Abstract
Plasminogen activator, urokinase (PLAU) is involved in cell migration, proliferation and tissue remodeling. PLAU upregulation is associated with an increase in aggressiveness, metastasis, and invasion of several cancer types, including breast cancer. In patients, this translates into decreased sensitivity to hormonal treatment, and poor prognosis. These clinical findings have led to the examination of PLAU as a biomarker for predicting breast cancer prognosis and therapy responses. In this study, we investigated the functional ability of PLAU to act as an oncogene in breast cancers by modulating its expression using CRISPR-deactivated Cas9 (CRISPR-dCas9) tools. Different effector domains (e.g., transcription modulators (VP64, KRAB)) alone or in combination with epigenetic writers (DNMT3A/3L, MSssI) were fused to dCas9 and targeted to the PLAU promoter. In MDA-MB-231 cells characterized by high PLAU expression downregulation of PLAU expression by CRISPR-dCas9-DNMT3A/3L-KRAB, resulted in decreased cell proliferation. Conversely, CRISPR-dCas9-VP64 induced PLAU upregulation in low PLAU expressing MCF-7 cells and significantly increased aggressiveness and invasion. In conclusion, modulation of PLAU expression affected metastatic related properties of breast cancer cells, thus further validating its oncogenic activity in breast cancer cells.
1. Introduction
Plasminogen activator, urokinase (PLAU) also known as urokinase-Type Plasminogen Activator (u-PA), encodes a protease that converts inactive plasminogen to the active serine protease plasmin [1]. Plasmin is involved in the breakdown of the extra-cellular matrix (ECM), and hence, plays an important role in favoring cell migration, cell proliferation and tissue remodeling. Based on the increased risk of metastasis that is associated with these functions, the oncogenic role of PLAU in several cancers, including breast cancer, has long been suggested [1,2,3,4,5,6,7]. In different studies it was shown that experimental inhibition of PLAU reduced the tumor growth, the aggressiveness and metastasis [1,8,9,10]. In one example, mice were inoculated with murine Lewis lung carcinoma cells and treated with anti-human antibodies directed towards PLAU. These antibodies cross-react with the murine PLAU and thereby inhibit PLAU activity. Injections with these antibodies resulted in significant inhibition of lung metastasis in a dose-dependent matter [5]. In a different study using a similar method, nude mice were inoculated with human squamous carcinoma cells, which express PLAU at high level and are known to metastasize to lungs and lymph nodes. Injection of these mice with anti-human PLAU antibodies abolished local invasion of the tumors [4]. Similarly, suppression of PLAU in human ovarian cancer cells by antisense phosphorothioate oligonucleotides delivered by liposomes, showed a reduced invasive capacity of the tumor when compared to untreated cells, and a reduction in intraperitoneal spread of the cancer cells in nude mice treated with PLAU antisense phosphorothioate oligonucleotides was also observed [6].
In breast cancer patients, increased levels of PLAU were associated with a worse prognosis and an increase in aggressiveness, metastasis, and invasion [11,12,13,14,15]. These clinical findings have led to the investigation of PLAU as a biomarker for predicting breast cancer prognosis and responsiveness to hormonal agents such as endocrine therapy [16,17,18]. Upfront determination of PLAU levels could help to classify patients, also of multiple other cancer types, for their risk for metastasis and treatment relapse, and thus to assist clinicians in designing individualized treatment strategies [11,12,13,14,19].
Since selective inhibition of PLAU is not yet clinically available, other methods to inhibit PLAU have been investigated [20,21,22,23]. One promising phytochemical compound is Withaferin A (WA). WA is a natural compound with wide-ranging pharmacological activities including cardio-protective, anti-inflammatory, immuno-modulatory, anti-angiogenesis, anti-metastasis and anti-carcinogenic effects [24,25,26]. WA decreases expression of genes encoding ECM-degrading proteases (including PLAU), as well as the expression of other genes involved in cell adhesion, inflammation, and metastasis in vitro and in vivo [20,24,27,28,29]. Treatment of the hormone-insensitive, aggressive MDA-MB-231 triple negative breast cancer cell line with WA led to a decreased PLAU expression and reversed its highly invasive metastatic phenotype. In fact, upon WA treatment, MDA-MB-231 cells displayed characteristics similar to those of the non-invasive, hormone-sensitive, estrogen receptor α positive luminal A MCF-7 breast cancer cells, which are derived from a milder and more treatable breast cancer subtype [20]. Moreover, Szarc vel Szic et al. found that WA treatment of MDA-MB-231 cells promoted PLAU promoter DNA hypermethylation resulting in phenotypes similar to those of the less aggressive luminal B stage breast cancer type [28]. Indeed, WA changed expression levels of several epigenetic DNA/histone methylation enzymes, which subsequently leads to reduced H3K4 me2/me3 histone methylation levels of WA-responsive genes, including PLAU, potentially explaining the observed changes in gene expression and phenotype [30,31,32]. Pakneshan et al. described how the methylation status of the PLAU promoter can be used as a prognostic marker in patients with breast carcinoma [33]. Since PLAU demethylation correlates with a bad prognosis [34], targeted methylation of PLAU is proposed as a potential novel therapy [35]. Guo Y et al., showed that methylation of the PLAU promoter inhibited Ets-1 transcription factor binding, thus blocking its transcription. In the same study it was shown that DNA methylation is the dominant mechanism involved in silencing PLAU gene expression [34]. Chik et al., investigated the effect of 5-azacytidine in combination with a DNA-methylating agent, showing that 5- azacytidine on its own could induce demethylation of pro-metastatic genes as well [35]. Technologies to induce gene-specific methylation of the PLAU target gene could thus present a solution to this problem. These observations prompted us to use CRISPR-dCas9 (deactivated CRISPR-Cas9) based Epigenetic Editing strategies for gene specific control of PLAU expression.
CRISPR-dCas9 can be coupled to epigenetic enzymes (“Epigenetic Editing”) and targeted specifically to modulate the expression of genes of interest [36,37,38]. Importantly, numerous in vivo approaches have indicated therapeutic potency [37,38,39]. By guiding the correct combinations of different effector domains (EDs), i.e., DNA and histone methylation state editors to the gene of interest, stable modulation of gene expression can be achieved [40,41,42,43,44]. Recently, Nunez et al. demonstrated that dCas9 fused to DNMT3A/L and KRAB (CRISPRoff-v2.1) led to long-lasting gene silencing for a wide spectrum of genes [45]. In addition to silencing resistance genes and modulating immune responses, long-lasting repression of genes involved in cancer progression by Epigenetic Editing might provide a novel approach to cancer therapy. In this study our aim was to test the oncogenic activity of PLAU in breast cancer cells, and to obtain further insight into PLAU as a potential therapeutic target in oncology.
2. Materials and Methods
2.1. Cell Culture and Chemical Reagents
Human embryonic kidney cells HEK293T (ATCC: CRL-3216), human estrogen receptor positive breast cancer cells MCF-7 (ATCC: HTB-22) and triple negative human breast cancer cells MDA-MB-231 (ATCC: CRM-HTB-26) were all cultured in DMEM medium (Lonza BioWhittaker #BE12-604F) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 50 μg/mL gentamycin sulfate. Cells were cultured in a humidified incubator with 5% CO2 at 37 °C. All cells were routinely tested for mycoplasma contamination.
