |||||||
| Country | ESBL | Enterobacteriaceae | Source | Prevalence | Reference | |
|---|---|---|---|---|---|---|
| 1 | Bangladesh | blaCTX-M-15 | E. coli | Urine | 80% | [83] |
| 2 | India | blaCTX-M-15 | E. coli | Skin and soft tissue | 70% | [84] |
| 3 | India | blaCTX-M-15 | E. coli | Urine, pus, extra intestinal clinical samples | 25% | [85] |
| 4 | Bangladesh | blaTEM | E. coli | Urine | 50% | [86] |
| 5 | Bangladesh | blaCTX-M-15 | E. coli | Rectal swabs | 48.2% | [87] |
| blaCTX-M-1 | 11.1% | |||||
| blaSHV-12 | 11.1% | |||||
| blaCTX-M-14 | 7.4% | |||||
| blaCTX-M-27 | 7.4% | |||||
| blaCTX-M-9 | 3.7% | |||||
| blaCTX-M-14b | 3.7% | |||||
| blaSHV-28 | 3.7% | |||||
| blaTEM-12 | 3.7% | |||||
| 6 | Bangladesh | blaCTX-M-1 | E. coli | Clinical specimens | 33.9% | [88] |
| blaCTX-M-1 | K. pneumoniae | 51.4% | ||||
| 7 | Bangladesh | Non-specific | E. coli | Urine | 25.84% | [81] |
| Non-specific | Klebsiella pneumoniae | 6.6% | ||||
| 8 | Bangladesh | blaTEM | E. coli | Urine | 22.7% | [89] |
| blaCTX-M | 24.2% | |||||
| blaSHV | 4.3% | |||||
| 9 | Bangladesh | Non-specific | K. pneumoniae | Tracheal swabs, sputum, wound swabs, pus, blood, urine | 50% | [90] |
| Non-specific | K. oxytoca | 25% | ||||
| 10 | Bangladesh | blaCTX-M-3 | Pseudomonas spp. | Urine, swab, pus | 78.0% | [91] |
| blaCTX-M- 14 | 80.0% | |||||
| 11 | Bangladesh | blaTEM | E. coli | Stool | 41% | [62] |
| blaCTX–M–group–1 | 96% | |||||
| 12 | India | blaCTX-M-15 | E. coli | Urine | 52% | [92] |
| blaOXA-2 | 8% | |||||
| 13 | India | Non-specific | E. coli | Pus | 9.8% | [93] |
| Urine | 82.6% | |||||
| 14 | North-East India | blaCTX-M | E. coli | Urine, sputum, vaginal discharge | 54.34% | [94] |
| blaTEM | 60.86 | |||||
| blaSHV | 63.04% | |||||
| 15 | South India | blaCTX-M-15 | E. coli | Urine, wound swab, sputum, pus, endotracheal secretions, bronchoalveolar lavage, bile fluid | 90% | [95] |
| 16 | Bihar, India | blaTEM | E. coli | Stool | 51.8% | [96] |
| blaSHV | 68% | |||||
| blaCTX-M | 86.1% | |||||
| 17 | India | blaSHV | Pseudomonas aeruginosa | Urine, blood, sputum, endotracheal aspirate | 15.1% | [97] |
| blaTEM | 57.1% | |||||
| 18 | Pakistan | blaCTX-M-15 | E. coli | Fecal samples | 86.2% | [98] |
| 19 | North-West Pakistan | Non-specific | P. aeruginosa | Burn patients | 35.85% | [99] |
| 20 | Lahore, Pakistan | blaCTX - M | E. coli, Klebsiella spp., Pseudomonas aeruginosa, Enterobacter spp., Acinetobacter spp. | Urine, pus, wound swabs | 76% | [100] |
| blaTEM | 28% | |||||
| blaSHV | 21% | |||||
| 21 | Faisalabad, Pakistan | blaCTX-M-1 | E. coli | Dog owners | 59% | [101] |
| Cat owners | 73.9% | |||||
| Veterinary professionals | 80% | |||||
| 22 | Pakistan | blaCTX-M1 | K. pneumoniae | Hospital waste | 71% | [102] |
| blaTEM | 53% | |||||
| blaSHV | 6% | |||||
| 23 | Lahore, Pakistan | blaCTX-M-I | E. coli | Clinical specimens | 72.1% | [103] |
| blaCTX-M-II | 8.5% | |||||
| 24 | Peshawar, Pakistan | blaCTX-M-15 | Pseudomonas aeruginosa | Clinical specimens | 19.71% | [104] |
