Chemical Transdifferentiation of Somatic Cells: Unleashing the Power of Small Molecules
Abstract
1. Introduction
2. Methods for Cell Transdifferentiation
2.1. Transcription Factor Overexpression
2.2. Chemical Small Molecules
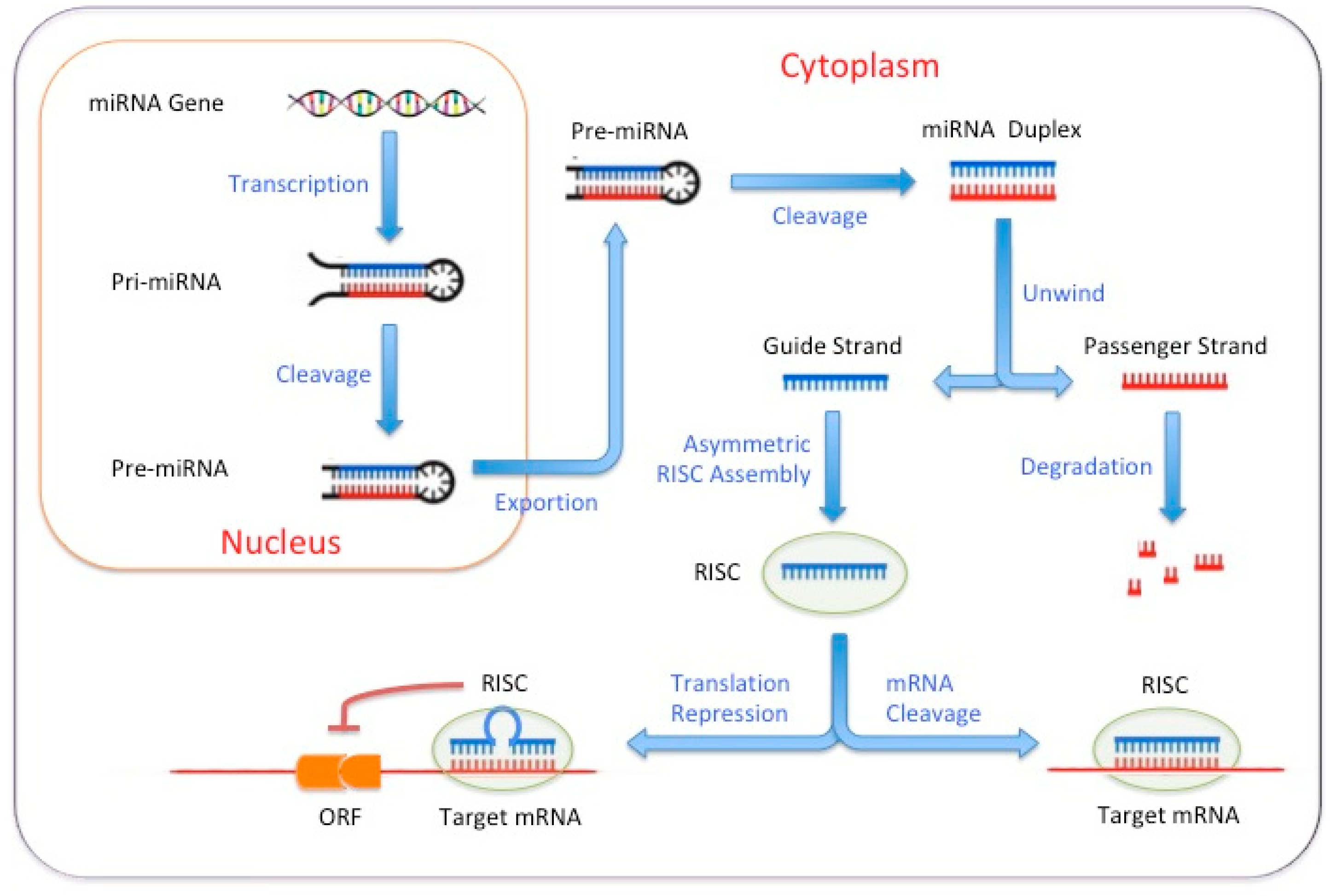
2.3. MicroRNA-Based Reprogramming
2.4. Extracellular Vesicle (EV)-Based Reprogramming
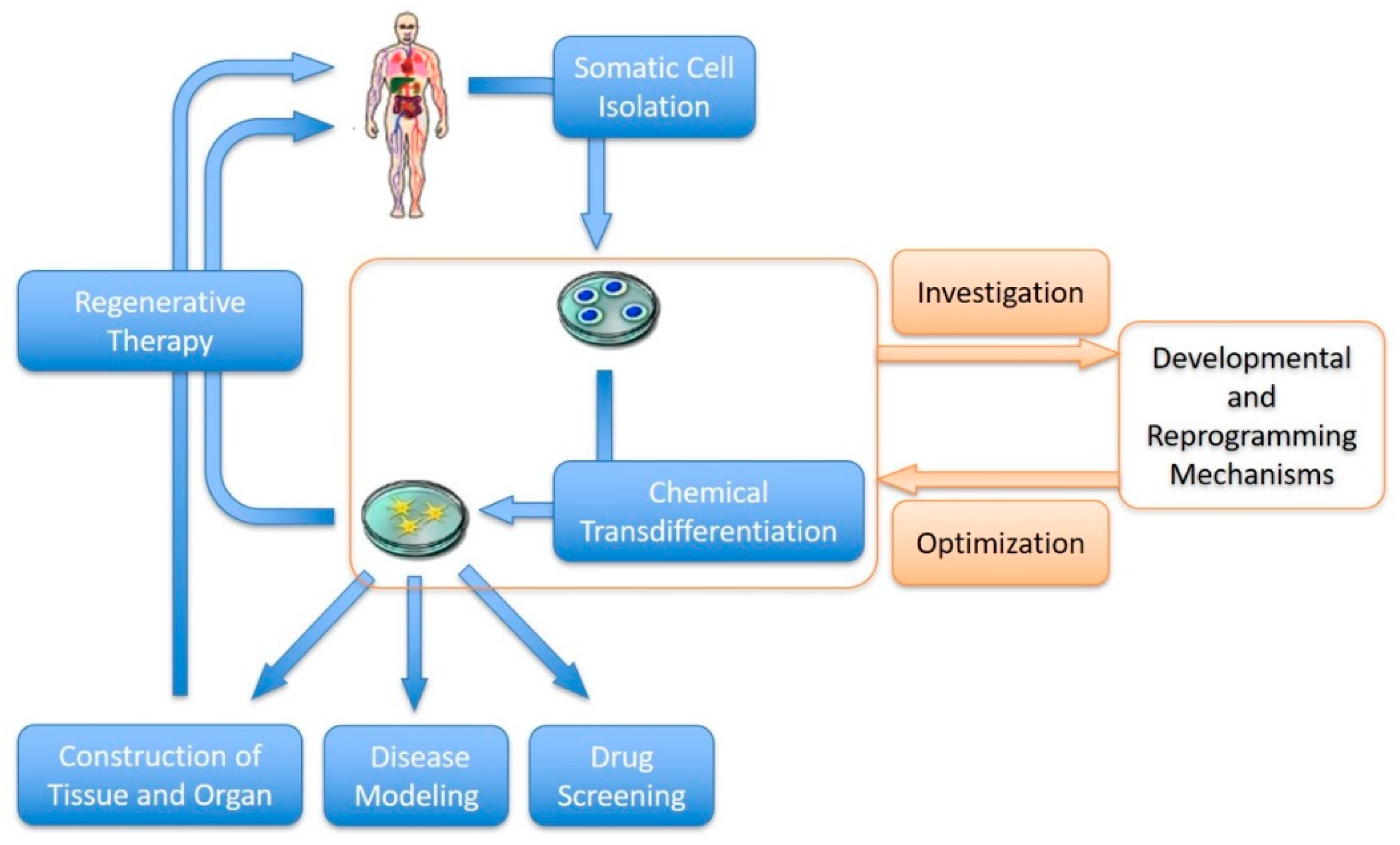
3. Chemical Transdifferentiation of Somatic Cells
3.1. The Power of Small Molecules: Chemical Transdifferentiation
3.2. Unlocking Potential: Transdifferentiation of Various Cell Types
3.3. Challenges: The Bottleneck of Chemical Transdifferentiation
4. Future Perspectives of Chemical Transdifferentiation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wong, W.T.; Cooke, J.P. Therapeutic transdifferentiation of human fibroblasts into endothelial cells using forced expression of lineage-specific transcription factors. J. Tissue Eng. 2016, 7, 2041731416628329. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Xie, M.; Laurent, T.; Ding, S. Progress in the reprogramming of somatic cells. Circ. Res. 2013, 112, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell 2012, 10, 678–684. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, R.; Cesano, A.; Kreider, B.L.; Lange, B.; Clark, S.C.; Nowell, P.C.; Finan, J.; Rovera, G.; Santoli, D. Growth factor-dependent differentiation along the myeloid and lymphoid lineages in an immature acute T lymphocytic leukemia. J. Immunol. 1990, 145, 3779–3787. [Google Scholar] [CrossRef]
- Rosenberg, L. In Vivo cell transformation: Neogenesis of beta cells from pancreatic ductal cells. Cell Transplant. 1995, 4, 371–383. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Y.; Liu, J.; Qian, L. Direct cell reprogramming: Approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 2021, 22, 410–424. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef]
- Li, W.; Cavelti-Weder, C.; Zhang, Y.; Clement, K.; Donovan, S.; Gonzalez, G.; Zhu, J.; Stemann, M.; Xu, K.; Hashimoto, T.; et al. Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells. Nat. Biotechnol. 2014, 32, 1223–1230. [Google Scholar] [CrossRef]
- Li, X.; Yu, Y.; Wei, R.; Li, Y.; Lv, J.; Liu, Z.; Zhang, Y. In Vitro and In Vivo study on angiogenesis of porcine induced pluripotent stem cell-derived endothelial cells. Differentiation 2021, 120, 10–18. [Google Scholar] [CrossRef]
