Circular RNAs: A New Approach to Multiple Sclerosis
Abstract
:1. General Considerations on Multiple Sclerosis
2. Non-Coding RNAs
2.1. Circular RNAs
2.1.1. circRNAs Identification Tools
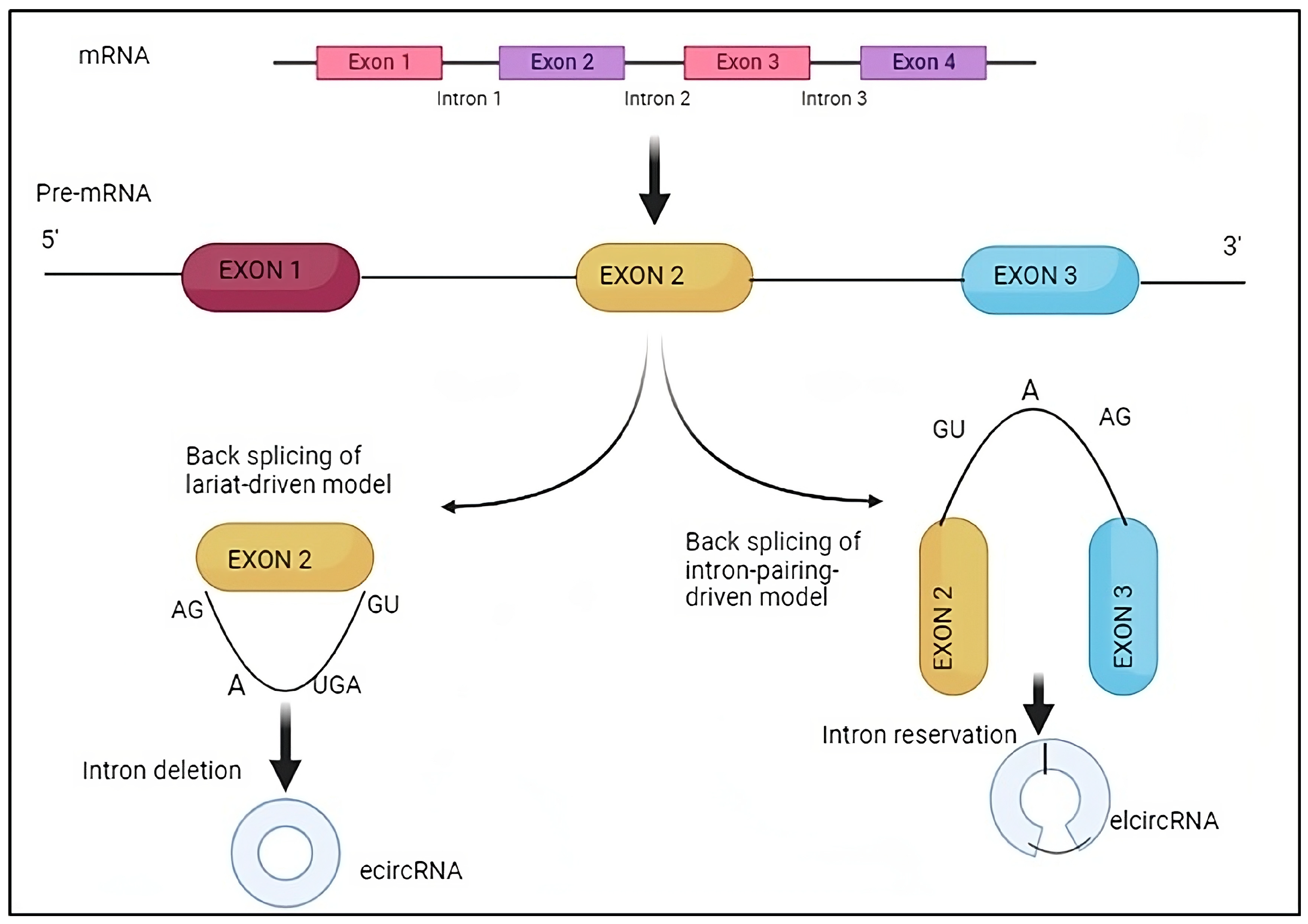
2.1.2. CircRNA Biogenesis
2.1.3. circRNAs Functions
3. Multiple Sclerosis and circRNAs
3.1. Epigenetic Mechanisms
3.2. circRNAs Biomarkers in MS
3.3. Extracellular Vesicles and circRNAs
3.4. circRNAs Genetic Variation in MS
3.5. circRNAs and B-Cell Function
3.6. Roles of circRNAs in MS
3.6.1. hsa_circ_0106803 of GSDMB Gene
3.6.2. circ_HECW2 and the Dysfunction of the Blood–Brain Barrier
3.6.3. hsa_circ_0106803 Modulates the Expression of ASIC1a mRNA
3.6.4. circINPP4B Regulates Th17 Cell Differentiation
3.6.5. hsa_circRNA_101145 and hsa_circRNA_001896 in Patients with Relapse-Remitting MS
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Browne, P.; Chandraratna, D.; Angood, C.; Tremlett, H.; Baker, C.; Taylor, B.V.; Thompson, A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology 2014, 83, 1022–1024. [Google Scholar] [CrossRef] [PubMed]
- GBD 2016 Neurology Collaborators. Global, regional, and national burden of neurological disorders, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 459–480. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, M.M. Multiple sclerosis review. Pharm. Ther. 2012, 37, 175–184. [Google Scholar]
- Lublin, F.D.; Reingold, S.C.; Cohen, J.A.; Cutter, G.R.; Sørensen, P.S.; Thompson, A.J.; Wolinsky, J.S.; Balcer, L.J.; Banwell, B.; Barkhof, F.; et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology 2014, 83, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Cree, B.A.C.; Arnold, D.L.; Chataway, J.; Chitnis, T.; Fox, R.J.; Pozo Ramajo, A.; Murphy, N.; Lassmann, H. Secondary Progressive Multiple Sclerosis: New Insights. Neurology 2021, 97, 378–388. [Google Scholar] [CrossRef]
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173. [Google Scholar] [CrossRef]
- Alcina, A.; Abad-Grau Mdel, M.; Fedetz, M.; Izquierdo, G.; Lucas, M.; Fernández, O.; Ndagire, D.; Catalá-Rabasa, A.; Ruiz, A.; Gayán, J.; et al. Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PLoS ONE 2012, 7, e29819. [Google Scholar] [CrossRef] [PubMed]
- Sintzel, M.B.; Rametta, M.; Reder, A.T. Vitamin D and Multiple Sclerosis: A Comprehensive Review. Neurol. Ther. 2018, 7, 59–85. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Jakimovski, D.; Ramanathan, M.; Weinstock-Guttman, B.; Zivadinov, R. The role of Epstein-Barr virus in multiple sclerosis: From molecular pathophysiology to in vivo imaging. Neural Regen. Res. 2019, 14, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Olsson, T.; Barcellos, L.F.; Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 2017, 13, 25–36. [Google Scholar] [CrossRef]
- Liu, R.; Du, S.; Zhao, L.; Jain, S.; Sahay, K.; Rizvanov, A.; Lezhnyova, V.; Khaibullin, T.; Martynova, E.; Khaiboullina, S.; et al. Autoreactive lymphocytes in multiple sclerosis: Pathogenesis and treatment target. Front. Immunol. 2022, 13, 996469. [Google Scholar] [CrossRef] [PubMed]