2.2. Plasmid Construction
2.2.1. Plasmids Encoding Transiently Expressed dCas9-Effector Domains
Plasmids pMLM3705 (dCas9-VP64) and MLM3636 (sgRNAs) were kind gifts from Keith Joung (Addgene plasmid #47754 and #43860, respectively), and CRISPRoff-v2.1 (OFF2.1) (DNMT3A/3L-dCas9-KRAB) from Jonathan S Weissman [45] (Addgene Plasmid #167981). We used dCas9-NED (no effector domain) (Addgene plasmid #109358) as negative control [46]. The catalytic domain of human histone methyltransferase PRDM9 was amplified from total cDNA of a testicular cancer cell line, and the catalytic domain of the histone methyltransferase DOT1L from human fibroblasts by Pfu DNA polymerase (Thermo Scientific, Leon-Rot, Germany) as described elsewhere [41]. These catalytic domains were inserted into dCas9-NED to create dCas9-PRDM9 and dCas9-DOT1L. The enzymatically inactive mutants were obtained by site-directed mutagenesis (Table S2).
2.2.2. Single Guide RNAs (sgRNAs)
Several sgRNAs targeting different regions of the PLAU gene (Figure 1) were cloned by inserting DNA oligonucleotides containing the 20-bp target region between the two BsmBI sites of MLM3636 [46]. As negative control, MLM3636, for transient transfection, or sgOUT(1–4), for lentiviral transduction, without inserted target sequence were used together with the effector domains (empty vector, EV). The sgOUT(1–4) plasmid was created to contain four sgRNA sequences (sgOUTall) in one plasmid. The MLM3636 plasmid was first digested with Acc65I to remove the original sgRNA expressing cassette. After self-re-circularization we had an empty vector which was digested with NheI and BamHI restriction enzymes. Additionally, the lenti sgRNA zeo backbone (Addgene#61427) was also digested with NheI and BamHI to cut out the coding sequence for sgRNA2.0. By ligating sgRNA2.0 with the digested empty vector plasmid a new plasmid (pMLM2.0) was obtained. To allow expression of multiple guides, an oligoduplex (AK497 and AK498) encoding restriction sites for StyI and Acc65I, respectively, (also restriction sites for BveI enzymes, which results in homologues ends after digestion with StyI and Acc65I) was inserted between the BamHI and Acc65I sites after the sgRNA2.0 sequence (pMLM2.0_T). Double digestion with StyI and Acc65I or with BveI results in compatible ends with NheI and Acc65I digested fragments. These restriction sites allowed us to sequentially clone different sgRNA expressing cassettes into one plasmid. The sgRNA tandem plasmid was created by inserting the 447-length NheI-Acc65I fragment encoding the sgRNA expressing cassette into pMLM2.0_T plasmid digested with StyI and Acc65I or BveI.
The plasmid lenti sgRNA (MS2) zeo backbone was a kind gift from Feng Zhang (Addgene plasmid # 61427). All four sgOUTall were cloned and inserted into the lentiviral sgRNA backbone in exactly the same way as for the transiently expressing MLM3636 sgRNA plasmid, by inserting DNA oligonucleotides containing the 20-bp target region between the two BsmBI sites of the lentiviral sgRNA plasmid (Table S2).
2.2.3. Lentiviral dCas9-Effector Domains
The lentiviral expression plasmid vector pHAGE EF1α dCas9-VP64 was a gift from Rene Maehr and Scot Wolfe (Addgene plasmid #50918). To create several stable cell lines, dCas9-EDs were made by replacing the VP64 gene with an oligonucleotide containing a MluI and an AsiSI site. By using AscI-PacI double digestion on previously obtained dCas9-EDs and transferring the EDs into pHAGE EF1α dCas9-NED, we created pHAGE EF1α dCas9-ED [46]. Briefly, pHAGE EF1α dCas9-M.SssI-Q147L and pHAGE EF1α dCas9-M.SssI-E186A were constructed by cutting out the M.SssI-Q147L and M.SssI-E186A genes with SgsI (AscI) and PacI restriction enzymes and cloning them in the pHAGE EF1α dCas9-NED vector between the AsiSI and MluI sites. The G9a catalytic domain and its mutant were digested out from pMX-ZF-IRES-GFP [47] with MluI and NotI and subcloned into the pHAGE EF1α dCas9- VP64, in which an additional multiple cloning site was added by replacing the coding sequence of the VP64 activator domain with a sequence containing a MluI restriction site. The Super KRAB (SKD) domain was subcloned by amplifying with primers containing MluI and NotI overhangs (Table S2).
2.3. Creation of Stable Cell Lines
Creation of MCF-7 or MDA-MB-231 cell lines stably expressing dCas9-ED was described elsewhere [46]. Briefly, lentiviral pHAGE-EF1α constructs encoding the dCas9-EDs were co-transfected with the second-generation packaging plasmids pCMVΔR8.91 and pCMV-VSV-G (#8454, Addgene, Watertown, MA, USA) on day one into HEK293T cells using PEI transfection reagents (#23966, Polysciences, Inc., Warrington, PA, USA) to produce lentiviral particles. The supernatant of HEK293T cells containing the virus was harvested 48 and 72 h after transfection. Host cells (MCF-7 or MDA-MB-231) were seeded in six-well plates and transduced on two consecutive days (day three and four) with 1.5 mL of the viral supernatant, supplemented with 8 µg/mL polybrene (Sigma-Aldrich, St. Louis, MO, USA). The transduced cells were selected on day seven in 8 µg/mL puromycin-supplemented medium for four days and subsequently cultured in 1 µg/mL puromycin-supplemented medium.
2.4. Lentiviral Transduction of Cells
For the lentiviral transductions of sgRNAs in stable MDA-MB-231 cells, the lentiviral sgRNA were co-transfected with the second-generation packaging plasmids pCMVΔR8.91 and pCMV-VSV-G on day one into HEK293T cells (seeded the previous day to reach a confluency of 70%) using PEI transfection reagents (#23966, Polysciences, Inc.) to produce lentiviral particles. The supernatant of HEK293T cells containing the virus was harvested at 48 and 72 h after transfection, supplemented with 8 µg/mL polybrene (Sigma-Aldrich) and frozen at −20 °C for a maximum of one month. MDA-MB-231 stably expressing dCas9-ED cell lines were seeded in normal medium (not supplemented with puromycine) on a 6-well plate at a concentration that would ensure 70% confluency on the next day. The viral supernatant that was harvested after 48 h, was thawed, supplemented again with 8 µg/mL polybrene and FBS and 1.5 mL of the pre-warmed and supplemented viral supernatant was applied to the stably expressing MDA-MB-231 cells at 24 h after seeding. After 12 h the medium containing the viral particles was refreshed with the viral supernatant that was harvested after 72 h, and prepared in the same way as before. The transduced cells were collected at 48 h after the first transduction (12 h after the second transduction) to assess their effect on gene expression.
2.5. Transient Transfection of Cells
Cells were seeded at a concentration that would ensure 70% confluency on the next day (day of transfection). MDA-MB-231 cells were plated in 24 well plate (1 × 105 cells/well), and transfected with 0.75 μg plasmid DNA (375 ng sgRNAs and 375 ng dCas9-ED) using 1.5 μL of Attractene Transfection Reagent (#301005, Qiagene, Venlo, The Netherlands). HEK293T and MCF-7 (5 × 105 cells/well), plated in 6-well plates, were transfected with 1 μg DNA (500 ng sgRNAs and 500 ng dCas9-ED) using SAINT (Synvolux Products & Therapeutics, Leiden, The Netherlands) in a 2:1 ratio for the wild-type cells or PEI in a 4:1 ratio for the stable cell lines. Twenty-four hours after transfection the medium was changed to normal growth medium, and 48 h after transfection cells were harvested to assess the short-term effect on gene expression. All transient transfection experiments were performed in triplicate.
To assess long-term effects on gene expression and proliferation capacity, cells were cultured for 5 or 12 days after transfection, and split when 80–90% confluent. Transient expression of sgRNAs allowed assessment of long-term effects on gene expression because the plasmids faded out with each cell division, even if the dCas9-ED was stably expressed (as in the case of the stable cell lines).