- Smith, Z.D.; Sindhu, C.; Meissner, A. Molecular features of cellular reprogramming and development. Nat. Rev. Mol. Cell Biol. 2016, 17, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Chakraborty, S.; Ji, H.; Kabadi, A.M.; Gersbach, C.A.; Christoforou, N.; Leong, K.W. A CRISPR/Cas9-based system for reprogramming cell lineagespecification. Stem Cell Rep. 2014, 3, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Annaluru, N.; Kandavelou, K.; Chandrasegaran, S. TALEN-mediated generation and genetic correction of disease-specific human induced pluripotent stem cells. Curr. Gene Ther. 2014, 14, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Cathomen, T.; Schambach, A. Zinc-finger nucleases meet iPS cells: Zinc positive: Tailored genome engineering meets reprogramming. Gene Ther. 2010, 17, 1–3. [Google Scholar] [CrossRef]
- Yu, C.; Liu, K.; Tang, S.; Ding, S. Chemical approaches to cell reprogramming. Curr. Opin. Genet. Dev. 2014, 28, 50–56. [Google Scholar] [CrossRef]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, H.; Yang, Q.; Yang, J.; Wang, H.; Xu, H.; Gao, W.Q. Chemical conversion of mouse fibroblasts into functional dopaminergic neurons. Exp. Cell Res. 2016, 347, 283–292. [Google Scholar] [CrossRef]
- Dai, P.; Harada, Y.; Takamatsu, T. Highly efficient direct conversion of human fibroblasts to neuronal cells by chemical compounds. J. Clin. Biochem. Nutr. 2015, 56, 166–170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated In Vitro and In Vivo direct reprogramming of cardiacfibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Camussi, G.; Deregibus, M.C.; Tetta, C. Paracrine/endocrine mechanism of stem cells on kidney repair: Role of microvesicle-mediated transfer of genetic information. Curr. Opin. Nephrol. Hypertens. 2010, 19, 7–12. [Google Scholar] [CrossRef]
- Alberti, G.; Russo, E.; Corrao, S.; Anzalone, R.; Kruzliak, P.; Miceli, V.; Conaldi, P.G.; Di Gaudio, F.; La Rocca, G. Current perspectives on adult mesenchymal stromal cell-derived extracellular vesicles: Biological features and clinical indications. Biomedicines 2022, 10, 2822. [Google Scholar] [CrossRef]
- Ramos, T.; Parekh, M.; Kaye, S.B.; Ahmad, S. Epithelial cell-derived extracellular vesicles trigger the differentiation of two epithelial cell lines. Int. J. Mol. Sci. 2022, 23, 1718. [Google Scholar] [CrossRef]
- Kim, H.; Song, B.W.; Park, S.J.; Choi, S.W.; Moon, H.; Hwang, K.C.; Kang, S.W.; Moon, S.H.; Yang, Y.; Kwon, I.C.; et al. Ultraefficient extracellular vesicle-guided direct reprogramming of fibroblasts into functional cardiomyocytes. Sci. Adv. 2022, 8, eabj6621. [Google Scholar] [CrossRef]
- López-Muneta, L.; Miranda-Arrubla, J.; Carvajal-Vergara, X. The future of direct cardiac reprogramming: Any GMT cocktail variety? Int. J. Mol. Sci. 2020, 21, 7950. [Google Scholar] [CrossRef]