- Mahad, D.H.; Trapp, B.D.; Lassmann, H. Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol. 2015, 14, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Martynova, E.; Khaibullin, T.; Salafutdinov, I.; Markelova, M.; Laikov, A.; Lopukhov, L.; Liu, R.; Sahay, K.; Goyal, M.; Baranwal, M.; et al. Seasonal Changes in Serum Metabolites in Multiple Sclerosis Relapse. Int. J. Mol. Sci. 2023, 24, 3542. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.D.; Paculova, H.; Kogut, S.; Heath, J.; Schjerven, H.; Frietze, S. Non-Coding RNA Signatures of B-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 2683. [Google Scholar] [CrossRef]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Chen, L.L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A guide to naming human non-coding RNA genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Dozmorov, M.G.; Giles, C.B.; Koelsch, K.A.; Wren, J.D. Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinform. 2013, 14 (Suppl. S14), S2. [Google Scholar] [CrossRef] [PubMed]
- Avenoso, A.; Campo, S.; Scuruchi, M.; Mania, M.; Innao, V.; D’Ascola, A.; Mandraffino, G.; Allegra, A.G.; Musolino, C.; Allegra, A. Quantitative polymerase Chain reaction profiling of microRNAs in peripheral lymph-monocytes from MGUS subjects. Pathol. Res. Pract. 2021, 218, 153317. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Meng, X.; Liao, Q.; Chen, M. Versatile interactions and bioinformatics analysis of noncoding RNAs. Brief. Bioinform. 2019, 20, 1781–1794. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Adachi, H.; Yu, Y.T. Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation. World J. Biol. Chem. 2014, 5, 398–408. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef]
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650. [Google Scholar] [CrossRef]
- Caserta, S.; Gangemi, S.; Murdaca, G.; Allegra, A. Gender Differences and miRNAs Expression in Cancer: Implications on Prognosis and Susceptibility. Int. J. Mol. Sci. 2023, 24, 11544. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Zamore, P.D. piRNAs. Curr. Biol. 2014, 24, R730–R733. [Google Scholar] [CrossRef] [PubMed]
- Musolino, C.; Oteri, G.; Allegra, A.; Mania, M.; D’Ascola, A.; Avenoso, A.; Innao, V.; Allegra, A.G.; Campo, S. Altered microRNA expression profile in the peripheral lymphoid compartment of multiple myeloma patients with bisphosphonate-induced osteonecrosis of the jaw. Ann. Hematol. 2018, 97, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Allegra, A.; Cicero, N.; Tonacci, A.; Musolino, C.; Gangemi, S. Circular RNA as a Novel Biomarker for Diagnosis and Prognosis and Potential Therapeutic Targets in Multiple Myeloma. Cancers 2022, 14, 1700. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995, 268, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [PubMed]
- Kos, A.; Dijkema, R.; Arnberg, A.C.; van der Meide, P.H.; Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Arnberg, A.C.; Van Ommen, G.J.; Grivell, L.A.; Van Bruggen, E.F.; Borst, P. Some yeast mitochondrial RNAs are circular. Cell 1980, 19, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Cocquerelle, C.; Daubersies, P.; Majérus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, F. Computational Strategies for Exploring Circular RNAs. Trends Genet. 2018, 34, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B. Improved circRNA Identification by Combining Prediction Algorithms. Front. Cell Dev. Biol. 2018, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157, Erratum in RNA 2013, 19, 426. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef]
