Prevalence and Virulence of Commensal Pseudomonas Aeruginosa Isolates from Healthy Individuals in Southern Vietnam (2018–2020)
Abstract
1. Introduction
2. Materials and Methods
2.1. Commensal P. aeruginosa Isolation
2.2. P. aeruginosa Identification
oprL—Specific Polymerase Chain Reaction and 16S rRNA Sequencing
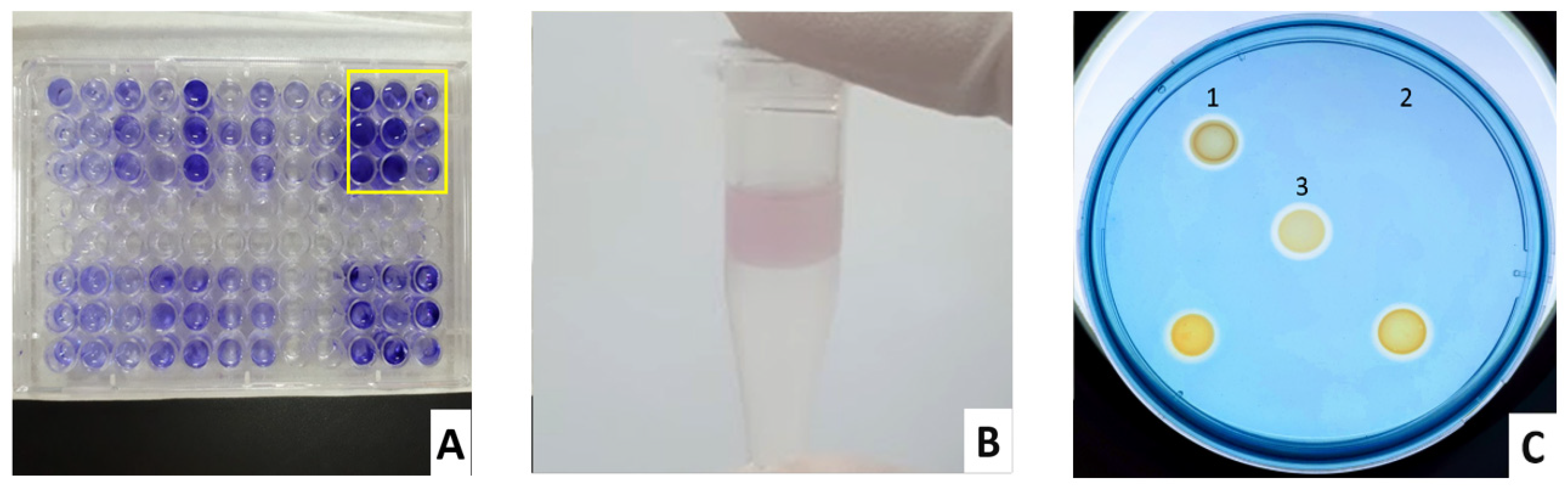
2.3. Virulence Testing
2.3.1. Biofilm
2.3.2. Pyocyanin
2.3.3. Siderophores, Lipase, Protease, and Gelatinase
2.4. Data Analysis
3. Results and Discussion
3.1. Prevalence of Commensal P. aeruginosa Isolates in Vietnamese Population
3.2. Relationship between Sex and Age to P. aeruginosa Colonization
3.3. Relationship of Sinusitis History to P. aeruginosa Colonization
3.4. Commensal P. aeruginosa Virulence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bergmans, D.; Bonten, M. Colonization and Infection with Pseudomonas aeruginosa in Intensive Care: Endogenous or Exogenous Origin? In Yearbook of Intensive Care and Emergency Medicine 1999; Vincent, J.-L., Ed.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 131–140. [Google Scholar]
- Proença, J.T.; Barral, D.C.; Gordo, I. Commensal-to-Pathogen Transition: One-Single Transposon Insertion Results in Two Pathoadaptive Traits in Escherichia Coli -Macrophage Interaction. Sci. Rep. 2017, 7, 4504. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa Infection: Lessons from a Versatile Opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.G.; Rosa, A.C.; Ferreira, A.S.; Moreira, L.M.; Proença, D.N.; Morais, P.V.; Cardoso, O. Virulence Factors and Infection Ability of Pseudomonas aeruginosa Isolates from a Hydropathic Facility and Respiratory Infections. J. Appl. Microbiol. 2014, 116, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Ołdak, E.; Trafny, E.A. Secretion of Proteases by Pseudomonas aeruginosa Biofilms Exposed to Ciprofloxacin. Antimicrob. Agents Chemother. 2005, 49, 3281–3288. [Google Scholar] [CrossRef] [PubMed]
- Prasad, A.S.B.; Shruptha, P.; Prabhu, V.; Srujan, C.; Nayak, U.Y.; Anuradha, C.K.R.; Ramachandra, L.; Keerthana, P.; Joshi, M.B.; Murali, T.S.; et al. Pseudomonas aeruginosa Virulence Proteins Pseudolysin and Protease IV Impede Cutaneous Wound Healing. Lab Investig. 2020, 100, 1532–1550. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Do, E.; Jung, W.H. Lipolytic Enzymes Involved in the Virulence of Human Pathogenic Fungi. Mycobiology 2013, 41, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Rosenau, F.; Isenhardt, S.; Gdynia, A.; Tielker, D.; Schmidt, E.; Tielen, P.; Schobert, M.; Jahn, D.; Wilhelm, S.; Jaeger, K.-E. Lipase LipC Affects Motility, Biofilm Formation and Rhamnolipid Production in Pseudomonas Aeruginosa. FEMS Microbiol. Lett. 2010, 309, 25–34. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence Factors of Pseudomonas aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front. Cell Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Pinna, A.; Usai, D.; Sechi, L.A.; Molicotti, P.; Zanetti, S.; Carta, A. Detection of Virulence Factors in Pseudomonas aeruginosa Strains Isolated from Contact Lens-Associated Corneal Ulcers. Cornea 2008, 27, 320–326. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Mah, T.F.; O’Toole, G.A. Mechanisms of Biofilm Resistance to Antimicrobial Agents. Trends. Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological Heterogeneity in Biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, T.; Vasconcelos, U. Colour Me Blue: The History and the Biotechnological Potential of Pyocyanin. Molecules 2021, 26, 927. [Google Scholar] [CrossRef] [PubMed]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The Role of Pyocyanin in Pseudomonas aeruginosa Infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef]
- Hassan, H.M.; Fridovich, I. Mechanism of the Antibiotic Action Pyocyanine. J. Bacteriol. 1980, 141, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial Iron Homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas aeruginosa Biofilm Formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef]
- Coffman, T.J.; Cox, C.D.; Edeker, B.L.; Britigan, B.E. Possible Role of Bacterial Siderophores in Inflammation. Iron Bound to the Pseudomonas Siderophore Pyochelin Can Function as a Hydroxyl Radical Catalyst. J. Clin. Investig. 1990, 86, 1030–1037. [Google Scholar] [CrossRef]
- Kluytmans, J.; van Belkum, A.; Verbrugh, H. Nasal Carriage of Staphylococcus aureus: Epidemiology, Underlying Mechanisms, and Associated Risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.-J.; Lavigne, J.-P.; Pitout, J.; Johnson, J.R. Intercontinental Emergence of Escherichia Coli Clone O25:H4-ST131 Producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef]
- De Vos, D.; Lim, A.; Pirnay, J.P.; Struelens, M.; Vandenvelde, C.; Duinslaeger, L.; Vanderkelen, A.; Cornelis, P. Direct Detection and Identification of Pseudomonas aeruginosa in Clinical Samples Such as Skin Biopsy Specimens and Expectorations by Multiplex PCR Based on Two Outer Membrane Lipoprotein Genes, OprI and OprL. J. Clin. Microbiol. 1997, 35, 1295–1299. [Google Scholar] [CrossRef] [PubMed]
- Enhanced Synergistic Effects of Xylitol and Isothiazolones for Inhibition of Initial Biofilm Formation by Pseudomonas Aeruginosa ATCC 9027 and Staphylococcus aureus ATCC 6538-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/31217374/ (accessed on 14 November 2022).