- Shi, Y.; Do, J.T.; Desponts, C.; Hahm, H.S.; Scholer, H.R.; Ding, S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2008, 2, 525–528. [Google Scholar] [CrossRef]
- Okitam, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Björklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef]
- Hawkins, R.D.; Hon, G.C.; Lee, L.K.; Ngo, Q.; Lister, R.; Pelizzola, M.; Edsall, L.E.; Kuan, S.; Luu, Y.; Klugman, S.; et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 2010, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Gascón, S.; Masserdotti, G.; Russo, G.L.; Götz, M. Direct neuronal reprogramming: Achievements, hurdles, and new roads to success. Cell Stem Cell 2017, 21, 18–34. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Hanna, J.; Zhang, X.; Ku, M.; Wernig, M.; Schorderet, P.; Bernstein, B.E.; Jaenisch, R.; Lander, E.S.; Meissner, A. Dissecting direct reprogramming through integrative genomic analysis. Nature 2008, 454, 49–55. [Google Scholar] [CrossRef]
- Mollinari, C.; Zhao, J.; Lupacchini, L.; Garaci, E.; Merlo, D.; Pei, G. Transdifferentiation: A new promise for neurodegenerative diseases. Cell Death Dis. 2018, 9, 830. [Google Scholar] [CrossRef]
- Chabrat, A.; Lacassagne, E.; Billiras, R.; Landron, S.; Pontisso-Mahout, A.; Darville, H.; Dupront, A.; Coge, F.; Schenker, E.; Piwnica, D.; et al. Pharmacological transdifferentiation of human nasal olfactory stem cells into dopaminergic neurons. Stem Cells Int. 2019, 2019, 2945435. [Google Scholar] [CrossRef]
- Thoma, E.C. Chemical conversion of human fibroblasts into functional schwann cells. Methods Mol. Biol. 2018, 1739, 127–136. [Google Scholar]
- Hu, W.; Qiu, B.; Guan, W.; Wang, Q.; Wang, M.; Li, W.; Gao, L.; Shen, L.; Huang, Y.; Xie, G.; et al. Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 2015, 17, 204–212. [Google Scholar] [CrossRef]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Huang, C.; Xu, X.; Gu, H.; Ye, Y.; Jiang, C.; Qiu, Z.; Xie, X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015, 25, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kang, K.; Lee, S.B.; Seo, D.; Yoon, S.; Kim, S.J.; Jang, K.; Jung, Y.K.; Lee, K.G.; Factor, V.M.; et al. Small molecule-mediated reprogramming of human hepatocytes into bipotent progenitor cells. J. Hepatol. 2019, 70, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Tang, W.; Yuan, Q.; Hui, L.; Wang, X.; Xie, X. Chemical cocktails enable hepatic reprogramming of mouse fibroblasts with a single transcription factor. Stem Cell Rep. 2017, 9, 499–512. [Google Scholar] [CrossRef]