2.6. Quantitative Real-Time PCR
Total RNA was isolated using TRIzol reagent (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Subsequently, cDNA was generated using the Revertaid cDNA synthesis kit with random hexamer primers (Thermo Scientific) or with M-MLV Reverse Transcriptase (Promega, Madison, WI, USA) and oligo (dT) primers. To assess gene expression, qRT-PCR was performed with 10 ng of cDNA input, using ABsolute qPCR SYBR Green (Thermo Scientific) or GoTas Green master Mix (Promega). PLAU expression was normalized to GAPDH, a house keeping gene, and all reactions were performed in triplicate using an ABI ViiA7 real-time PCR system (Applied Biosystems, Waltham, MA, USA) for 45 cycles or Rotor-Gene Q (Qiagen). Primer sequences are provided in Table S1. Ct values were obtained, and quantitative analysis was performed using the cycle threshold (ΔΔCt) method after normalization to GAPDH expression. Fold change was calculated relative to control samples.
2.7. Migration Assay
The migration capacity of cells was evaluated using the xCELLigence real-time cell analysis (RTCA) system (Roche, Penzberg, Germany) as previously described [20]. Briefly, 160 µL and 30 µL of media was added to the lower and upper chambers of modified 16-well plates (CIM-16, Roche), respectively. The lower chambers either contained FBS supplemented or FBS free medium to assess chemotactic and background migration. CIM-16 plates were subsequently placed in the RTCA DP instrument at 37 °C for 1 h to measure background signal. Serum-deprived stable MCF-7 cells were harvested using TrypLE ExpressTM (Invitrogen, Waltham, MA, USA), resuspended in serum-free medium, and seeded into the upper chambers of the CIM-16 plates at a density of 3 × 104 cells/well. After adding the cells, CIM-16 plates were incubated in the laminar flow hood for 30 min at room temperature allowing cells to settle before placing them in the RTCA DP instrument at 37 °C. Chemotactic migration values were obtained by subtracting background migration signals. At each condition measurements were performed in triplicate and analyzed for 12 h.
2.8. Proliferation Assay
Effects of modulation of PLAU expression on cells proliferation were evaluated by Trypan Blue Cell Analysis [48]. After 48 h of transient transfection, the MDA-MB-231 cells were harvested, counted, and re-plated in 1:4 ratio in a 24 well plate. Cell count determinations were repeated every 3–4 days until 144 h after transfection.
2.9. TCGA Analysis
The PLAU gene expression analysis was performed using R (version 4.2.1) and R studio (version 2022.07.2). RNA-Seq samples, from The Cancer Genome Atlas (TCGA), corresponding to TCGA-BRCA [49] dataset was obtained using R package TCGAbiolinks [50] (version 2.24.3). From the dataset, only samples that were categorized as “primary tumor” or “solid tissue normal” were used for analysis. In total, 1219 samples, comprising of 1106 “primary tumor” and 113 “solid tissue normal” samples were subjected to the analysis. The RNA-seq counts and the patient data were stored as DGEList() object of the edgeR package [51](version 3.38.4) to enable the use of different packages for the gene expression quantification. Subsequently, the RNA-seq counts were filtered and normalized using filterByExpr() and calcNormFactors() functions of the edgeR package (version 3.38.4). The filtered and normalized counts were further processed using voom() function of limma package [52] (version 3.52.4) to assess the expression of PLAU gene in primary tumor and solid tissue normal samples. The expression level of PLAU in the two sample categories were extracted as table, to perform statistical analysis and plot graphs using GraphPad prism (version 8.4.2). The higher expression of PLAU in primary tumor samples was statistically significant as determined by the Mann–Whitney test in GraphPad Prism (version 8.4.2).
2.10. Statistics
Statistical tests were performed using Graphpad Prism 7 software. Comparison between target conditions and controls were investigated with an unpaired two-tailed t-test. Differences were considered statistically significant if the p-value was <0.05. All data are presented as the mean ± S.D. of three independent, biological replicates, unless stated differently.
3. Results
3.1. Screening of sgRNAs for PLAU
The Cancer Genome Atlas (TCGA) data demonstrated a higher PLAU gene expression for 1095 breast cancer samples compared to 113 healthy breast tissue (Figure S1). To test the oncogenic activity of PLAU in breast cancer cells, twelve sgRNAs targeting the PLAU promoter were designed around the two alternative transcription start sites (TSSs) (see schematic representation in Figure 1), which are highly controlled by DNA methylation and histone methylation [28]. The sgOUTall (OUT1, 2, 3, 4) group is located outside of the CGI and upstream of the two alternative TSSs; the sgTSSall (TSS1, 2, 3, 4) group is located around TSS1; the sgINall (IN1, 2, 3, 4) group is located downstream of the TSSs and completely inside the CGI. Two additional mixtures were tested: sgMIX1 (TSS1, TSS3, IN2, IN4) and sgMIX2 (TSS2, TSS4, IN1, IN3) to more widely cover the promoter region.
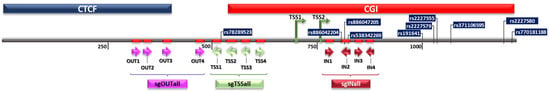
Figure 1.
Position of sgRNAs designed to modulate PLAU gene expression.
These different sgRNAs were compared for their efficacy to modulate PLAU gene expression in transient transfection experiments in either MDA-MB-231 cells characterized by high PLAU levels, and in HEK293T and MCF-7 cell lines, which express PLAU at low levels (Figure 2). MDA-MB-231 cells were transfected with dCas9-SKD and CRISPRoff-v2.1 plasmids (Figure S2), whereas HEK293T and MCF-7 cells were transfected using dCas9-VP64 (Figure 2).
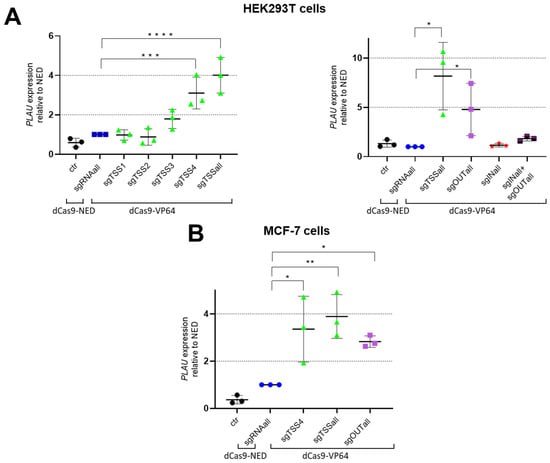
Figure 2.
sgRNA-induced upregulation of PLAU expression in (A) HEK293T, and in (B) MCF-7 cells. (A) qRT-PCR results of HEK293T cells tested for VP64-induced upregulation using sgTSS4, individually and as mix, sgOUTall and/or sgINall; (B) qRT-PCR results of MCF-7 cells tested for VP64-induced upregulation using sgTSS4, sgTSSall and sgOUTall. Data are represented as means of 3 independent experiments, relative to GAPDH and normalised to transfected to cells with dCas9-NED. Significance is presented as compared to the dCas9-NED condition; * p < 0.05, ** p < 0.01, *** p < 0.005, **** p < 0.001.
In MDA-MB-231 cells, sgTSS4, sgTSSall, sgMIX1 and sgMIX2 were tested two and five days after transfection. Only the sgTSSall group combined with dCas9-SKD or CRISPRoff-v2.1 resulted in reduced PLAU expression compared to dCas9-NED five days after transfection (Figure S2).
In HEK293T cells, in which the PLAU promoter is unmethylated, expression of dCas9-VP64 together with sgTSS4, or with the combination of four sgRNAs (sgTSSall) resulted in a 3.1-fold (p = 0.034) and 4.0-fold (p = 0.012) induction of the PLAU gene, respectively, relative to cells treated to express dCas9-NED (Figure 2A, left graph). A separate set of experiments confirmed the dCas9-VP64 induction with sgTSSall, leading to a 8.2-fold induction relative to dCas9-NED transfected cells (p = 0.021) (Figure 2A, right graph). In this set of experiments, the sgOUTall group resulted in a 4.8-fold induction of the PLAU gene relative to dCas9-NED (p = 0.022). No significant induction was obtained for the sgINall group or for a combination of sgINall and sgOUTall group.