- Pseudomonas Aeruginosa (Schroeter) Migula-9027|ATCC. Available online: https://www.atcc.org/products/9027 (accessed on 14 November 2022).
- Schulte, G.; Bohne, L.; Winkler, U. Glycogen and Various Other Polysaccharides Stimulate the Formation of Exolipase by Pseudomonas aeruginosa. Can. J. Microbiol. 1982, 28, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Vfr or CyaB Promote the Expression of the Pore-Forming Toxin ExlBA Operon in Pseudomonas Aeruginosa ATCC 9027 without Increasing Its Virulence in Mice|Microbiology Society. Available online: https://www.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.001083 (accessed on 14 November 2022).
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and Characterization of Genes for a Second Anthranilate Synthase in Pseudomonas aeruginosa: Interchangeability of the Two Anthranilate Synthases and Evolutionary Implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Gahlout, M.; Prajapati, H.; Chauhan, P.; Patel, N.; Solanki, D. Isolation and Screening of Pyocyanin Producing Pseudomonas Spp. from Soil. Int. J. Adv. Res. Biol. Sci. 2017, 4, 147–152. [Google Scholar] [CrossRef]
- Louden, B.C.; Haarmann, D.; Lynne, A.M. Use of Blue Agar CAS Assay for Siderophore Detection. J. Microbiol. Biol. Educ. 2011, 12, 51–53. [Google Scholar] [CrossRef]
- Manjunatha, H.; Naik, M.; Rangeshwaran, R. Identification of Fluorescent Pseudomonas Isolates with Potential Biocontrol Activity from the Rhizosphere of Crops. J. Pure Appl. Microbio. 2017, 11, 1487–1495. [Google Scholar] [CrossRef]
- Matthijs, S.; Coorevits, A.; Gebrekidan, T.T.; Tricot, C.; Wauven, C.V.; Pirnay, J.-P.; De Vos, P.; Cornelis, P. Evaluation of OprI and OprL Genes as Molecular Markers for the Genus Pseudomonas and Their Use in Studying the Biodiversity of a Small Belgian River. Res. Microbiol. 2013, 164, 254–261. [Google Scholar] [CrossRef]
- Abdullahi, R.; Lihan, S.; Carlos, B.; Maurice Bilung, L.; Michelle, K.; Collick, F. Detection of OprL Gene and Antibiotic Resistance of Pseudomonas Aeruginosa from Aquaculture Environment. Eur. J. Exp. Biol. 2013, 3, 148–152. [Google Scholar]
- Hoang, S.; Georget, A.; Asselineau, J.; Venier, A.-G.; Leroyer, C.; Rogues, A.M.; Thiébaut, R. Risk Factors for Colonization and Infection by Pseudomonas aeruginosa in Patients Hospitalized in Intensive Care Units in France. PLoS ONE 2018, 13, e0193300. [Google Scholar] [CrossRef]
- Berthelot, P.; Grattard, F.; Mahul, P.; Pain, P.; Jospé, R.; Venet, C.; Carricajo, A.; Aubert, G.; Ros, A.; Dumont, A.; et al. Prospective Study of Nosocomial Colonization and Infection Due to Pseudomonas aeruginosa in Mechanically Ventilated Patients. Intensive Care Med. 2001, 27, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Pasteur, M.C.; Helliwell, S.M.; Houghton, S.J.; Webb, S.C.; Foweraker, J.E.; Coulden, R.A.; Flower, C.D.; Bilton, D.; Keogan, M.T. An Investigation into Causative Factors in Patients with Bronchiectasis. Am. J. Respir. Crit. Care Med. 2000, 162, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Casetta, A.; Audibert, F.; Brivet, F.; Boutros, N.; Boithias, C.; Lebrun, L. Emergence of Nosocomial Pseudomonas aeruginosa Colonization/Infection in Pregnant Women with Preterm Premature Rupture of Membranes and in Their Neonates. J. Hosp. Infect. 2003, 54, 158–160. [Google Scholar] [CrossRef]