- Tang, W.; Guo, R.; Shen, S.J.; Zheng, Y.; Lu, Y.T.; Jiang, M.M.; Cui, X.; Jiang, C.Z.; Xie, X. Chemical cocktails enable hepatic reprogramming of human urine-derived cells with a single transcription factor. Acta Pharmacol. Sin. 2019, 40, 620–629. [Google Scholar] [CrossRef]
- Sayed, N.; Wong, W.T.; Ospino, F.; Meng, S.; Lee, J.; Jha, A.; Dexheimer, P.; Aronow, B.J.; Cooke, J.P. Transdifferentiation of human fibroblasts to endothelial cells: Role of innate immunity. Circulation 2015, 131, 300–309. [Google Scholar] [CrossRef]
- Liu, C.; Medina, P.; Thomas, D.; Chen, I.Y.; Sallam, K.; Sayed, D.; Sayed, N. A protocol for transdifferentiation of human cardiac fibroblasts into endothelial cells via activation of innate immunity. STAR Protoc. 2021, 2, 100556. [Google Scholar] [CrossRef]
- Ubil, E.; Duan, J.; Pillai, I.C.; Rosa-Garrido, M.; Wu, Y.; Bargiacchi, F.; Lu, Y.; Stanbouly, S.; Huang, J.; Rojas, M.; et al. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature 2014, 514, 585–590. [Google Scholar] [CrossRef]
- Zhu, S.; Russ, H.A.; Wang, X.; Zhang, M.; Ma, T.; Xu, T.; Tang, S.; Hebrok, M.; Ding, S. Human pancreatic beta-like cells converted from fibroblasts. Nat. Commun. 2016, 7, 10080. [Google Scholar] [CrossRef]
- Kunisada, Y.; Tsubooka-Yamazoe, N.; Shoji, M.; Hosoya, M. Small molecules induce efficient differentiation into insulin-producing cells from human induced pluripotent stem cells. Stem Cell Res. 2012, 8, 274–284. [Google Scholar] [CrossRef]
- Bramswig, N.C.; Everett, L.J.; Schug, J.; Dorrell, C.; Liu, C.; Luo, Y.; Streeter, P.R.; Naji, A.; Grompe, M.; Kaestner, K.H. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Investig. 2013, 123, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Alvarez-Perez, J.C.; Felsenfeld, D.P.; Liu, H.; Sivendran, S.; Bender, A.; Kumar, A.; Sanchez, R.; Scott, D.K.; Garcia-Ocaña, A.; et al. A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication. Nat. Med. 2015, 21, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Streeter, P.R. Targeted delivery of harmine to xenografted human pancreatic islets promotes robust cell proliferation. Sci. Rep. 2022, 12, 19127. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhang, L.; Wu, Z.; Chen, Y.; Wang, F.; Chen, G. In Vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 2014, 14, 188–202. [Google Scholar] [CrossRef]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Björklund, A.; Grealish, S.; Parmar, M. Generation of induced neurons via direct conversion In Vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Dennison, R.; Usuga, E.; Chen, H.; Paul, J.Z.; Arbelaez, C.A.; Teng, Y.D. Direct cell reprogramming and phenotypic conversion: An analysis of experimental attempts to transform astrocytes into neurons in adult animals. Cells 2023, 12, 618. [Google Scholar] [CrossRef]
