Molecular Subtyping in Muscle-Invasive Bladder Cancer on Predicting Survival and Response of Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and RNA Isolation
2.2. Gene Expression Study
2.3. Clustering and Differential Expression Analysis
2.4. Validation with TCGA Dataset
3. Results
3.1. Patient Information and Clinical Characteristics
3.2. Transcriptome Profiling and Classification of Thai MIBC
3.3. ROC Analysis of 37 Differentially Expressed Genes Found in MIBC Tissues
3.4. Clinical Characteristics and Molecular Subtypes of MIBC Associated with Treatment Outcome and the Response of Perioperative Chemotherapy
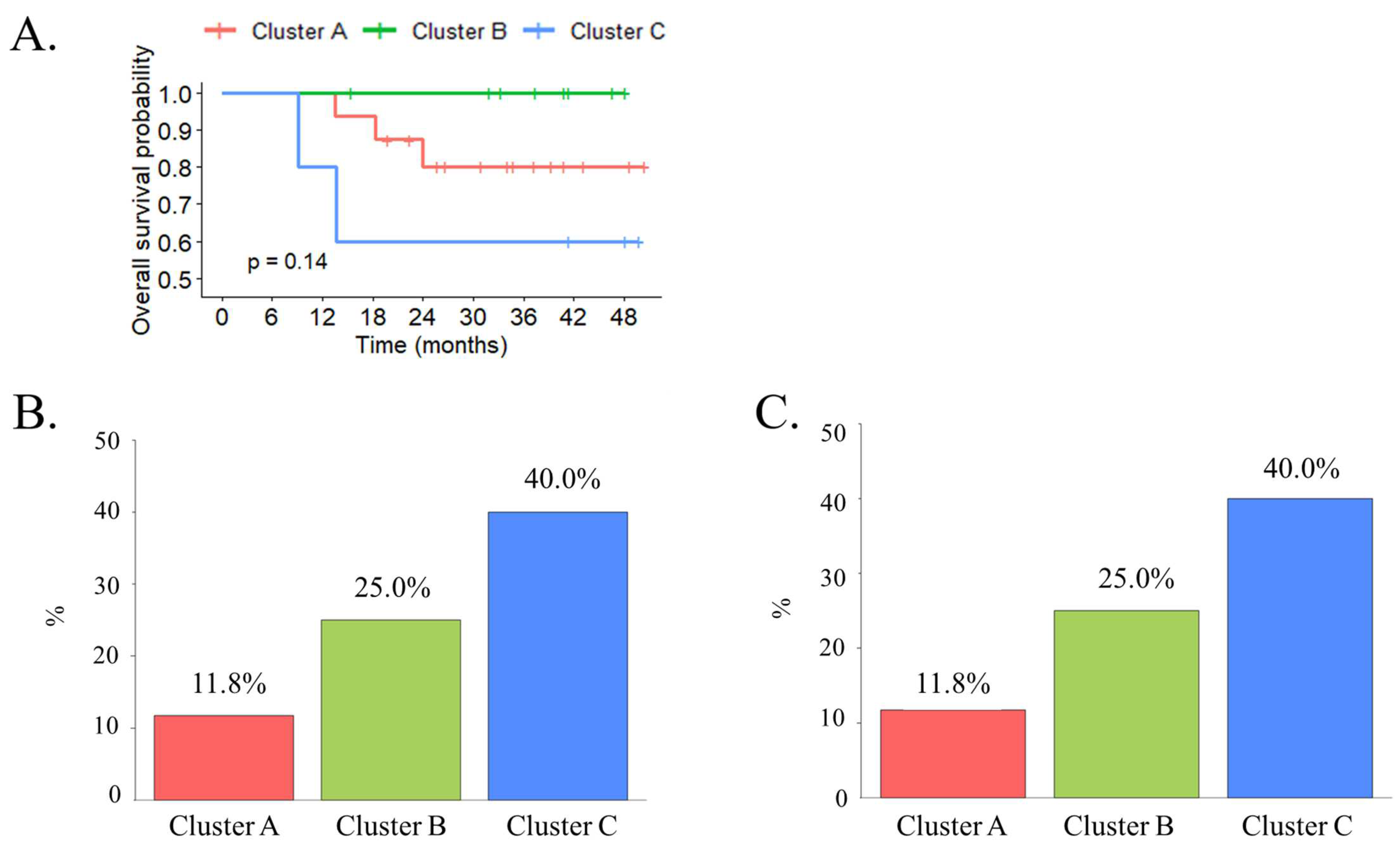
3.5. The Transcriptomic Classification Using PCA Analysis of Tissue Samples with the TCGA Data Provided a Significant Prognostic Value of MIBC Overall Survival
3.6. Certain Signaling Pathways Were Associated with Each Type of MIBC Cluster
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef] [PubMed]
- NICE Guidance. Bladder Cancer: Diagnosis and Management of Bladder Cancer: © NICE (2015) Bladder Cancer: Diagnosis and Management of Bladder Cancer. BJU Int. 2017, 120, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Milowsky, M.I.; Rumble, R.B.; Booth, C.M.; Gilligan, T.; Eapen, L.J.; Hauke, R.J.; Boumansour, P.; Lee, C.T. Guideline on Muscle-Invasive and Metastatic Bladder Cancer (European Association of Urology Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement. J. Clin. Oncol. 2016, 34, 1945–1952. [Google Scholar] [CrossRef] [PubMed]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.W.G.; Winters, B.; Douglas, J.; Van Kessel, K.E.; Fransen van de Putte, E.E.; Sommerlad, M.; Wang, N.Q.; et al. Impact of Molecular Subtypes in Muscle-Invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef]
- Pa, J.; Mj, D. Pathways of Development and Progression in Bladder Cancer: New Correlations between Clinical Observations and Molecular Mechanisms. Semin. Urol. 1993, 11, 177–192. [Google Scholar]
- Dobruch, J.; Oszczudłowski, M. Bladder Cancer: Current Challenges and Future Directions. Medicina 2021, 57, 749. [Google Scholar] [CrossRef]
- Knowles, M.A.; Hurst, C.D. Molecular Biology of Bladder Cancer: New Insights into Pathogenesis and Clinical Diversity. Nat. Rev. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef]
- Tan, T.Z.; Rouanne, M.; Tan, K.T.; Huang, R.Y.-J.; Thiery, J.-P. Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-Cohort Analysis of 2411 Tumors. Eur. Urol. 2019, 75, 423–432. [Google Scholar] [CrossRef]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic Subtypes of High-Grade Bladder Cancer Reflect the Hallmarks of Breast Cancer Biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.