- Gómez-Zorrilla, S.; Camoez, M.; Tubau, F.; Cañizares, R.; Periche, E.; Dominguez, M.A.; Ariza, J.; Peña, C. Prospective Observational Study of Prior Rectal Colonization Status as a Predictor for Subsequent Development of Pseudomonas aeruginosa Clinical Infections. Antimicrob. Agents Chemother. 2015, 59, 5213–5219. [Google Scholar] [CrossRef]
- Iglewski, B.H. Pseudomonas. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Rodríguez-Martínez, J.-M.; Poirel, L.; al Naiemi, N.; Debets-Ossenkopp, Y.J.; Nordmann, P. Characterization of Fluoroquinolone Resistance in a Clinical Isolate of Pseudomonas stutzeri. J. Antimicrob. Chemother. 2010, 65, 366–367. [Google Scholar] [CrossRef] [PubMed]
- Al-Zaidi, J.R. Antibiotic Susceptibility Patterns of Pseudomonas aeruginosa Isolated from Clinical and Hospital Environmental Samples in Nasiriyah, Iraq. Afr. J. Microbiol. Res. 2016, 10, 844–849. [Google Scholar] [CrossRef][Green Version]
- Chotirmall, S.H.; Smith, S.G.; Gunaratnam, C.; Cosgrove, S.; Dimitrov, B.D.; O’Neill, S.J.; Harvey, B.J.; Greene, C.M.; McElvaney, N.G. Effect of Estrogen on Pseudomonas Mucoidy and Exacerbations in Cystic Fibrosis. N. Engl. J. Med. 2012, 366, 1978–1986. [Google Scholar] [CrossRef]
- Pieters, A.; Bakker, M.; Hoek, R.A.S.; Altenburg, J.; van Westreenen, M.; Aerts, J.G.J.V.; van der Eerden, M.M. Predicting Factors for Chronic Colonization of Pseudomonas aeruginosa in Bronchiectasis. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2299–2304. [Google Scholar] [CrossRef]
- Baig, U.; Laxmi, V.; Ojha, A.; Watve, M. Geriatric Infections: Decreased Immunity or Evolved Opportunists? J. Biosci. 2020, 45, 57. [Google Scholar] [CrossRef]
- Paling, F.P.; Wolkewitz, M.; Depuydt, P.; de Bus, L.; Sifakis, F.; Bonten, M.J.M.; Kluytmans, J.A.J.W. P. Aeruginosa Colonization at ICU Admission as a Risk Factor for Developing P. Aeruginosa ICU Pneumonia. Antimicrob. Resist. Infect. Control 2017, 6, 38. [Google Scholar] [CrossRef][Green Version]
- Niederman, M.S. Gram-Negative Colonization of the Respiratory Tract: Pathogenesis and Clinical Consequences. Semin. Respir. Infect 1990, 5, 173–184. [Google Scholar] [PubMed]
- Shapiro, E.D.; Milmoe, G.J.; Wald, E.R.; Rodnan, J.B.; Bowen, A.D. Bacteriology of the Maxillary Sinuses in Patients with Cystic Fibrosis. J. Infect. Dis. 1982, 146, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Aanæs, K. Bacterial Sinusitis Can Be a Focus for Initial Lung Colonisation and Chronic Lung Infection in Patients with Cystic Fibrosis. J. Cyst. Fibros. 2013, 12, S1–S20. [Google Scholar] [CrossRef]