- Sripathi, S.R.; Hu, M.W.; Turaga, R.C.; Mikeasky, R.; Satyanarayana, G.; Cheng, J.; Duan, Y.; Maruotti, J.; Wahlin, K.J.; Berlinicke, C.A.; et al. IKKβ inhibition attenuates epithelial mesenchymal transition of human stem cell-derived retinal pigment epithelium. Cells 2023, 12, 1155. [Google Scholar] [CrossRef]
- Roy, S.; Amin, S.; Roy, S. Retinal fibrosis in diabetic retinopathy. Exp. Eye Res. 2016, 142, 71–75. [Google Scholar] [CrossRef]
- Sloan, L.C.; Funk, K.M.; Tamiya, S.; Song, Z.H. Effect of N-oleoyl dopamine on myofibroblast trans-differentiation of retinal pigment epithelial cells. Biochem. Biophys. Res. Commun. 2023, 667, 127–131. [Google Scholar] [CrossRef]
- Napolitano, F.; Rapakoulia, T.; Annunziata, P.; Hasegawa, A.; Cardon, M.; Napolitano, S.; Vaccaro, L.; Iuliano, A.; Wanderlingh, L.G.; Kasukawa, T.; et al. Automatic identification of small molecules that promote cell conversion and reprogramming. Stem. Cell Rep. 2021, 16, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Trokovic, R.; Weltner, J.; Noisa, P.; Raivio, T.; Otonkoski, T. Combined negative effect of donor age and time in culture on the reprogramming efficiency into induced pluripotent stem cells. Stem Cell Res. 2015, 15, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.; Sudha, T.; Darwish, N.H.E.; Eghotny, S.; E-Dief, A.; Hassaan, P.S.; Mousa, S.A. In Vitro differentiation of human bone marrow stromal cells into neural precursor cells using small molecules. J. Neurosci. Methods 2021, 363, 109340. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, C.; Daley, T.P.; Wang, F.; Cao, W.S.; Bhate, S.; Lin, X.; Still, C., 2nd; Liu, H.; Zhao, D.; et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 2018, 23, 758–771. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Z.; Welch, J.D.; Gao, X.; Wang, L.; Garbutt, T.; Keepers, B.; Ma, H.; Prins, J.F.; Shen, W.; et al. Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell 2019, 25, 149–164. [Google Scholar] [CrossRef]
- Treutlein, B.; Lee, Q.Y.; Camp, J.G.; Mall, M.; Koh, W.; Shariati, S.A.; Sim, S.; Neff, N.F.; Skotheim, J.M.; Wernig, M.; et al. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 2016, 534, 391–395. [Google Scholar] [CrossRef]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef]
- Bar-Nur, O.; Russ, H.A.; Efrat, S.; Benvenisty, N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet Beta cells. Cell Stem Cell 2011, 9, 17–23. [Google Scholar] [CrossRef]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R.; et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 2020, 4, 446–462. [Google Scholar] [CrossRef]