-L.; et al. Identification of Distinct Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer with Different Sensitivities to Frontline Chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Akbani, R.; Broom, B.M.; Wang, W.; Verhaak, R.G.W.; McConkey, D.; Lerner, S.; Morgan, M.; Creighton, C.J.; Smith, C.; et al. Comprehensive Molecular Characterization of Urothelial Bladder Carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Dadhania, V.; Zhang, M.; Zhang, L.; Bondaruk, J.; Majewski, T.; Siefker-Radtke, A.; Guo, C.C.; Dinney, C.; Cogdell, D.E.; Zhang, S.; et al. Meta-Analysis of the Luminal and Basal Subtypes of Bladder Cancer and the Identification of Signature Immunohistochemical Markers for Clinical Use. EBioMedicine 2016, 12, 105–117. [Google Scholar] [CrossRef] [PubMed]
- McConkey, D.J.; Choi, W. Molecular Subtypes of Bladder Cancer. Curr. Oncol. Rep. 2018, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef] [PubMed]
- Marzouka, N.-A.-D.; Eriksson, P.; Rovira, C.; Liedberg, F.; Sjödahl, G.; Höglund, M. A Validation and Extended Description of the Lund Taxonomy for Urothelial Carcinoma Using the TCGA Cohort. Sci. Rep. 2018, 8, 3737. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; de Reyniès, A.; Allory, Y.; Sjödahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-Invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Sjödahl, G.; Lauss, M.; Lövgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Fernö, M.; Ringnér, M.; et al. A Molecular Taxonomy for Urothelial Carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef] [PubMed]
- Dyrskjøt, L. Molecular Subtypes of Bladder Cancer: Academic Exercise or Clinical Relevance? Eur. Urol. 2019, 75, 433–434. [Google Scholar] [CrossRef]
- Faltas, B.M.; Prandi, D.; Tagawa, S.T.; Molina, A.M.; Nanus, D.M.; Sternberg, C.; Rosenberg, J.; Mosquera, J.M.; Robinson, B.; Elemento, O.; et al. Clonal Evolution of Chemotherapy-Resistant Urothelial Carcinoma. Nat. Genet. 2016, 48, 1490–1499. [Google Scholar] [CrossRef]
- Ochoa, A.E.; Choi, W.; Su, X.; Siefker-Radtke, A.; Czerniak, B.; Dinney, C.; McConkey, D.J. Specific Micro-RNA Expression Patterns Distinguish the Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer. Oncotarget 2016, 7, 80164–80174. [Google Scholar] [CrossRef]
- Rinaldetti, S.; Rempel, E.; Worst, T.S.; Eckstein, M.; Steidler, A.; Weiss, C.A.; Bolenz, C.; Hartmann, A.; Erben, P. Subclassification, Survival Prediction and Drug Target Analyses of Chemotherapy-Naïve Muscle-Invasive Bladder Cancer with a Molecular Screening. Oncotarget 2018, 9, 25935–25945. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Ochoa, A.; McConkey, D.J.; Aine, M.; Höglund, M.; Kim, W.Y.; Real, F.X.; Kiltie, A.E.; Milsom, I.; Dyrskjøt, L.; et al. Genetic Alterations in the Molecular Subtypes of Bladder Cancer: Illustration in the Cancer Genome Atlas Dataset. Eur. Urol. 2017, 72, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Aung, C.S.; Kruger, W.A.; Poronnik, P.; Roberts-Thomson, S.J.; Monteith, G.R. Plasma Membrane Ca2+-ATPase Expression during Colon Cancer Cell Line Differentiation. Biochem. Biophys. Res. Commun. 2007, 355, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Grice, D.M.; Faddy, H.M.; Nguyen, N.; Leitch, S.; Wang, Y.; Muend, S.; Kenny, P.A.; Sukumar, S.; Roberts-Thomson, S.J.; et al. Store-Independent Activation of Orai1 by SPCA2 in Mammary Tumors. Cell 2010, 143, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Maynard, J.P.; Lee, J.-S.; Sohn, B.H.; Yu, X.; Lopez-Terrada, D.; Finegold, M.J.; Goss, J.A.; Thevananther, S. P2X3 Purinergic Receptor Overexpression Is Associated with Poor Recurrence-Free Survival in Hepatocellular Carcinoma Patients. Oncotarget 2015, 6, 41162–41179. [Google Scholar] [CrossRef]
- Bellacosa, A.; Kumar, C.C.; Di Cristofano, A.; Testa, J.R. Activation of AKT Kinases in Cancer: Implications for Therapeutic Targeting. Adv. Cancer Res. 2005, 94, 29–86. [Google Scholar] [CrossRef]
- Kobayashi, I.; Semba, S.; Matsuda, Y.; Kuroda, Y.; Yokozaki, H. Significance of Akt Phosphorylation on Tumor Growth and Vascular Endothelial Growth Factor Expression in Human Gastric Carcinoma. Pathobiology 2006, 73, 8–17. [Google Scholar] [CrossRef]
- Altomare, D.A.; Testa, J.R. Perturbations of the AKT Signaling Pathway in Human Cancer. Oncogene 2005, 24, 7455–7464. [Google Scholar] [CrossRef]
- Lee, S.; Rauch, J.; Kolch, W. Targeting MAPK Signaling in Cancer: Mechanisms of Drug Resistance and Sensitivity. Int. J. Mol. Sci. 2020, 21, 1102. [Google Scholar] [CrossRef]
- Sever, R.; Brugge, J.S. Signal Transduction in Cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a006098. [Google Scholar] [CrossRef]
- Brown, B.C.; Bray, N.L.; Pachter, L. Expression Reflects Population Structure. PLoS Genet. 2018, 14, e1007841. [Google Scholar] [CrossRef] [PubMed]
- Sjödahl, G.; Eriksson, P.; Lövgren, K.; Marzouka, N.-A.-D.; Bernardo, C.; Nordentoft, I.; Dyrskjøt, L.; Liedberg, F.; Höglund, M. Discordant Molecular Subtype Classification in the Basal-Squamous Subtype of Bladder Tumors and Matched Lymph-Node Metastases. Mod. Pathol. 2018, 31, 1869–1881. [Google Scholar] [CrossRef] [PubMed]