- Liang, D.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.L.; Cherry, S.; Wilusz, J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, Y.; Wang, X.; Shen, J.; An, W. Advances in Circular RNA and Its Applications. Int. J. Med. Sci. 2022, 19, 975–985. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.L.; Yang, L.; Chen, L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef]
- Aktaş, T.; Avşar Ilık, İ.; Maticzka, D.; Bhardwaj, V.; Pessoa Rodrigues, C.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017, 544, 115–119. [Google Scholar] [CrossRef]
- Yu, C.Y.; Kuo, H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, Y.; Han, B.; Bai, Y.; Zhou, R.; Gan, G.; Chao, J.; Hu, G.; Yao, H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 2017, 13, 1722–1741, Erratum in Autophagy 2020, 18, 234; Erratum in Autophagy 2020, 16, 1553; Erratum in Autophagy 2020, 16, 2117–2118. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X.; Yang, B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Stoll, L.; Rodríguez-Trejo, A.; Guay, C.; Brozzi, F.; Bayazit, M.B.; Gattesco, S.; Menoud, V.; Sobel, J.; Marques, A.C.; Venø, M.T.; et al. A circular RNA generated from an intron of the insulin gene controls insulin secretion. Nat. Commun. 2020, 11, 5611. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015, 21, 172–179. [Google Scholar] [CrossRef]
- Li, H.M.; Ma, X.L.; Li, H.G. Intriguing circles: Conflicts and controversies in circular RNA research. Wiley Interdiscip Rev. RNA 2019, 10, e1538. [Google Scholar] [CrossRef]
- Farh, K.K.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef]
- Zheleznyakova, G.Y.; Piket, E.; Marabita, F.; Pahlevan Kakhki, M.; Ewing, E.; Ruhrmann, S.; Needhamsen, M.; Jagodic, M.; Kular, L. Epigenetic research in multiple sclerosis: Progress, challenges, and opportunities. Physiol. Genom. 2017, 49, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.L.; Garg, P.; Thin, T.H.; Yoo, S.; Dutta, R.; Trapp, B.D.; Haroutunian, V.; Zhu, J.; Donovan, M.J.; Sharp, A.J.; et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains. Nat. Neurosci. 2014, 17, 121–130. [Google Scholar] [CrossRef]
- Bos, S.D.; Page, C.M.; Andreassen, B.K.; Elboudwarej, E.; Gustavsen, M.W.; Briggs, F.; Quach, H.; Leikfoss, I.S.; Bjølgerud, A.; Berge, T.; et al. Genome-wide DNA methylation profiles indicate CD8+ T cell hypermethylation in multiple sclerosis. PLoS ONE 2015, 10, e0117403. [Google Scholar] [CrossRef]
- Ferreira, H.J.; Davalos, V.; de Moura, M.C.; Soler, M.; Perez-Salvia, M.; Bueno-Costa, A.; Setien, F.; Moran, S.; Villanueva, A.; Esteller, M. Circular RNA CpG island hypermethylation-associated silencing in human cancer. Oncotarget 2018, 9, 29208–29219. [Google Scholar] [CrossRef]
- Shayevitch, R.; Askayo, D.; Keydar, I.; Ast, G. The importance of DNA methylation of exons on alternative splicing. RNA 2018, 24, 1351–1362. [Google Scholar] [CrossRef]
- Xu, T.; Wang, L.; Jia, P.; Song, X.; Zhao, Z. An Integrative Transcriptomic and Methylation Approach for Identifying Differentially Expressed Circular RNAs Associated with DNA Methylation Change. Biomedicines 2021, 9, 657. [Google Scholar] [CrossRef]
- Cardamone, G.; Paraboschi, E.M.; Soldà, G.; Liberatore, G.; Rimoldi, V.; Cibella, J.; Airi, F.; Tisato, V.; Cantoni, C.; Gallia, F.; et al. The circular RNA landscape in multiple sclerosis: Disease-specific associated variants and exon methylation shape circular RNA expression profile. Mult. Scler. Relat. Disord. 2023, 69, 104426. [Google Scholar] [CrossRef]