In MCF-7 cells, where the PLAU promoter is hypermethylated (https://www.encodeproject.org/experiments/ENCSR000CPT/), sgTSS4 (the most effective guide from the sgTSSall group based on HEK293T results), sgTSSall, and sgOUTall, were tested for gene induction using dCas9-VP64 (Figure 2B). Both groups as well as the individual sgTSS4 resulted in a significant induction of the PLAU gene (3.9-fold for sgTSSall, 2.8-fold for sgOUTall and 3.4-fold for sgTSS4), relative to cells treated to express dCas9-NED. CRISPR-dCas9 off-targets effects were evaluated in MCF-7 upon transfections with plasmids transferring sgRNAs targeting a different gene (KDM4A) using dCas-VP64. No off-target regulation of PLAU was observed for dCas-VP64 in these cells (Figure S3).
3.2. Downregulation of the PLAU Gene
Based on the obtained sgRNA screening results described above, we further investigated the functional effects of PLAU repression in MDA-MB-231. Co-transfecting sgPLAUtss1 with dCas9-SKD or with CRISPRoff-v2.1, did not result in significant difference after two days compared to the negative control (dCas9-NED) (Figure 3A left). Additionally, five days after transfection, sgTSSall-SKD and sgTSSall—OFF2.1 weakly reduced PLAU expression (means of 0.82 and 0.86-fold expression left, respectively, compared to cells expressing sgTSSall and dCas9-NED (Figure 3A, right panel)). Despite the variable, non-statistically significant reduction at mRNA level, we observed a clear decrease in cell proliferation for all CRISPRoff-v2.1 experiments at 144 h (Figure 3B). A complete block in cell growth was observed until 96 h for both sgTSSall-SKD and sgTSSall-OFFv2.1 expressing cells, but only CRISPRoff-v2.1, that contains both KRAB and DNMT3A/3L domains, was able to maintain reduced cell proliferation. These data indicate that mild PLAU downregulation can reduce the aggressiveness of the triple negative breast cancer cell.
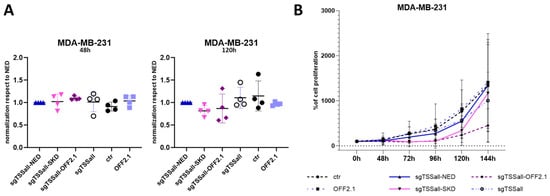
Figure 3.
PLAU down-regulation and its functional effect in MDA-MB-231 cells. (A) qRT-PCR results of PLAU mRNA expression in MDA-MB-231 cells transfected to transiently express dCas9-SKD or CRISPRoff-v2.1 (OFF2.1) and sgTSSall at 2 (48 h, left panel) and 5 days (120 h) after transfection (right panel), compared to dCas9-NED. As negative controls, cells were transfected to express sgRNA only (sgTSSall) or CRISPRoff-v2.1 (OFF2.1) only. (B) Proliferation assay results of the cells transfected with plasmids shown in (A). Data are represented as means of 4 independent experiments.
In parallel, we adopted another approach to increase efficiency of PLAU gene repression. Stable MDA-MB-231 cells, engineered to express dCas9-NED, dCas9-G9A or its mutant, or -M.SssI-Q147L, a M.SssI variant with reduced DNA binding, or its catalytic inactive E186A mutant [53], were transduced a second time with lentiviral constructs containing either the four sgOUT(1–4), or a sgRNA without DNA recognizing insert (empty vector (EV)) as a control.
Even though a trend towards PLAU downregulation was achieved in MDA-MB-231 cells stably expressing -M.SssI-Q147L and G9A wild tipe and mutant for sgOUTall, no significant repression of PLAU could be observed (Figure S4).
3.3. Epigenetic Editing to Induce PLAU Upregulation
Next to growth inhibition upon repression, we set out to demonstrate the oncogenic activity of PLAU in cells in which PLAU is expressed at low levels by inducing PLAU expression using dCas9-VP64. Moreover, to investigate whether Epigenetic Editing could lead to long-term PLAU induction, the epigenetic enzymes PRDM9 (writing H3K4me3) and DOT1L (writing H3K79me), fused to CRISPR-dCas9, were tested using sgTSS4 and sgOUTall, in these cells (HEK293T and MCF-7 cells; Figure S5).
In all sets of experiments, PLAU induction using dCas9-VP64 was confirmed to be in the same range as shown in Figure 2 after 48 h (Figure S5A,B). Since VP64 is an artificial transcription factor without any enzymatic activity on its own, a transient effect was expected. Indeed, after 10 days of transfection the induction disappeared and PLAU expression became similar to control conditions (ctr and dCas9-NED).
Relative to cells treated to express dCas9-NED and the combination of guides, neither one of the epigenetic enzymes, nor a combination of both, was able to accomplish a significant induction of the PLAU gene, not in the short-term (48 h after transfection) nor in the long-term (10 days after transfection) experiments (Figure S5).
3.4. Functional Effects of PLAU Induction
As PLAU is known to play an important role in breast cancer cell migration, it was investigated whether inducing PLAU expression alone was indeed sufficient to provoke functional changes in MCF-7 cells. To this end we analyzed the migratory capacities of the MCF-7 cells stably expressing dCas9-VP64 (MCF-7-VP64) after transient transfection with the sgOUT(1–4) plasmid, which expresses all four individual sgOUTall from a single plasmid. As negative controls, the MCF-7-NED stable cell line was transfected with the sgOUT(1–4) plasmid or empty guide vector (EV), which was also used to transfect the MCF-7-VP64 stable cells. After 48 h a 5.3-fold induction of PLAU expression was observed in MCF-7-VP64 cells transfected with sgOUT(1–4) compared to MCF-7-NED cells transfected with EV (p = 0.052) (Figure 4A). Twelve hours later, MCF-7-VP64 stable cells transfected with sgPLAU tandem displayed a 1.9-fold (p < 0.001) increase in migration compared to the MCF-7-NED stable cell line transfected with EV (Figure 4B) as measured by real-time analysis of transwell migration. These data indicate that our CRISPR-dCas9 system was able to influence the migratory capability of MCF-7 cells through upregulation of PLAU expression.
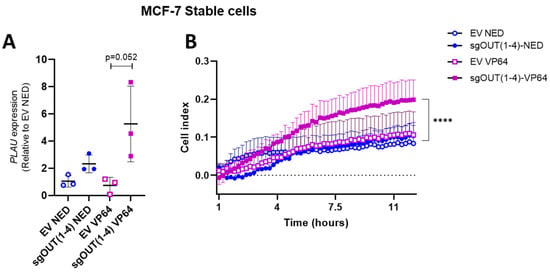
Figure 4.
PLAU upregulation and its functional effect in stable MCF-7 cells. (A) qRT-PCR results of PLAU mRNA expression in stable MCF-7 cells using the sgOUT(1–4) or EV as negative control (B) Migration assay results of the cells transfected in (A). Data are represented as means of 3 independent experiments, relative to GAPDH and normalized to MCF-7-NED + EV; **** p < 0.001.
4. Discussion
Overexpression of PLAU has been identified as a promising therapeutic target for inhibition in (co)treatment of triple negative breast cancer in multiple studies [1,8,9,10], including ours [20,28]. Based on epigenetic studies, a role for DNA methylation has been suggested in inducing inhibition of PLAU expression [33,34,35]. Using CRISPR-dCas9 targeting tools, we aimed to reprogram PLAU gene expression in MDA-MB-231 and MCF-7 cells to validate its effects on the invasive proliferation and migratory properties. The versatility of the CRISPR-dCas9 platform to upregulate PLAU expression in cells expressing low levels, while repressing expression in PLAU overexpressing cells, strengthened our conclusion on the oncogenic function of PLAU and its involvement in breast tumor aggressiveness.