- Becker, J.S.; Nicetto, D.; Zaret, K.S. H3K9me3-dependent heterochromatin: Barrier to cell fate changes. Trends Genet. 2016, 32, 29–41. [Google Scholar] [CrossRef]
- Kuang, J.; Huang, T.; Pei, D. The art of reprogramming for regenerative medicine. Front. Cell Dev. Biol. 2022, 10, 927555. [Google Scholar] [CrossRef] [PubMed]
- Das, R.N.; Tevet, Y.; Safriel, S.; Han, Y.; Moshe, N.; Lambiase, G.; Bassi, I.; Nicenboim, J.; Brückner, M.; Hirsch, D.; et al. Generation of specialized blood vessels via lymphatic transdifferentiation. Nature 2022, 606, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Tiwari, V.K. Epigenetic reprogramming of cell identity: Lessons from development for regenerative medicine. Clin. Epigenet. 2021, 13, 144. [Google Scholar] [CrossRef] [PubMed]
| Method | Advantages | Shortcomings |
|---|---|---|
| Transcription Factor Overexpression | - Direct and specific reprogramming of cells - Well-established techniques - High efficiency in some cases | - Requires genetic manipulation - Limited to cell types with known transcription factor combinations - Potential risk of tumorigenicity |
| Small Molecules | - Non-genetic approach - Can be easily delivered to target cells - Versatile and modifiable | - Less efficient compared to transcription factor-based approaches - Requires optimization and identification of specific molecules |
| MicroRNA-Based Reprogramming | - Non-genetic approach - Fine-tuned regulation of gene expression | - Complex interaction networks between microRNAs and target genes - Limited efficiency |
| EV-Based Conversion | - Utilizes environmental cues and signaling factors - Mimics developmental processes | - Limited efficiency and reproducibility - May require complex and expensive culture conditions |
| Three-Dimensional Culture Systems | - Provides a more physiological context - Better recapitulation of tissue architecture and function | - Complexity in establishing and maintaining 3D cultures - Variability in differentiation outcomes |
| Transdifferentiation | Small Molecules | Mechanism | References |
|---|---|---|---|
| Fibroblasts to Neurons | Valproic acid (VPA) | Histone deacetylase (HDAC) inhibitor | [40] |
| Forskolin | Activator of the cyclic adenosine monophosphate (cAMP) pathway | [41] [40] | |
| ISX9 | Stimulates neurogenesis | [41] | |
| CHIR99021 | Inhibitor of glycogen synthase kinase 3 beta (GSK3β) | [41] [40] | |
| Repsox | Inhibitor of transforming growth factor-beta (TGF-β) receptor | [40] | |
| Fibroblasts to Cardiomyocytes | CHIR99021 | Inhibitor of GSK3β | [19] [42] |