- Kular, L.; Liu, Y.; Ruhrmann, S.; Zheleznyakova, G.; Marabita, F.; Gomez-Cabrero, D.; James, T.; Ewing, E.; Lindén, M.; Górnikiewicz, B.; et al. DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun. 2018, 9, 2397. [Google Scholar] [CrossRef]
- Wang, Y.; Murakami, Y.; Yasui, T.; Wakana, S.; Kikutani, H.; Kinoshita, T.; Maeda, Y. Significance of glycosylphosphatidylinositol-anchored protein enrichment in lipid rafts for the control of autoimmunity. J. Biol. Chem. 2013, 288, 25490–25499. [Google Scholar] [CrossRef]
- Murakami, H.; Wang, Y.; Hasuwa, H.; Maeda, Y.; Kinoshita, T.; Murakami, Y. Enhanced response of T lymphocytes from Pgap3 knockout mouse: Insight into roles of fatty acid remodeling of GPI anchored proteins. Biochem. Biophys. Res. Commun. 2012, 417, 1235–1241. [Google Scholar] [CrossRef]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef]
- El-Abd, N.; Fawzy, A.; Elbaz, T.; Hamdy, S. Evaluation of annexin A2 and as potential biomarkers for hepatocellular carcinoma. Tumour Biol. 2016, 37, 211–216. [Google Scholar] [CrossRef]
- Cañas, F.; Simonin, L.; Couturaud, F.; Renaudineau, Y. Annexin A2 autoantibodies in thrombosis and autoimmune diseases. Thromb. Res. 2015, 135, 226–230. [Google Scholar] [CrossRef]
- Iparraguirre, L.; Muñoz-Culla, M.; Prada-Luengo, I.; Castillo-Triviño, T.; Olascoaga, J.; Otaegui, D. Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum. Mol. Genet. 2017, 26, 3564–3572. [Google Scholar] [CrossRef]
- Fang, W.; Fa, Z.Z.; Xie, Q.; Wang, G.Z.; Yi, J.; Zhang, C.; Meng, G.X.; Gu, J.L.; Liao, W.Q. Complex Roles of Annexin A2 in Host Blood-Brain Barrier Invasion by Cryptococcus neoformans. CNS Neurosci. Ther. 2017, 23, 291–300. [Google Scholar] [CrossRef]
- Lopez-Ramirez, M.A.; Wu, D.; Pryce, G.; Simpson, J.E.; Reijerkerk, A.; King-Robson, J.; Kay, O.; de Vries, H.E.; Hirst, M.C.; Sharrack, B.; et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 2014, 28, 2551–2565. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, Y.; Cui, W.; Li, M.; Li, B.; Guo, L. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2014, 266, 56–63. [Google Scholar] [CrossRef]
- Hagiwara, K.; Katsuda, T.; Gailhouste, L.; Kosaka, N.; Ochiya, T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015, 589 Pt B, 4071–4078. [Google Scholar] [CrossRef]
- Iparraguirre, L.; Alberro, A.; Sepúlveda, L.; Osorio-Querejeta, I.; Moles, L.; Castillo-Triviño, T.; Hansen, T.B.; Muñoz-Culla, M.; Otaegui, D. RNA-Seq profiling of leukocytes reveals a sex-dependent global circular RNA upregulation in multiple sclerosis and 6 candidate biomarkers. Hum. Mol. Genet. 2020, 29, 3361–3372. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Huang, Y.; Zhang, H.; Lu, H.; Zheng, J.C. Exosomal miRNAs in central nervous system diseases: Biomarkers, pathological mediators, protective factors and therapeutic agents. Prog. Neurobiol. 2019, 183, 101694. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Selmaj, I.; Cichalewska, M.; Namiecinska, M.; Galazka, G.; Horzelski, W.; Selmaj, K.W.; Mycko, M.P. Global exosome transcriptome profiling reveals biomarkers for multiple sclerosis. Ann. Neurol. 2017, 81, 703–717. [Google Scholar] [CrossRef]