First, we showed that reduction in PLAU expression could be achieved in invasive hormone resistant MDA-MB-231 cells by CRISPRoff-v2.1, which decreased cancer cell proliferation. Conversely, an increase in PLAU expression was obtained in non-invasive, hormone-sensitive, ΕRα positive luminal A MCF-7 breast cancer cells by dCas9-VP64-sg TSSall leading to a higher migratory capacity of the cells.
Whilst WA treatment has been shown to provide promising therapeutic effects associated with a downregulation of PLAU [20], treatment with this phytopharmaceutical compound modulates the expression of multiple genes [24,27,28]. Gene targeted approaches such as CRISPR-dCas9-directed Epigenetic Editing thus assist in unravelling the contribution of individual WA-responsive genes in suppressing aggressive and invasive breast cancer phenotypes. For example, studies have shown effects of WA on FOXO3a and BIM mediated apoptosis, inhibition and induction of ROS [54,55,56,57]. An extensive microarray transcriptome profiling of MCF-7 and MDA-MB-231 cells identified multiple common, as well as cell line-specific target genes of WA, that were present in multiple pathways [20]. By targeting these genes, individually and in combination with CRISPR-dCas9-EDs, their specific involvement can be further investigated. This can ultimately assist in identification of potent therapeutic targets and contribute to more personalized treatment options. CRISPR-dCas9 targeting tools are well suited to target multiple genes at the same time, which is beneficial in conditions such as cancer where multiple genes are involved.
Although dCas9-VP64 is useful to up-regulate gene expression, it generally induces transient effects. Targeting of epigenetic writers or erasers to genes aims to achieve sustained gene expression effects. Moreover, such epigenetic editing tools can target multiple EDs to the same gene, which can result in additive, and potentially long-lasting effects. One example by Cano-Rodriguez et al. shows the additive effects of a combination of enzymes (PRDM9 (writing H3K4me3) and DOT1L (writing H3K79me)) in effectively re-expressing silenced genes. These effects were shown to be sustained when the promoter was hypomethylated [41]. Here, we used the same combination of enzymes to evaluate whether Epigenetic Editing could lead to long-term PLAU induction. However, no increase in PLAU expression was achieved when targeting PRDM9 or DOT1. The lack of gene induction observed when using sgTSS4 or sgOUTall in combination with PRDM9 and/or DOT1L as well as the limited repressive effects in MDA-MB-231 cells could be due to various factors, which become more pronounced for epigenetic effectors which likely have weaker transcriptional effects than the strong VP64 activator. Some of these factors are related to the intrinsic characteristics of the PLAU promoter (Figure 1). Data retrieved from Ensembl and USCS databases show that a CCCTC-binding factor (CTCF) binding site is present upstream of TSS1 which could affect binding of sgOUT1–4 from the sgOUTall group. These sites are well known transcription factor binding sites that can block the communication between enhancers and promoters [58], which could affect the efficiency of binding of sgRNAs. Alternatively, in Figure 1A phenotype-associated SNPs (Rs numbers) are depicted that are present in the PLAU promoter region. These are all regularly occurring SNPs, and it could be that specific SNPs in regions of sgRNA binding cause loss of binding affinity. Moreover, there are five possible splice-variants of PLAU, of which three are protein coding. The qRT-PCR primers were directed to the most validated and characterized isoform, which is also highly expressed in MDA-MB-231 cells. However, we did not investigate the impact of our epigenetic constructs on different transcript isoforms, which might be differentially affected, explaining the more pronounced functional effect in CRISPR-offv2.1 induced growth inhibition.
Interestingly, although targeting the recently described three-domain prolonged repressive dCas9 version (CRISPRoff-v2.1) did not improve the PLAU repression at the level of gene activity compared to targeting the KRAB domain alone on day 3, a functional improvement was obtained in cell growth inhibition measured after 6 days. In transiently transfected cells, PLAU expression reduction only occurred in the cells which express CRISPRoff-v2.1. It is conceivable that the failure of maintaining the reduced cell proliferation can be attributed to the cell fraction, in which the silencing of PLAU was not complete. Partial silencing is such that it does not induce cell death, but only a decrease in proliferation [59]. Another important factor that impacts the efficiency of Epigenetic Editing is delivery of large CRISPR-dCas9 constructs to the cells [60]. To efficiently deliver the CRISPR-dCas9 system to the cells, researchers make use of a variety of approaches. One option is to use lentiviruses, which integrate into the genome of the host cells, and in this way continuously express the desired constructs [61]. This approach can be useful to investigate mechanistic effects in vitro, however the phenomenon of integration, which can cause severe side effects such as cancer, limits its clinical use [62,63]. Another successful technique to deliver the Epigenetic Editing constructs into the cells are non-viral ligand-directed targeting approaches that target cancer cell specific surface receptors. These approaches, which might have therapeutic potential, make use of liposomes and other nanoparticles, that are more efficiently taken up by the cells, and are protected from degradation by the biological environment [62,64]. Further, as this method holds the possibility to selectively target disease tissues [65], it is more suitable to use in the clinic as side-effects caused by off-target effects are lowered and the uptake by the patient is improved compared to, e.g., viral-based methods [66]. However, a possible disadvantage is the shorter in vivo life-time some of these extracellular vesicles can have compared to adeno-associated virus (AAV) -mediated delivery [67,68]. This latter delivery method is the preferred option in clinical trials with gene editing therapies at the moment [60]. AAV is a non-pathogenic virus with a very mild immune response capable of delivering constructs in a preferred tissue, with a high efficiency and without integrating into the host genome [69,70]. Epigenetic Editing studies have successfully delivered CRISPR-dCas9-EDs in vivo using AAV delivery, although the limiting packaging capacity of these AAV vectors (~4.5 kb) and the large sizes of Epigenetic Editing constructs require optimization [71]. Extracellular vesicles (EVs) or ribonucleoprotein (RNP), based on the direct delivery of sgRNA and dCas protein, have been shown to be an interesting approach for therapeutic Epigenetic Editing [72].
Despite the many developments with regard to using CRISPR-dCas9 in gene expression modulation, the success of future clinical applications relies largely on safety and efficiency of delivery. The recent founding of various companies to translate Epigenetic Editing to the clinic will provide important insights on issues to be overcome to progress with Epigenetic Editing for cancer and beyond [72].
Indeed, our PLAU-modulation platform using dCas9-VP64 and CRISPRoff-v2.1 could be exploited further for use in a broad variety of pathological conditions that would benefit from PLAU modulation such as ischemic brain injury [73,74,75], lung fibrosis [76], male infertility [77], type 2 diabetes mellitus [78] and diabetic keratopathy [79], besides breast cancer [3,22]. The use of Epigenetic Editing tools, together with the optimization of in-patient delivery techniques, will open novel avenues to manipulate specific genes such as PLAU to treat aggressive breast cancer, and many other diseases.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/biomedicines11010102/s1, Figure S1: Analysis of TCGA data assessing PLAU mRNA gene expression; Figure S2: Screening of sgRNAs to induce repression of PLAU expression in MDA-MB-231 cells; Figure S3: PLAU gene expression in MCF-7 cells transfected to express sgRNA targeting an irrelevant gene (KDM4A); Figure S4: Assessment of PLAU expression in MDA-MB-231 stable cell lines expressing repressing dCas9-EDs; Figure S5: Epigenetic Editing of PLAU in HEK293T cells and MCF-7 cells; Table S1: Primer sequences as used in the quantitative real-time PCR; Table S2: Plasmids used for transient and lentiviral delivery.