- Frederick, J.; Braude, A.I. Anaerobic Infection of the Paranasal Sinuses. N. Engl. J. Med. 1974, 290, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.L.D.C.; Alves, L.R.; Da Paz, J.N.P.; Rabelo, M.A.; Maciel, M.A.V.; De Morais, M.M.C. Analysis of Biofilm Production by Clinical Isolates of Pseudomonas aeruginosa from Patients with Ventilator-Associated Pneumonia. Rev. Bras Ter. Intensiva. 2017, 29, 310–316. [Google Scholar] [CrossRef]
- Perez, L.R.R.; Costa, M.C.N.; Freitas, A.L.P.; Barth, A.L. Evaluation of Biofilm Production by Pseudomonas aeruginosa Isolates Recovered from Cystic Fibrosis and Non-Cystic Fibrosis Patients. Braz. J. Microbiol. 2011, 42, 476. [Google Scholar] [CrossRef][Green Version]
- Guembe, M.; Alonso, B.; Lucio, J.; Pérez-Granda, M.J.; Cruces, R.; Sánchez-Carrillo, C.; Fernández-Cruz, A.; Bouza, E. Biofilm Production Is Not Associated with Poor Clinical Outcome in 485 Patients with Staphylococcus aureus Bacteraemia. Clin. Microbiol. Infect. 2018, 24, 659.e1–659.e3. [Google Scholar] [CrossRef]
- Jefferson, K.K. What Drives Bacteria to Produce a Biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef]
- Gupte, A.; Jyot, J.; Ravi, M.; Ramphal, R. High Pyocyanin Production and Non-Motility of Pseudomonas aeruginosa Isolates Are Correlated with Septic Shock or Death in Bacteremic Patients. PLoS ONE 2021, 16, e0253259. [Google Scholar] [CrossRef]
- Sajeed Ali, S.; Vidhale, N. Evaluation of Siderophore Produced by Different Clinical Isolate Pseudomonas aeruginosa. Int. J. Microbiol. Res. 2011, 3, 131–135. [Google Scholar] [CrossRef]
- Georgescu, M.; Gheorghe, I.; Curutiu, C.; Lazar, V.; Bleotu, C.; Chifiriuc, M.-C. Virulence and Resistance Features of Pseudomonas aeruginosa Strains Isolated from Chronic Leg Ulcers. BMC Infect. Dis. 2016, 16, 92. [Google Scholar] [CrossRef] [PubMed]
- Naughton, S.; Parker, D.; Seemann, T.; Thomas, T.; Turnbull, L.; Rose, B.; Bye, P.; Cordwell, S.; Whitchurch, C.; Manos, J. Pseudomonas aeruginosa AES-1 Exhibits Increased Virulence Gene Expression during Chronic Infection of Cystic Fibrosis Lung. PLoS ONE 2011, 6, e24526. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ramezanpour, M.; Fong, S.A.; Cooksley, C.; Murphy, J.; Suzuki, M.; Psaltis, A.J.; Wormald, P.J.; Vreugde, S. Pseudomonas aeruginosa Exoprotein-Induced Barrier Disruption Correlates with Elastase Activity and Marks Chronic Rhinosinusitis Severity. Front. Cell. Infect. Microbiol. 2019, 9, 38. [Google Scholar] [CrossRef]
- Liu, C.M.; Price, L.B.; Hungate, B.A.; Abraham, A.G.; Larsen, L.A.; Christensen, K.; Stegger, M.; Skov, R.; Andersen, P.S. Staphylococcus aureus and the Ecology of the Nasal Microbiome. Sci. Adv. 2015, 1, e1400216. [Google Scholar] [CrossRef]
- Davies, G.; Wells, A.U.; Doffman, S.; Watanabe, S.; Wilson, R. The Effect of Pseudomonas aeruginosa on Pulmonary Function in Patients with Bronchiectasis. Eur. Respir. J. 2006, 28, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Solano, L.; Macia, M.D.; Fajardo, A.; Oliver, A.; Martinez, J.L. Chronic Pseudomonas aeruginosa Infection in Chronic Obstructive Pulmonary Disease. Clin. Infect. Dis. 2008, 47, 1526–1533. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Number Overall (n = 609) | |