- Fanale, D.; Taverna, S.; Russo, A.; Bazan, V. Circular RNA in Exosomes. Adv. Exp. Med. Biol. 2018, 1087, 109–117. [Google Scholar] [CrossRef]
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013, 4, 307. [Google Scholar] [CrossRef]
- Iparraguirre, L.; Alberro, A.; Hansen, T.B.; Castillo-Triviño, T.; Muñoz-Culla, M.; Otaegui, D. Profiling of Plasma Extracellular Vesicle Transcriptome Reveals That circRNAs Are Prevalent and Differ between Multiple Sclerosis Patients and Healthy Controls. Biomedicines 2021, 9, 1850. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Iranifar, E.; Seresht, B.M.; Momeni, F.; Fadaei, E.; Mehr, M.H.; Ebrahimi, Z.; Rahmati, M.; Kharazinejad, E.; Mirzaei, H. Exosomes and microRNAs: New potential therapeutic candidates in Alzheimer disease therapy. J. Cell Physiol. 2019, 234, 2296–2305. [Google Scholar] [CrossRef]
- Mattingly, J.; Li, Y.; Bihl, J.C.; Wang, J. The promise of exosome applications in treating central nervous system diseases. CNS Neurosci. Ther. 2021, 27, 1437–1445. [Google Scholar] [CrossRef]
- Paraboschi, E.M.; Cardamone, G.; Soldà, G.; Duga, S.; Asselta, R. Interpreting Non-coding Genetic Variation in Multiple Sclerosis Genome-Wide Associated Regions. Front. Genet. 2018, 9, 647. [Google Scholar] [CrossRef]
- Brucklacher-Waldert, V.; Stuerner, K.; Kolster, M.; Wolthausen, J.; Tolosa, E. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain 2009, 132 Pt 12, 3329–3341. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Wen, Z.J.; Xu, H.M.; Zhang, Y.; Zhang, Y.F. Exosomal noncoding RNAs in central nervous system diseases: Biological functions and potential clinical applications. Front. Mol. Neurosci. 2022, 15, 1004221. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zuo, X.; Deng, H.; Liu, X.; Liu, L.; Ji, A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 2013, 97, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Krishna Swaroop, A.; Krishnan Namboori, P.K.; Esakkimuthukumar, M.; Praveen, T.K.; Nagarjuna, P.; Patnaik, S.K.; Selvaraj, J. Leveraging decagonal in-silico strategies for uncovering IL-6 inhibitors with precision. Comput. Biol. Med. 2023, 163, 107231. [Google Scholar] [CrossRef] [PubMed]
- Najafi, S.; Aghaei Zarch, S.M.; Majidpoor, J.; Pordel, S.; Aghamiri, S.; Fatih Rasul, M.; Asemani, Y.; Vakili, O.; Mohammadi, V.; Movahedpour, A.; et al. Recent insights into the roles of circular RNAs in human brain development and neurologic diseases. Int. J. Biol. Macromol. 2023, 225, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.; Rubner, P.; Schautzer, F.; Egg, R.; Ulmer, H.; Mayringer, I.; Dilitz, E.; Deisenhammer, F.; Reindl, M. Antimyelin antibodies as a predictor of clinically definite multiple sclerosis after a first demyelinating event. N. Engl. J. Med. 2003, 349, 139–145. [Google Scholar] [CrossRef]
- Allegra, A.; Caserta, S.; Genovese, S.; Pioggia, G.; Gangemi, S. Gender Differences in Oxidative Stress in Relation to Cancer Susceptibility and Survival. Antioxidants 2023, 12, 1255. [Google Scholar] [CrossRef] [PubMed]