Author Contributions
Conceptualization, W.V.B. and M.G.R.; methodology, A.E.N., C.D., M.K., A.K. and E.L.; validation, F.S. and D.G.; formal analysis, F.S., D.G. and E.L.; investigation, F.S., D.G., E.L. and M.G.S.R.; resources, M.K. and A.K.; writing—original draft preparation, D.G., E.L. and M.G.R.; writing—review and editing, F.S., A.K., P.J.V., E.L. and L.A.; supervision, W.V.B., P.J.V. and M.G.R.; project administration, A.E.N.; funding acquisition, P.J.V. and M.G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by European Union H2020-MSCA-ITN-2014-ETN 642691 EpiPredict; Foundation Against Cancer (Belgium), grant number 7872; Research Foundation Flanders (Belgium), grant number FWO G1179120N, G059713N.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to acknowledge COST Action EpiChemBio CM1406 for networking activities and short-term scientific missions. We thank Jim Jacob for the TCGA data analysis. The results shown in Figure S1 are in whole based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga (accessed on 5 December 2022).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
| Bp | base pair |
| CGI | CpG island |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| DMEM | Dulbecco’s modified Eagle medium |
| DNA | deoxyribonucleic acid |
| ECM | extracellular matrix |
| ED | effector domain |
| FBS | fetal bovine serum |
| HEK293T | human embryonic kidney cells |
| MCS | multicloning site |
| NED | no effector domain |
| PEI | polyethylenimine |
| PLAU | plasminogen activator, urokinase |
| sgRNA | single guide ribonucleic acid |
| SKD | super KRAB domain |
| u-PA | plasminogen activator |
| WA | withaferin A |
References
- McMahon, B.J.; Kwaan, H.C. Components of the Plasminogen-Plasmin System as Biologic Markers for Cancer. Adv. Exp. Med. Biol. 2015, 867, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Mazar, A.P.; Ahn, R.W.; O’Halloran, T.V. Development of novel therapeutics targeting the urokinase plasminogen activator receptor (uPAR) and their translation toward the clinic. Curr. Pharm. Des. 2011, 17, 1970–1978. [Google Scholar] [CrossRef]
- Mahmood, N.; Mihalcioiu, C.; Rabbani, S.A. Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front. Oncol. 2018, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Moquet-Torcy, G.; Tolza, C.; Piechaczyk, M.; Jariel-Encontre, I. Transcriptional complexity and roles of Fra-1/AP-1 at the uPA/Plau locus in aggressive breast cancer. Nucleic Acids Res. 2014, 42, 11011–11024. [Google Scholar] [CrossRef] [PubMed]
- Santibanez, J.F. Transforming growth factor-Beta and urokinase-type plasminogen activator: Dangerous partners in tumorigenesis-implications in skin cancer. ISRN Dermatol. 2013, 2013, 597927. [Google Scholar] [CrossRef]
- Duffy, M.J.; Duggan, C.; Mulcahy, H.E.; McDermott, E.W.; O’Higgins, N.J. Urokinase plasminogen activator: A prognostic marker in breast cancer including patients with axillary node-negative disease. Clin. Chem. 1998, 44, 1177–1183. [Google Scholar] [CrossRef]
- Duffy, M.J.; O’Grady, P.; Devaney, D.; O’Siorain, L.; Fennelly, J.J.; Lijnen, H.J. Urokinase-plasminogen activator, a marker for aggressive breast carcinomas. Preliminary report. Cancer 1988, 62, 531–533. [Google Scholar] [CrossRef]
- Ossowski, L.; Russo-Payne, H.; Wilson, E.L. Inhibition of urokinase-type plasminogen activator by antibodies: The effect on dissemination of a human tumor in the nude mouse. Cancer Res. 1991, 51, 274–281. [Google Scholar]
- Kobayashi, H.; Gotoh, J.; Shinohara, H.; Moniwa, N.; Terao, T. Inhibition of the metastasis of Lewis lung carcinoma by antibody against urokinase-type plasminogen activator in the experimental and spontaneous metastasis model. Thromb. Haemost. 1994, 71, 474–480. [Google Scholar] [CrossRef]
- Wilhelm, O.; Schmitt, M.; Hohl, S.; Senekowitsch, R.; Graeff, H. Antisense inhibition of urokinase reduces spread of human ovarian cancer in mice. Clin. Exp. Metastasis 1995, 13, 296–302. [Google Scholar] [CrossRef]
- Gouri, A.; Dekaken, A.; El Bairi, K.; Aissaoui, A.; Laabed, N.; Chefrour, M.; Ciccolini, J.; Milano, G.; Benharkat, S. Plasminogen Activator System and Breast Cancer: Potential Role in Therapy Decision Making and Precision Medicine. Biomark. Insights 2016, 11, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Look, M.P.; van Putten, W.L.; Duffy, M.J.; Harbeck, N.; Christensen, I.J.; Thomssen, C.; Kates, R.; Spyratos, F.; Ferno, M.; Eppenberger-Castori, S.; et al. Pooled analysis of prognostic impact of urokinase-type plasminogen activator and its inhibitor PAI-1 in 8377 breast cancer patients. J. Natl. Cancer Inst. 2002, 94, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Janicke, F.; Prechtl, A.; Thomssen, C.; Harbeck, N.; Meisner, C.; Untch, M.; Sweep, C.G.; Selbmann, H.K.; Graeff, H.; Schmitt, M.; et al. Randomized adjuvant chemotherapy trial in high-risk, lymph node-negative breast cancer patients identified by urokinase-type plasminogen activator and plasminogen activator inhibitor type 1. J. Natl. Cancer Inst. 2001, 93, 913–920. [Google Scholar] [CrossRef][Green Version]
- Harbeck, N.; Schmitt, M.; Meisner, C.; Friedel, C.; Untch, M.; Schmidt, M.; Sweep, C.G.; Lisboa, B.W.; Lux, M.P.; Beck, T.; et al. Ten-year analysis of the prospective multicentre Chemo-N0 trial validates American Society of Clinical Oncology (ASCO)-recommended biomarkers uPA and PAI-1 for therapy decision making in node-negative breast cancer patients. Eur. J. Cancer 2013, 49, 1825–1835. [Google Scholar] [CrossRef]
- Duffy, M.J.; McGowan, P.M.; Harbeck, N.; Thomssen, C.; Schmitt, M. uPA and PAI-1 as biomarkers in breast cancer: Validated for clinical use in level-of-evidence-1 studies. Breast Cancer Res. 2014, 16, 428. [Google Scholar] [CrossRef] [PubMed]
- Eastman, B.M.; Jo, M.; Webb, D.L.; Takimoto, S.; Gonias, S.L. A transformation in the mechanism by which the urokinase receptor signals provides a selection advantage for estrogen receptor-expressing breast cancer cells in the absence of estrogen. Cell. Signal. 2012, 24, 1847–1855. [Google Scholar] [CrossRef]
- Foekens, J.A.; Look, M.P.; Peters, H.A.; van Putten, W.L.; Portengen, H.; Klijn, J.G. Urokinase-type plasminogen activator and its inhibitor PAI-1: Predictors of poor response to tamoxifen therapy in recurrent breast cancer. J. Natl. Cancer Inst. 1995, 87, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Meijer-van Gelder, M.E.; Look, M.P.; Peters, H.A.; Schmitt, M.; Brunner, N.; Harbeck, N.; Klijn, J.G.; Foekens, J.A. Urokinase-type plasminogen activator system in breast cancer: Association with tamoxifen therapy in recurrent disease. Cancer Res. 2004, 64, 4563–4568. [Google Scholar] [CrossRef]