|---|---|---|
| Sex | Male | 299 (49.10%) |
| Female | 310 (50.90%) | |
| Age | 0–17 | 65 (10.67%) |
| 18-59 | 514 (84.40%) | |
| ≥60 | 30 (4.93%) | |
| Health status | With sinusitis history | 129 (21.18%) |
| Without sinusitis history | 480 (78.82%) | |
| Characteristics | P. aeruginosa Carriers | Non-P. aeruginosa Carriers | OR | p-Value (Fisher’s Exact Test) | |
|---|---|---|---|---|---|
| Sex | Female (n = 310) | 12 (3.87%) | 298 (96.13%) | 1.68 | 0.29 |
| Male (n = 299) | 7 (2.34%) | 292 (97.66%) | 1.00 | ||
| Health status | With Sinusitis history (n = 129) | 14 (10.85%) | 115 (89.15%) | 11.57 | <0.0001 |
| Without sinusitis history (n = 480) | 5 (1.04%) | 475 (98.96%) | 1.00 | ||
| Age groups, years | 0–17 (n = 65) | 1 (1.56%) | 64 (98.46%) | 1.00 | |
| 18–59 (n = 514) | 16 (3.11%) | 498 (96.89%) | 2.02 | 0.0496 | |
| ≥60 (n = 30) | 2 (6.67%) | 28 (93.33%) | 6.75 | 0.125 | |
| Virulence Factors | Average Virulence Value ± Standard Deviation | |
|---|---|---|
| Commensal P. aeruginosa Isolates | P. aeruginosa ATCC 9027 | |
| Biofilm (OD550 nm) | 0.11 ± 0.06 | 0.72 ± 0.10 |
| Pyocyanin (µg/mL) | 0.61 ± 0.40 | 0.60 ± 0.23 |
| Siderophores (mm) | 1.31 ± 0.34 | 1.33 ± 0.29 |
| Lipase (mm) | 0.18 ± 0.04 | 0.17 ± 0.03 |
| Protease (mm) | 0.28 ± 0.09 | 0.17 ± 0.03 |
| Gelatinase (mm) | 0.29 ± 0.06 | 0.22 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, N.B.V.; Truong, Q.M.; Nguyen, L.Q.A.; Nguyen, N.M.H.; Tran, Q.H.; Dinh, T.T.P.; Hua, V.S.; Nguyen, V.D.; Lambert, P.A.; Nguyen, T.T.H. Prevalence and Virulence of Commensal Pseudomonas Aeruginosa Isolates from Healthy Individuals in Southern Vietnam (2018–2020). Biomedicines 2023, 11, 54. https://doi.org/10.3390/biomedicines11010054
Tran NBV, Truong QM, Nguyen LQA, Nguyen NMH, Tran QH, Dinh TTP, Hua VS, Nguyen VD, Lambert PA, Nguyen TTH. Prevalence and Virulence of Commensal Pseudomonas Aeruginosa Isolates from Healthy Individuals in Southern Vietnam (2018–2020). Biomedicines. 2023; 11(1):54. https://doi.org/10.3390/biomedicines11010054
Chicago/Turabian StyleTran, Nguyen Bao Vy, Quang Minh Truong, Lam Que Anh Nguyen, Ngoc My Huong Nguyen, Quang Hung Tran, Thi Tuyet Phuong Dinh, Vinh Son Hua, Van Dung Nguyen, Peter A. Lambert, and Thi Thu Hoai Nguyen. 2023. "Prevalence and Virulence of Commensal Pseudomonas Aeruginosa Isolates from Healthy Individuals in Southern Vietnam (2018–2020)" Biomedicines 11, no. 1: 54. https://doi.org/10.3390/biomedicines11010054
APA StyleTran, N. B. V., Truong, Q. M., Nguyen, L. Q. A., Nguyen, N. M. H., Tran, Q. H., Dinh, T. T. P., Hua, V. S., Nguyen, V. D., Lambert, P. A., & Nguyen, T. T. H. (2023). Prevalence and Virulence of Commensal Pseudomonas Aeruginosa Isolates from Healthy Individuals in Southern Vietnam (2018–2020). Biomedicines, 11(1), 54. https://doi.org/10.3390/biomedicines11010054