- Lindskrog, S.V.; Prip, F.; Lamy, P.; Taber, A.; Groeneveld, C.S.; Birkenkamp-Demtröder, K.; Jensen, J.B.; Strandgaard, T.; Nordentoft, I.; Christensen, E.; et al. An Integrated Multi-Omics Analysis Identifies Prognostic Molecular Subtypes of Non-Muscle-Invasive Bladder Cancer. Nat. Commun. 2021, 12, 2301. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Babă, D.-F.; de Cobelli, O.; Musi, G.; Lucarelli, G.; Terracciano, D.; Porreca, A.; Busetto, G.M.; Del Giudice, F.; Soria, F.; et al. Neutrophil Percentage-to-Albumin Ratio Predicts Mortality in Bladder Cancer Patients Treated with Neoadjuvant Chemotherapy Followed by Radical Cystectomy. Future Sci. 2021, 7, FSO709. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Chen, K.; Babbra, R.K.; Feng, W.; Pejovic, N.; Nallicheri, A.; Harris, P.K.; Dienstbach, K.; Atkocius, A.; Maguire, L.; et al. Urine Tumor DNA Detection of Minimal Residual Disease in Muscle-Invasive Bladder Cancer Treated with Curative-Intent Radical Cystectomy: A Cohort Study. PLoS Med. 2021, 18, e1003732. [Google Scholar] [CrossRef]
- Crocetto, F.; Barone, B.; Ferro, M.; Busetto, G.M.; La Civita, E.; Buonerba, C.; Di Lorenzo, G.; Terracciano, D.; Schalken, J.A. Liquid Biopsy in Bladder Cancer: State of the Art and Future Perspectives. Crit. Rev. Oncol./Hematol. 2022, 170, 103577. [Google Scholar] [CrossRef]



| Thai Patient Dataset | Percentage | TCGA Dataset | Percentage | |
|---|---|---|---|---|
| Samples | 30 | 231 | ||
| Average age (range) | 67.5 (52–92) | 69 (46–90) | ||
| Gender Male Female | 26 4 | 86.2 13.8 | 169 62 | 73.16 26.64 |
| ECOG 0 1 | 21 9 | 70 30 | 158 73 | 68.5 31.5 |
| T stage T 2 T 3 T 4 | 24 6 0 | 80 20 0 | 75 123 33 | 32.47 53.25 14.28 |
| N stage N 0 N 1 N 2 N 3 N x | 22 7 1 0 0 | 73.3 23.3 3.3 0 0 | 143 28 42 4 14 | 58.01 10.68 18.2 1.94 6.06 |
| M stage M 0 M 1 Not available | 30 0 0 | 100 0 0 | 116 5 110 | 50.22 2.16 47.62 |
| Genes | Cluster A | Cluster B | Cluster C |
|---|---|---|---|
| CCL2 (C-C Motif Chemokine Ligand 2) | 0.7680 | 0.941 | 0.859 |
| FGF2 (Fibroblast growth factor 2) | 0.6940 | 0.947 | 0.886 |
| RYR3 (Ryanodine receptor 3) | 0.5200 | 0.721 | 0.735 |
| MYLK (Myosin light chain kinase) | 0.8040 | 0.955 | 0.914 |
| EDNRA (Endothelin receptor type A) | 0.8110 | 0.951 | 0.833 |
| FGF7 (Fibroblast growth factor 7) | 0.7890 | 0.976 | 0.927 |
| BDKRB1 (Bradykinin receptor B1) | 0.7030 | 0.872 | 0.727 |
| PDGFRB (Platelet-derived growth factor receptor beta) | 0.8380 | 0.988 | 0.838 |
| HGF (Hepatocyte Growth Factor) | 0.7310 | 0.945 | 0.867 |
| AVPR1A (Arginine vasopressin receptor 1A) | 0.6200 | 0.774 | 0.708 |
| NGF (Nerve growth factor) | 0.6650 | 0.893 | 0.783 |
| PTGFR (Prostaglandin F receptor) | 0.7230 | 0.947 | 0.912 |
| PDE1A (Phosphodiesterase 1A) | 0.6170 | 0.916 | 0.86 |
| PTGER3 (Prostaglandin E receptor 3) | 0.7790 | 0.951 | 0.821 |
| CAMK2A (Calcium/calmodulin-dependent protein kinase 2 Alpha) | 0.8200 | 0.971 | 0.862 |
| NTRK3 (Neurotrophic receptor tyrosine kinase 3) | 0.5380 | 0.828 | 0.831 |
| P2RX1 (Purinergic receptor P2X 1) | 0.7710 | 0.96 | 0.907 |