- Banys-Paluchowski, M.; Witzel, I.; Aktas, B.; Fasching, P.A.; Hartkopf, A.; Janni, W.; Kasimir-Bauer, S.; Pantel, K.; Schon, G.; Rack, B.; et al. The prognostic relevance of urokinase-type plasminogen activator (uPA) in the blood of patients with metastatic breast cancer. Sci. Rep. 2019, 9, 2318. [Google Scholar] [CrossRef]
- Szarc vel Szic, K.; Op de Beeck, K.; Ratman, D.; Wouters, A.; Beck, I.M.; Declerck, K.; Heyninck, K.; Fransen, E.; Bracke, M.; De Bosscher, K.; et al. Pharmacological levels of Withaferin A (Withania somnifera) trigger clinically relevant anticancer effects specific to triple negative breast cancer cells. PLoS ONE 2014, 9, e87850. [Google Scholar] [CrossRef]
- Jiang, J.; Jedinak, A.; Sliva, D. Ganodermanontriol (GDNT) exerts its effect on growth and invasiveness of breast cancer cells through the down-regulation of CDC20 and uPA. Biochem. Biophys. Res. Commun. 2011, 415, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Han, K.Y.; Chen, P.N.; Hong, M.C.; Hseu, Y.C.; Chen, K.M.; Hsu, L.S.; Chen, W.J. Naringenin Attenuated Prostate Cancer Invasion via Reversal of Epithelial-to-Mesenchymal Transition and Inhibited uPA Activity. Anticancer Res. 2018, 38, 6753–6758. [Google Scholar] [CrossRef] [PubMed]
- Hseu, Y.C.; Lin, Y.C.; Rajendran, P.; Thigarajan, V.; Mathew, D.C.; Lin, K.Y.; Way, T.D.; Liao, J.W.; Yang, H.L. Antrodia salmonea suppresses invasion and metastasis in triple-negative breast cancer cells by reversing EMT through the NF-kappaB and Wnt/beta-catenin signaling pathway. Food Chem. Toxicol. 2019, 124, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Hassannia, B.; Logie, E.; Vandenabeele, P.; Vanden Berghe, T.; Vanden Berghe, W. Withaferin A: From ayurvedic folk medicine to preclinical anti-cancer drug. Biochem. Pharmacol. 2020, 173, 113602. [Google Scholar] [CrossRef]
- Kupchan, S.M.; Anderson, W.K.; Bollinger, P.; Doskotch, R.W.; Smith, R.M.; Renauld, J.A.; Schnoes, H.K.; Burlingame, A.L.; Smith, D.H. Tumor inhibitors. XXXIX. Active principles of Acnistus arborescens. Isolation and structural and spectral studies of withaferin A and withacnistin. J. Org. Chem. 1969, 34, 3858–3866. [Google Scholar] [CrossRef]
- Mohan, R.; Hammers, H.J.; Bargagna-Mohan, P.; Zhan, X.H.; Herbstritt, C.J.; Ruiz, A.; Zhang, L.; Hanson, A.D.; Conner, B.P.; Rougas, J.; et al. Withaferin A is a potent inhibitor of angiogenesis. Angiogenesis 2004, 7, 115–122. [Google Scholar] [CrossRef]
- Vanden Berghe, W.; Sabbe, L.; Kaileh, M.; Haegeman, G.; Heyninck, K. Molecular insight in the multifunctional activities of Withaferin A. Biochem. Pharmacol. 2012, 84, 1282–1291. [Google Scholar] [CrossRef]
- Szarc Vel Szic, K.; Declerck, K.; Crans, R.A.J.; Diddens, J.; Scherf, D.B.; Gerhauser, C.; Vanden Berghe, W. Epigenetic silencing of triple negative breast cancer hallmarks by Withaferin A. Oncotarget 2017, 8, 40434–40453. [Google Scholar] [CrossRef]
- Hassannia, B.; Wiernicki, B.; Ingold, I.; Qu, F.; Van Herck, S.; Tyurina, Y.Y.; Bayir, H.; Abhari, B.A.; Angeli, J.P.F.; Choi, S.M.; et al. Nano-targeted induction of dual ferroptotic mechanisms eradicates high-risk neuroblastoma. J. Clin. Investig. 2018, 128, 3341–3355. [Google Scholar] [CrossRef]
- Royston, K.J.; Paul, B.; Nozell, S.; Rajbhandari, R.; Tollefsbol, T.O. Withaferin A and sulforaphane regulate breast cancer cell cycle progression through epigenetic mechanisms. Exp. Cell Res. 2018, 368, 67–74. [Google Scholar] [CrossRef]
- Royston, K.J.; Udayakumar, N.; Lewis, K.; Tollefsbol, T.O. A Novel Combination of Withaferin A and Sulforaphane Inhibits Epigenetic Machinery, Cellular Viability and Induces Apoptosis of Breast Cancer Cells. Int. J. Mol. Sci. 2017, 18, 1092. [Google Scholar] [CrossRef] [PubMed]
- Mirza, S.; Sharma, G.; Parshad, R.; Gupta, S.D.; Pandya, P.; Ralhan, R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J. Breast Cancer 2013, 16, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Pakneshan, P.; Tetu, B.; Rabbani, S.A. Demethylation of urokinase promoter as a prognostic marker in patients with breast carcinoma. Clin. Cancer Res. 2004, 10, 3035–3041. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Pakneshan, P.; Gladu, J.; Slack, A.; Szyf, M.; Rabbani, S.A. Regulation of DNA methylation in human breast cancer. Effect on the urokinase-type plasminogen activator gene production and tumor invasion. J. Biol. Chem. 2002, 277, 41571–41579. [Google Scholar] [CrossRef] [PubMed]
- Chik, F.; Machnes, Z.; Szyf, M. Synergistic anti-breast cancer effect of a combined treatment with the methyl donor S-adenosyl methionine and the DNA methylation inhibitor 5-aza-2’-deoxycytidine. Carcinogenesis 2014, 35, 138–144. [Google Scholar] [CrossRef] [PubMed]
- De Groote, M.L.; Verschure, P.J.; Rots, M.G. Epigenetic Editing: Targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012, 40, 10596–10613. [Google Scholar] [CrossRef]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef]
- Cortés-Mancera, F.M.; Sarno, F.; Goubert, D.; Rots, M.G. Gene-Targeted DNA Methylation: Towards Long-Lasting Reprogramming of Gene Expression? In DNA Methyltransferases—Role and Function; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Gomez, J.A.; Beitnere, U.; Segal, D.J. Live-Animal Epigenome Editing: Convergence of Novel Techniques. Trends Genet. 2019, 35, 527–541. [Google Scholar] [CrossRef]
- Amabile, A.; Migliara, A.; Capasso, P.; Biffi, M.; Cittaro, D.; Naldini, L.; Lombardo, A. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell 2016, 167, 219–232.e14. [Google Scholar] [CrossRef]
- Cano-Rodriguez, D.; Gjaltema, R.A.; Jilderda, L.J.; Jellema, P.; Dokter-Fokkens, J.; Ruiters, M.H.; Rots, M.G. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun. 2016, 7, 12284. [Google Scholar] [CrossRef]
- Song, J.; Cano-Rodriquez, D.; Winkle, M.; Gjaltema, R.A.; Goubert, D.; Jurkowski, T.P.; Heijink, I.H.; Rots, M.G.; Hylkema, M.N. Targeted epigenetic editing of SPDEF reduces mucus production in lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L334–L347. [Google Scholar] [CrossRef]
- Stolzenburg, S.; Beltran, A.S.; Swift-Scanlan, T.; Rivenbark, A.G.; Rashwan, R.; Blancafort, P. Stable oncogenic silencing in vivo by programmable and targeted de novo DNA methylation in breast cancer. Oncogene 2015, 34, 5427–5435. [Google Scholar] [CrossRef] [PubMed]
- Mlambo, T.; Nitsch, S.; Hildenbeutel, M.; Romito, M.; Muller, M.; Bossen, C.; Diederichs, S.; Cornu, T.I.; Cathomen, T.; Mussolino, C. Designer epigenome modifiers enable robust and sustained gene silencing in clinically relevant human cells. Nucleic Acids Res. 2018, 46, 4456–4468. [Google Scholar] [CrossRef] [PubMed]