- Magliozzi, R.; Howell, O.; Vora, A.; Serafini, B.; Nicholas, R.; Puopolo, M.; Reynolds, R.; Aloisi, F. Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain 2007, 130 Pt 4, 1089–1104. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.L.; Bar-Or, A.; Cohen, J.A.; Comi, G.; Correale, J.; Coyle, P.K.; Cross, A.H.; de Seze, J.; Leppert, D.; Montalban, X.; et al. Ofatumumab versus Teriflunomide in Multiple Sclerosis. N. Engl. J. Med. 2020, 383, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Montalban, X.; Hauser, S.L.; Kappos, L.; Arnold, D.L.; Bar-Or, A.; Comi, G.; de Seze, J.; Giovannoni, G.; Hartung, H.P.; Hemmer, B.; et al. Ocrelizumab versus Placebo in Primary Progressive Multiple Sclerosis. N. Engl. J. Med. 2017, 376, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Zurawska, A.E.; Mycko, M.P.; Selmaj, I.; Raine, C.S.; Selmaj, K.W. Multiple Sclerosis: circRNA Profile Defined Reveals Links to B-Cell Function. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e1041. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Alazami, A.M.; Jaber, F.; Hoyos-Bachiloglu, R.; Jones, J.; Weeks, S.; Alosaimi, M.F.; Bainter, W.; Cangemi, B.; Badran, Y.R.; et al. Hypomorphic variants in AK2 reveal the contribution of mitochondrial function to B-cell activation. J. Allergy Clin. Immunol. 2020, 146, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Heizmann, B.; Kastner, P.; Chan, S. The Ikaros family in lymphocyte development. Curr. Opin. Immunol. 2018, 51, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Keshari, P.K.; Harbo, H.F.; Myhr, K.M.; Aarseth, J.H.; Bos, S.D.; Berge, T. Allelic imbalance of multiple sclerosis susceptibility genes IKZF3 and IQGAP1 in human peripheral blood. BMC Genet. 2016, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Cortés, M.; Georgopoulos, K. Aiolos is required for the generation of high affinity bone marrow plasma cells responsible for long-term immunity. J. Exp. Med. 2004, 199, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Avitahl, N.; Cariappa, A.; Friedrich, C.; Ikeda, T.; Renold, A.; Andrikopoulos, K.; Liang, L.; Pillai, S.; Morgan, B.A.; et al. Aiolos regulates B cell activation and maturation to effector state. Immunity 1998, 9, 543–553. [Google Scholar] [CrossRef] [PubMed]
- van Wijnen, A.J.; Bagheri, L.; Badreldin, A.A.; Larson, A.N.; Dudakovic, A.; Thaler, R.; Paradise, C.R.; Wu, Z. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone 2021, 143, 115659. [Google Scholar] [CrossRef] [PubMed]
- Marinho, S.; Custovic, A.; Marsden, P.; Smith, J.A.; Simpson, A. 17q12-21 variants are associated with asthma and interact with active smoking in an adult population from the United Kingdom. Ann. Allergy Asthma. Immunol. 2012, 108, 402–411.e9. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Clayton, D.G.; Concannon, P.; Akolkar, B.; Cooper, J.D.; Erlich, H.A.; Julier, C.; Morahan, G.; Nerup, J.; Nierras, C.; et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009, 41, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Tanaka, S.; Fujii, T.; Aoki, A.; Komiyama, H.; Ezawa, K.; Sumiyama, K.; Sagai, T.; Shiroishi, T. Members of a novel gene family, Gsdm, are expressed exclusively in the epithelium of the skin and gastrointestinal tract in a highly tissue-specific manner. Genomics 2007, 89, 618–629. [Google Scholar] [CrossRef]