| TNC (Tenascin C) | 0.8530 | 0.947 | 0.746 |
| COL4A4 (Collagen type IV alpha 4 chain) | 0.5890 | 0.852 | 0.785 |
| COL6A2 (Collagen type VI alpha 2 Chain) | 0.8640 | 0.988 | 0.886 |
| ITGA8 (Integrin subunit alpha 8) | 0.6030 | 0.72 | 0.799 |
| COL6A3 (Collagen type VI alpha 3 chain) | 0.8430 | 0.979 | 0.849 |
| CREB5 (CAMP-responsive element binding protein 5) | 0.8200 | 0.946 | 0.79 |
| TNXB (Tenascin XB) | 0.6460 | 0.875 | 0.815 |
| ANGPT1 (Angiopoietin 1) | 0.6820 | 0.826 | 0.686 |
| IGF1 (Insulin-like growth factor 1) | 0.689 | 0.919 | 0.879 |
| COL1A1 (Collagen type I alpha 1 chain) | 0.869 | 0.95 | 0.736 |
| COL1A2 (Collagen type I alpha 2 chain) | 0.857 | 0.967 | 0.787 |
| ITGA11 (Integrin subunit alpha 11) | 0.824 | 0.915 | 0.67 |
| NPR1 (Natriuretic peptide receptor 1) | 0.628 | 0.885 | 0.808 |
| KCNMB1 (Potassium calcium-activated channel subfamily M regulatory beta subunit 1) | 0.749 | 0.957 | 0.936 |
| ADORA1 (Adenosine A1 receptor) | 0.761 | 0.89 | 0.762 |
| PRKG1 (Protein kinase cGMP-dependent 1) | 0.777 | 0.964 | 0.889 |
| ATP1B2 (ATPase Na+/K+ transporting subunit beta 2) | 0.542 | 0.743 | 0.751 |
| ADRA2A (Adrenoceptor alpha 2A) | 0.582 | 0.915 | 0.845 |
| KCNMA1 (Potassium calcium-activated channel subfamily M alpha 1) | 0.799 | 0.954 | 0.88 |
| GNAO1 (G Protein subunit alpha O1) | 0.739 | 0.939 | 0.85 |
| Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% Cl | p Value | HR | 95% Cl | p Value | |
| T stage | ||||||
| 2 | Ref | |||||
| 3–4 | 6.62 | 1.09–40.1 | 0.041 | 25.64 | 2.31–284.04 | 0.006 |
| N stage | ||||||
| 0 | Ref | 0.036 | ||||
| 1 | 4.05 | 0.45–5.46 | ||||
| 2 | 6.46 | 0.35–6.86 | ||||
| Age (years) | ||||||
| ≤65 | Ref | |||||
| >65 | 2.42 | 0.27–21.67 | 0.392 | |||
| Lymph node metastasis | ||||||
| negative | Ref | |||||
| positive | 2.39 | 1.29–4.41 | 0.01 | |||
| LVI | ||||||
| negative | Ref | |||||
| positive | 1.08 | 0.18–6.5 | 0.929 | |||
| Ureteric margin | ||||||
| negative | Ref | |||||
| positive | 5.32 | 0.59–48.17 | 0.212 | |||
| Cluster | ||||||
| Cluster A | Ref | |||||
| Cluster B | 0 | 1.6–4.94 | 0.108 | |||
| Cluster C | 2.63 | 0.44–15.79 | 0.291 | |||
| No. | Term | Overlap | p-Value | Adjusted p-Value |
|---|---|---|---|---|
| Cluster A | ||||
| 1 | Chemokine signaling pathway | 44/192 | 2.24 × 10−10 | 9.16 × 10−9 |
| 2 | Calcium signaling pathway | 49/240 | 1.50 × 10−9 | 5.38 × 10−8 |
| 3 | JAK-STAT signaling pathway | 32/162 | 2.14 × 10−6 | 4.08 × 10−5 |
| 4 | PI3K-Akt signaling pathway | 55/354 | 2.45 × 10−6 | 4.38 × 10−5 |
| 5 | B cell receptor signaling pathway | 18/81 | 7.13 × 10−5 | 7.03 × 10−4 |
| 6 | Ras signaling pathway | 35/232 | 2.80 × 10−4 | 2.28 × 10−3 |
| 7 | T cell receptor signaling pathway | 19/104 | 6.66 × 10−4 | 4.76 × 10−3 |