| 25 | Lahore, Pakistan | Non-specific | E. coli | Healthy individuals | 57.0% | [105] |
| Patients | 53.0% | |||||
| 26 | Faisalabad, Pakistan | blaCTXM-1 | E. coli | Urine | 70%, | [106] |
| blaTEM-1 | 74.4% | |||||
| blaCTXM-15 | 49% |
| Country | ESBL | Enterobacteriaceae | Species | Source | Prevalence | Reference | |
|---|---|---|---|---|---|---|---|
| 1 | Bangladesh | blaTEM | E. coli | Chicken | Droppings | 78% | [86] |
| 2 | India | blaCTX-M-15 | E. coli | Poultry | Meat | 17% | [107] |
| 3 | Pakistan | blaCTX-M-15 | E. coli | Migratory birds | Fecal samples | 92.3% | [61] |
| 4 | Bangladesh | blaTEM | E. coli | Chicken | Meat | 86% | [108] |
| 5 | India | blaCTX-M-15 | E. coli | Piglets | Fecal samples | 2.94% | [109] |
| blaTEM | 6.47% | ||||||
| 6 | India | blaCTX-M-1 | E. coli | Piglets | Fecal samples | 55.55% | [110] |
| 7 | West Bengal, India | blaCTX-M | Klebsiella spp. | Broiler | Cloacal swabs | 10.7% | [111] |
| blaSHV | 51.5% | ||||||
| blaTEM | 48.5% | ||||||
| 8 | West Bengal, India | blaCTX-M | E. coli | Cattle | Milk | 54.54% | [112] |
| 9 | Assam and Meghalaya | blaCTX-M | E. coli, Salmonella. | Pigs | Fecal samples | 0.67% | [113] |
| blaTEM | 2.76% | ||||||
| 10 | Faisalabad, Pakistan | blaCTX-M-1 | E. coli | Dogs | Fecal samples | 81.8% | [101] |
| Cats | 73.9% | ||||||
| 11 | Pakistan | blaCTX-M-15 | E. coli | Wild birds | Fecal samples | 92.3% | [61] |
| 12 | Punjab, Pakistan | blaTEM-1 | Salmonella enterica serovar Infantis | Poultry | Post mortem specimens | 44·4% | [114] |
| 13 | Lahore, Pakistan | Non-specific | E. coli | Cattle | Feces | 66.0% | [105] |
| Chicken | Feces | 59.0% | |||||
| Cattle, Chicken | Raw meat | 70.0% | |||||
| 14 | Pakistan | blaCTX-M-15 | E. coli | Cows | Mastitic milk samples | 63.04% | [115] |
| blaCTX-M-55, blaCTX-M-14 | 8.69% | ||||||
| blaCTX-M-3, blaCTX-M-1 | 2.17% | ||||||
| blaTEM | 47.82% | ||||||
| blaSHV | 17.39% | ||||||
| 15 | Pakistan | blaCTX-MblaTEM | E. coli | Backyard chicken | Cloacal swabs | 45.1% | [116] |
| 16 | Bangladesh | blaSHV | E. coli | Broiler | Raw meat swabs | 12.8% | [117] |
| Layer | 7.61% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines 2023, 11, 2937. https://doi.org/10.3390/biomedicines11112937
Husna A, Rahman MM, Badruzzaman ATM, Sikder MH, Islam MR, Rahman MT, Alam J, Ashour HM. Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines. 2023; 11(11):2937. https://doi.org/10.3390/biomedicines11112937
Chicago/Turabian StyleHusna, Asmaul, Md. Masudur Rahman, A. T. M. Badruzzaman, Mahmudul Hasan Sikder, Mohammad Rafiqul Islam, Md. Tanvir Rahman, Jahangir Alam, and Hossam M. Ashour. 2023. "Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities" Biomedicines 11, no. 11: 2937. https://doi.org/10.3390/biomedicines11112937
APA StyleHusna, A., Rahman, M. M., Badruzzaman, A. T. M., Sikder, M. H., Islam, M. R., Rahman, M. T., Alam, J., & Ashour, H. M. (2023). Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines, 11(11), 2937. https://doi.org/10.3390/biomedicines11112937