- Evsyukova, I.; Somarelli, J.A.; Gregory, S.G.; Garcia-Blanco, M.A. Alternative splicing in multiple sclerosis and other autoimmune diseases. RNA Biol. 2010, 7, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, B.; Zhang, Y.; Bai, Y.; Chao, J.; Hu, G.; Yao, H. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy 2018, 14, 404–418, Erratum in Autophagy 2022, 18, 234; Erratum in Autophagy 2020, 16, 2119. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Fan, X.; Wang, Z.; Zhang, L.; Guo, S. Circ_HECW2 functions as a miR-30e-5p sponge to regulate LPS-induced endothelial-mesenchymal transition by mediating NEGR1 expression. Brain Res. 2020, 1748, 147114. [Google Scholar] [CrossRef]
- Cardamone, G.; Paraboschi, E.M.; Rimoldi, V.; Duga, S.; Soldà, G.; Asselta, R. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int. J. Mol. Sci. 2017, 18, 576. [Google Scholar] [CrossRef] [PubMed]
- Paraboschi, E.M.; Rimoldi, V.; Soldà, G.; Tabaglio, T.; Dall’Osso, C.; Saba, E.; Vigliano, M.; Salviati, A.; Leone, M.; Benedetti, M.D.; et al. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum. Mol. Genet. 2014, 23, 6746–6761. [Google Scholar] [CrossRef]
- Anonymous. Alternative Splicing of GSDMB Modulates Pyroptosis in Cancer Cells. Cancer Discov. 2023, 13, OF10. [Google Scholar] [CrossRef]
- Schmiedel, B.J.; Seumois, G.; Samaniego-Castruita, D.; Cayford, J.; Schulten, V.; Chavez, L.; Ay, F.; Sette, A.; Peters, B.; Vijayanand, P. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat. Commun. 2016, 7, 13426. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Miller, M.; Beppu, A.K.; Mueller, J.; McGeough, M.D.; Vuong, C.; Karta, M.R.; Rosenthal, P.; Chouiali, F.; Doherty, T.A.; et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc. Natl. Acad. Sci. USA 2016, 113, 13132–13137. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, G.; Hosseini Quchani, S.; Sahraian, M.A.; Abolhassani, F.; Sadighi Gilani, M.A.; Dehghan Tarzjani, M.; Atoof, F. Leukocyte Gene Expression and Plasma Concentration in Multiple Sclerosis: Alteration of Transforming Growth Factor-βs, Claudin-11, and Matrix Metalloproteinase-2. Cell. Mol. Neurobiol. 2016, 36, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, F.; Di Marco, R.; Patti, F.; Reggio, E.; Nicoletti, A.; Zaccone, P.; Stivala, F.; Meroni, P.L.; Reggio, A. Blood levels of transforming growth factor-beta 1 (TGF-beta1) are elevated in both relapsing remitting and chronic progressive multiple sclerosis (MS) patients and are further augmented by treatment with interferon-beta 1b (IFN-beta1b). Clin. Exp. Immunol. 1998, 113, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Lamkanfi, M.; Vande Walle, L.; Kanneganti, T.D. Deregulated inflammasome signaling in disease. Immunol. Rev. 2011, 243, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host cell death and inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116, Erratum in Nature 2016, 540, 150. [Google Scholar] [CrossRef]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Han, J.; Zhang, Y.; Wang, X.; Fan, H.; Hua, F.; Yao, R. TLR4-RelA-miR-30a signal pathway regulates Th17 differentiation during experimental autoimmune encephalomyelitis development. J. Neuroinflamm. 2019, 16, 183. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.L.; Kumar, G.; Agasing, A.; Ko, R.M.; Axtell, R.C. Role of TFH Cells in Promoting T Helper 17-Induced Neuroinflammation. Front. Immunol. 2018, 9, 382. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Zhou, J.; Wang, T.; Han, J.; Ma, L.; Yu, H.; Geng, D.; Fan, H.; Zhang, Q.; Hua, F.; et al. MiR-30a inhibits Th17 differentiation and demyelination of EAE mice by targeting the IL-21R. Brain Behav. Immun. 2016, 57, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Sun, D.; Guan, Y.; Wang, Z.; Sang, D.; Liu, M.; Pu, Y.; Fang, X.; Wang, D.; Huang, A.; et al. Disulfiram and Diphenhydramine Hydrochloride Upregulate miR-30a to Suppress IL-17-Associated Autoimmune Inflammation. J. Neurosci. 2016, 36, 9253–9266. [Google Scholar] [CrossRef]