| 8 | cGMP-PKG signaling pathway | 26/167 | 1.00 × 10−3 | 6.85 × 10−3 |
| 9 | Rap1 signaling pathway | 29/210 | 3.48 × 10−3 | 2.16 × 10−2 |
| 10 | NF-kappa B signaling pathway | 17/04 | 4.24 × 10−3 | 2.58 × 10−2 |
| Cluster B | ||||
| 1 | Calcium signaling pathway | 94/240 | 2.89 × 10−16 | 1.73 × 10−14 |
| 2 | PI3K-Akt signaling pathway | 112/354 | 1.09 × 10−11 | 4.11 × 10−10 |
| 3 | Chemokine signaling pathway | 71/192 | 2.95 × 10−8 | 9.77 × 10−10 |
| 4 | cGMP-PKG signaling pathway | 57/167 | 6.91 × 10−8 | 9.45 × 10−6 |
| 5 | AGE-RAGE signaling pathway in diabetic complications | 39/100 | 1.58 × 10−7 | 1.98 × 10−6 |
| 6 | Rap1 signaling pathway | 62/210 | 5.47 × 10−6 | 4.71 × 10−5 |
| 7 | cAMP signaling pathway | 63/126 | 7.13 × 10−6 | 5.80 × 10−5 |
| 8 | Ras signaling pathway | 65/232 | 2.14 × 10−5 | 1.61 × 10−4 |
| 9 | Apelin signaling pathway | 41/137 | 1.43 × 10−4 | 9.18 × 10−4 |
| 10 | Phospholipase D signaling pathway | 43/148 | 2.08 × 10−4 | 1.31 × 10−3 |
| Cluster C | ||||
| 1 | cGMP-PKG signaling pathway | 32/167 | 2.30 × 10−9 | 1.25 × 10−7 |
| 2 | Calcium signaling pathway | 38/240 | 1.96 × 10−8 | 5.92 × 10−7 |
| 3 | PI3K-Akt signaling pathway | 46/354 | 3.13 × 10−4 | 8.52 × 10−6 |
| 4 | Oxytocin signaling pathway | 21/154 | 2.53 × 10−4 | 4.06 × 10−3 |
| 5 | MAPK signaling pathway | 33/294 | 2.54 × 10−4 | 4.06 × 10−3 |
| 6 | cAMP signaling pathway | 26/216 | 3.81 × 10−4 | 5.46 × 10−3 |
| 7 | Apelin signaling pathway | 18/137 | 1.05 × 10−3 | 1.25 × 10−2 |
| 8 | Relaxin signaling pathway | 17/129 | 1.39 × 10−3 | 1.53 × 10−2 |
| 9 | AGE-RAGE signaling pathway in diabetic complications | 14/100 | 2.00 × 10−3 | 2.01 × 10−2 |
| 10 | Ras signaling pathway | 25/232 | 2.37 × 10−3 | 2.22 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bejrananda, T.; Saetang, J.; Sangkhathat, S. Molecular Subtyping in Muscle-Invasive Bladder Cancer on Predicting Survival and Response of Treatment. Biomedicines 2023, 11, 69. https://doi.org/10.3390/biomedicines11010069
Bejrananda T, Saetang J, Sangkhathat S. Molecular Subtyping in Muscle-Invasive Bladder Cancer on Predicting Survival and Response of Treatment. Biomedicines. 2023; 11(1):69. https://doi.org/10.3390/biomedicines11010069
Chicago/Turabian StyleBejrananda, Tanan, Jirakrit Saetang, and Surasak Sangkhathat. 2023. "Molecular Subtyping in Muscle-Invasive Bladder Cancer on Predicting Survival and Response of Treatment" Biomedicines 11, no. 1: 69. https://doi.org/10.3390/biomedicines11010069
APA StyleBejrananda, T., Saetang, J., & Sangkhathat, S. (2023). Molecular Subtyping in Muscle-Invasive Bladder Cancer on Predicting Survival and Response of Treatment. Biomedicines, 11(1), 69. https://doi.org/10.3390/biomedicines11010069