| A83-01 | Inhibitor of TGF-β type I receptor | [19] | |
| LIF | Leukemia inhibitory factor | [42] | |
| PD0325901 | Inhibitor of MEK1/2 | [42] | |
| Y-27632 | ROCK inhibitor | [19] | |
| AS8351 | Histone demethylase (HDM) inhibitor | [19] | |
| SU16F | PDGFRβ inhibitor | [19] | |
| Fibroblasts to Hepatocytes | A83-01 | Inhibitor of TGF-β type I receptor | [43] |
| VPA | HDAC inhibitor | [44] [45] | |
| CHIR99021 | Inhibitor of GSK3β | [44] [45] [43] | |
| HGF | Hepatocyte growth factor | [43] | |
| EGF | Epidermal growth factor | [43] | |
| TTNPB | Retinoic acid (RA) receptor agonist | [44] [45] | |
| Dznep | Histone methyltransferase EZH2 inhibitor | [44] [45] | |
| Fibroblasts to Endothelial Cells | VEGF | Vascular endothelial growth factor | [46] [47] |
| SB431542 | Inhibitor of TGF-β receptor | [46] [47] | |
| bFGF | Basic fibroblast growth factor | [46] [47] | |
| BMP4 | Bone morphogenetic protein 4 | [46] [47] | |
| PolyI:C | Toll-like receptor 3 (TLR3) agonist | [46] [47] | |
| RITA | Inhibitor of the p53-MDM2 interaction | [48] | |
| 8-Br-cAMP | Activator of cyclic AMP-dependent protein kinase | [47] | |
| Fibroblasts to Pancreatic Beta Cells | Activin A | Member of TGF- β superfamily | [49] |
| Forskolin | Activator of the cAMP pathway | [50] [49] | |
| GDC-0449 | Antagonist of sonic hedgehog | [49] | |
| Nicotinamide | Vitamin B3 or niacin | [50] [49] | |
| Sodium butyrate (NaB) | Inhibitor of histone deacetylase | [51] [49] | |
| RG108 | Inhibitor of DNA methylase | [51] [49] | |
| Compound-E | Inhibitor of Notch signaling | [49] | |
| Harmine | Inhibitor of DYRK1A | [52] [53] | |
| Dexamethasone | Agonist of glucocorticoid receptor | [50] [49] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Li, X.; Xing, J.; Zhou, J.; Li, H. Chemical Transdifferentiation of Somatic Cells: Unleashing the Power of Small Molecules. Biomedicines 2023, 11, 2913. https://doi.org/10.3390/biomedicines11112913
Zhang Y, Li X, Xing J, Zhou J, Li H. Chemical Transdifferentiation of Somatic Cells: Unleashing the Power of Small Molecules. Biomedicines. 2023; 11(11):2913. https://doi.org/10.3390/biomedicines11112913
Chicago/Turabian StyleZhang, Yu, Xuefeng Li, Jianyu Xing, Jinsong Zhou, and Hai Li. 2023. "Chemical Transdifferentiation of Somatic Cells: Unleashing the Power of Small Molecules" Biomedicines 11, no. 11: 2913. https://doi.org/10.3390/biomedicines11112913
APA StyleZhang, Y., Li, X., Xing, J., Zhou, J., & Li, H. (2023). Chemical Transdifferentiation of Somatic Cells: Unleashing the Power of Small Molecules. Biomedicines, 11(11), 2913. https://doi.org/10.3390/biomedicines11112913