- Nunez, J.K.; Chen, J.; Pommier, G.C.; Cogan, J.Z.; Replogle, J.M.; Adriaens, C.; Ramadoss, G.N.; Shi, Q.; Hung, K.L.; Samelson, A.J.; et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 2021, 184, 2503–2519 e2517. [Google Scholar] [CrossRef] [PubMed]
- Goubert, D.; Koncz, M.; Kiss, A.; Rots, M.G. Establishment of Cell Lines Stably Expressing dCas9-Fusions to Address Kinetics of Epigenetic Editing. In Epigenome Editing; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1767, pp. 395–415. [Google Scholar] [CrossRef]
- Falahi, F.; Huisman, C.; Kazemier, H.G.; van der Vlies, P.; Kok, K.; Hospers, G.A.; Rots, M.G. Towards sustained silencing of HER2/neu in cancer by epigenetic editing. Mol. Cancer Res. 2013, 11, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Louis, K.S.; Siegel, A.C. Cell viability analysis using trypan blue: Manual and automated methods. In Mammalian Cell Viability; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2011; Volume 740, pp. 7–12. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Darii, M.V.; Cherepanova, N.A.; Subach, O.M.; Kirsanova, O.V.; Rasko, T.; Slaska-Kiss, K.; Kiss, A.; Deville-Bonne, D.; Reboud-Ravaux, M.; Gromova, E.S. Mutational analysis of the CG recognizing DNA methyltransferase SssI: Insight into enzyme-DNA interactions. Biochim. Biophys. Acta 2009, 1794, 1654–1662. [Google Scholar] [CrossRef]
- Stan, S.D.; Hahm, E.R.; Warin, R.; Singh, S.V. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res. 2008, 68, 7661–7669. [Google Scholar] [CrossRef] [PubMed]
- Hahm, E.R.; Singh, S.V. Withaferin A-induced apoptosis in human breast cancer cells is associated with suppression of inhibitor of apoptosis family protein expression. Cancer Lett. 2013, 334, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Stan, S.D.; Zeng, Y.; Singh, S.V. Ayurvedic medicine constituent withaferin a causes G2 and M phase cell cycle arrest in human breast cancer cells. Nutr. Cancer 2008, 60 (Suppl. S1), 51–60. [Google Scholar] [CrossRef] [PubMed]
- Widodo, N.; Priyandoko, D.; Shah, N.; Wadhwa, R.; Kaul, S.C. Selective killing of cancer cells by Ashwagandha leaf extract and its component Withanone involves ROS signaling. PLoS ONE 2010, 5, e13536. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yu, N.K.; Kaang, B.K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 2015, 47, e166. [Google Scholar] [CrossRef]
- Pavet, V.; Shlyakhtina, Y.; He, T.; Ceschin, D.G.; Kohonen, P.; Perala, M.; Kallioniemi, O.; Gronemeyer, H. Plasminogen activator urokinase expression reveals TRAIL responsiveness and supports fractional survival of cancer cells. Cell Death Dis. 2014, 5, e1043. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, F.; Gao, G. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Elegheert, J.; Behiels, E.; Bishop, B.; Scott, S.; Woolley, R.E.; Griffiths, S.C.; Byrne, E.F.X.; Chang, V.T.; Stuart, D.I.; Jones, E.Y.; et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat. Protoc. 2018, 13, 2991–3017. [Google Scholar] [CrossRef]
- Wang, M.; Glass, Z.A.; Xu, Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017, 24, 144–150. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Guo, P.; Yang, J.; Huang, J.; Auguste, D.T.; Moses, M.A. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc. Natl. Acad. Sci. USA 2019, 116, 18295–18303. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Xu, J.L.; Jia, Y.Y.; Zhang, Y.X.; Wang, W.; Li, C.; He, W.; Zhou, S.Y.; Zhang, B.L. Enhance transgene responses through improving cellular uptake and intracellular trafficking by bio-inspired non-viral vectors. J. Nanobiotechnol. 2020, 18, 26. [Google Scholar] [CrossRef] [PubMed]
- Goran Ronquist, K. Extracellular vesicles and energy metabolism. Clin. Chim. Acta 2019, 488, 116–121. [Google Scholar] [CrossRef]
- Van Gestel, M.A.; Boender, A.J.; de Vrind, V.A.; Garner, K.M.; Luijendijk, M.C.; Adan, R.A. Recombinant adeno-associated virus: Efficient transduction of the rat VMH and clearance from blood. PLoS ONE 2014, 9, e97639. [Google Scholar] [CrossRef]
- Liao, H.K.; Hatanaka, F.; Araoka, T.; Reddy, P.; Wu, M.Z.; Sui, Y.; Yamauchi, T.; Sakurai, M.; O’Keefe, D.D.; Nunez-Delicado, E.; et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 2017, 171, 1495–1507.e15. [Google Scholar] [CrossRef]
- Naso, M.F.; Tomkowicz, B.; Perry, W.L., 3rd; Strohl, W.R. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs 2017, 31, 317–334. [Google Scholar] [CrossRef]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef]
- DeFrancesco, L. Chroma Medicine and Tune Therapeutics: Two companies take up epigenome editing. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Cho, E.; Lee, K.J.; Seo, J.W.; Byun, C.J.; Chung, S.J.; Suh, D.C.; Carmeliet, P.; Koh, J.Y.; Kim, J.S.; Lee, J.Y. Neuroprotection by urokinase plasminogen activator in the hippocampus. Neurobiol. Dis. 2012, 46, 215–224. [Google Scholar] [CrossRef]
- Diaz, A.; Merino, P.; Manrique, L.G.; Ospina, J.P.; Cheng, L.; Wu, F.; Jeanneret, V.; Yepes, M. A Cross Talk between Neuronal Urokinase-type Plasminogen Activator (uPA) and Astrocytic uPA Receptor (uPAR) Promotes Astrocytic Activation and Synaptic Recovery in the Ischemic Brain. J. Neurosci. 2017, 37, 10310–10322. [Google Scholar] [CrossRef] [PubMed]
- Merino, P.; Yepes, M. Urokinase-type Plasminogen Activator Induces Neurorepair in the Ischemic Brain. J. Neurol. Exp. Neurosci. 2018, 4, 24–29. [Google Scholar] [CrossRef][Green Version]
- Horowitz, J.C.; Tschumperlin, D.J.; Kim, K.K.; Osterholzer, J.J.; Subbotina, N.; Ajayi, I.O.; Teitz-Tennenbaum, S.; Virk, A.; Dotson, M.; Liu, F.; et al. Urokinase Plasminogen Activator Overexpression Reverses Established Lung Fibrosis. Thromb. Haemost. 2019, 119, 1968–1980. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Liu, Y.; Xiong, Z.; Hu, L.; Xiong, C.L. Tissue-specific inhibition of urokinase-type plasminogen activator expression in the testes of mice by inducible lentiviral RNA interference causes male infertility. Reprod. Fertil. Dev. 2017, 29, 2149–2156. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Z.; Ou, S.H.; Chang, L.C.; Lin, Y.F.; Pei, D.; Chen, J.S. Deficiency of Urokinase Plasminogen Activator May Impair beta Cells Regeneration and Insulin Secretion in Type 2 Diabetes Mellitus. Molecules 2019, 24, 4208. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Mi, X.; Gao, N.; Yan, C.; Yu, F.S. Hyperglycemia-suppressed expression of Serpine1 contributes to delayed epithelial wound healing in diabetic mouse corneas. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3383–3392. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).