- Mycko, M.P.; Zurawska, A.E.; Selmaj, I.; Selmaj, K.W. Impact of Diminished Expression of circRNA on Multiple Sclerosis Pathomechanisms. Front. Immunol. 2022, 13, 875994. [Google Scholar] [CrossRef]
- Casili, G.; Lanza, M.; Campolo, M.; Siracusa, R.; Paterniti, I.; Ardizzone, A.; Scuderi, S.A.; Cuzzocrea, S.; Esposito, E. Synergic Therapeutic Potential of PEA-Um Treatment and NAAA Enzyme Silencing in the Management of Neuroinflammation. Int. J. Mol. Sci. 2020, 21, 7486. [Google Scholar] [CrossRef] [PubMed]
- Baecher-Allan, C.; Kaskow, B.J.; Weiner, H.L. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 2018, 97, 742–768. [Google Scholar] [CrossRef] [PubMed]
- Hemmer, B.; Kerschensteiner, M.; Korn, T. Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol. 2015, 14, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Cencioni, M.T.; Mattoscio, M.; Magliozzi, R.; Bar-Or, A.; Muraro, P.A. B cells in multiple sclerosis—From targeted depletion to immune reconstitution therapies. Nat. Rev. Neurol. 2021, 17, 399–414. [Google Scholar] [CrossRef]
- Habek, M. Immune and autonomic nervous system interactions in multiple sclerosis: Clinical implications. Clin. Auton. Res. 2019, 29, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.N.; Watson, C.M.; Owen, S.J.; Calder, V.L. Nervous and immune system disorders in multiple sclerosis. Brain Behav. Immun. 1988, 2, 335–340. [Google Scholar] [CrossRef] [PubMed]


| circRNA | Gene | References |
|---|---|---|
| hsa_circ_0106803 | GSDMB | Cardamone et al. [127] |
| hsa_circ_0007990 | PGAP3 | Cardamone et al. [75] |
| circ_0005402 circ_0035560 | ANXA2 | Iparraguirre et al. [83] |
| hsa_circ_0043813 | STAT3 | Paraboschi et al. [101] |
| hsa_circRNA_101348 hsa_circRNA_102611 hsa_circRNA_104361 | AK2 IKZF3 CBX5 | Zurawska et al. [114] |
| circ_HECW2 | HECW2 | Yang et al. [125] Dong et al. [126] |
| hsa_circ_0106803 | ASIC1a | Cardamone et al. [127] Xia et al. [89] |
| circ_3998 | INPP4B | Han et al. [35] |
| hsa_circRNA_101145 hsa_circRNA_001896 | NOS1 NOPCHAP1 DSE | Mycko et al. [144] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sciaccotta, R.; Murdaca, G.; Caserta, S.; Rizzo, V.; Gangemi, S.; Allegra, A. Circular RNAs: A New Approach to Multiple Sclerosis. Biomedicines 2023, 11, 2883. https://doi.org/10.3390/biomedicines11112883
Sciaccotta R, Murdaca G, Caserta S, Rizzo V, Gangemi S, Allegra A. Circular RNAs: A New Approach to Multiple Sclerosis. Biomedicines. 2023; 11(11):2883. https://doi.org/10.3390/biomedicines11112883
Chicago/Turabian StyleSciaccotta, Raffaele, Giuseppe Murdaca, Santino Caserta, Vincenzo Rizzo, Sebastiano Gangemi, and Alessandro Allegra. 2023. "Circular RNAs: A New Approach to Multiple Sclerosis" Biomedicines 11, no. 11: 2883. https://doi.org/10.3390/biomedicines11112883
APA StyleSciaccotta, R., Murdaca, G., Caserta, S., Rizzo, V., Gangemi, S., & Allegra, A. (2023). Circular RNAs: A New Approach to Multiple Sclerosis. Biomedicines, 11(11), 2883. https://doi.org/10.3390/biomedicines11112883







