Clinical Performance of Self-Collected Nasal Swabs and Antigen Rapid Tests for SARS-CoV-2 Detection in Resource-Poor Settings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Setting and Participants
2.2. Sample Collection
2.3. Sample Size
2.4. Antigen Rapid Test (Ag-RDT) Testing
2.5. Real-Time PCR Testing
2.6. Data Analysis
3. Results
3.1. Characteristics of Study Population
3.2. Performance of Nasal Swabs for SARS-CoV-2 Detection
3.3. Performance of Nasal Swab-Based Antigen Rapid Tests for SARS-CoV-2 Detection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low Performance of Rapid Antigen Detection Test as Frontline Testing for COVID-19 Diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, I.N.; Emeribe, A.U.; Akande, A.O.; Ghamba, P.E.; Adekola, H.A.; Ibrahim, Y.; Dangana, A. Roles and Challenges of Coordinated Public Health Laboratory Response against COVID-19 Pandemic in Africa. J. Infect Dev. Ctries 2020, 14, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Favresse, J.; Gillot, C.; Oliveira, M.; Cadrobbi, J.; Elsen, M.; Eucher, C.; Laffineur, K.; Rosseels, C.; Van Eeckhoudt, S.; Nicolas, J.-B.; et al. Head-to-Head Comparison of Rapid and Automated Antigen Detection Tests for the Diagnosis of SARS-CoV-2 Infection. JCM 2021, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Bohn, M.K.; Loh, T.P.; Wang, C.-B.; Mueller, R.; Koch, D.; Sethi, S.; Rawlinson, W.D.; Clementi, M.; Erasmus, R.; Leportier, M.; et al. IFCC Interim Guidelines on Serological Testing of Antibodies against SARS-CoV-2. Clin. Chem. Lab. Med. 2020, 58, 2001–2008. [Google Scholar] [CrossRef]
- Pollock, N.R.; Lee, F.; Ginocchio, C.C.; Yao, J.D.; Humphries, R.M. Considerations for Assessment and Deployment of Rapid Antigen Tests for Diagnosis of Coronavirus Disease 2019. Open Forum Infect. Dis. 2021, 8, ofab110. [Google Scholar] [CrossRef]
- Jani, I.V.; Peter, T.F. Nucleic Acid Point-of-Care Testing to Improve Diagnostic Preparedness. Clin. Infect. Dis. 2022, 75, 723–728. [Google Scholar] [CrossRef]
- Hanson, K.E.; Barker, A.P.; Hillyard, D.R.; Gilmore, N.; Barrett, J.W.; Orlandi, R.R.; Shakir, S.M. Self-Collected Anterior Nasal and Saliva Specimens versus Health Care Worker-Collected Nasopharyngeal Swabs for the Molecular Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01824-20. [Google Scholar] [CrossRef]
- CDC Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed on 20 June 2022).
- WHO WHO COVID-19: Case Definitions. WHO 2019-NCoV Surveillance Case Definition 2020.2. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-Surveillance_Case_Definition-2020.2 (accessed on 25 April 2021).
- Gaur, R.; Verma, D.K.; Mohindra, R.; Goyal, K.; Gupta, S.; Singla, V.; Sahni, V.; Ghosh, A.; Soni, R.K.; Bhalla, A.; et al. Buccal Swabs as Non-Invasive Specimens for Detection of Severe Acute Respiratory Syndrome Coronavirus-2. J. Int. Med. Res. 2021, 49, 030006052110169. [Google Scholar] [CrossRef]
- Vlek, A.L.M.; Wesselius, T.S.; Achterberg, R.; Thijsen, S.F.T. Combined Throat/Nasal Swab Sampling for SARS-CoV-2 Is Equivalent to Nasopharyngeal Sampling. Eur. J. Clin. Microbiol. Infect Dis. 2021, 40, 193–195. [Google Scholar] [CrossRef]
- Wang, X.; Tan, L.; Wang, X.; Liu, W.; Lu, Y.; Cheng, L.; Sun, Z. Comparison of Nasopharyngeal and Oropharyngeal Swabs for SARS-CoV-2 Detection in 353 Patients Received Tests with Both Specimens Simultaneously. Int. J. Infect. Dis. 2020, 94, 107–109. [Google Scholar] [CrossRef]
- Jamal, A.J.; Mozafarihashjin, M.; Coomes, E.; Anceva-Sami, S.; Barati, S.; Crowl, G.; Faheem, A.; Farooqi, L.; Kandel, C.E.; Khan, S.; et al. Sensitivity of Midturbinate versus Nasopharyngeal Swabs for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Infect. Control Hosp. Epidemiol. 2021, 42, 1001–1003. [Google Scholar] [CrossRef] [PubMed]
- Abdulrahman, A.; Mustafa, F.; AlAwadhi, A.I.; Alansari, Q.; AlAlawi, B.; AlQahtani, M. Comparison of SARS-CoV-2 Nasal Antigen Test to Nasopharyngeal RT-PCR in Mildly Symptomatic Patients; Infectious Diseases (except HIV/AIDS). medRxiv 2020. [Google Scholar] [CrossRef]
- Palmas, G.; Moriondo, M.; Trapani, S.; Ricci, S.; Calistri, E.; Pisano, L.; Perferi, G.; Galli, L.; Venturini, E.; Indolfi, G.; et al. Nasal Swab as Preferred Clinical Specimen for COVID-19 Testing in Children. Pediatric Infect. Dis. J. 2020, 39, e267–e270. [Google Scholar] [CrossRef] [PubMed]
- Abongo, T.; Ulo, B.; Karanja, S. Community Health Volunteers’ Contribution to Tuberculosis Patients Notified to National Tuberculosis Program through Contact Investigation in Kenya. BMC Public Health 2020, 20, 1184. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration Guidance for Industry and FDA Staff—Statistical Guidance on Reporting Results from Studies Evaluating Diagnostic Tests 2007. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/statistical-guidance-reporting-results-studies-evaluating-diagnostic-tests-guidance-industry-and-fda (accessed on 15 August 2022).
- WHO Global Influenza Surveillance Network: Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; World Health Organization: Geneva, Switzerland, 2011.
- Kohmer, N.; Eckermann, L.; Böddinghaus, B.; Götsch, U.; Berger, A.; Herrmann, E.; Kortenbusch, M.; Tinnemann, P.; Gottschalk, R.; Hoehl, S.; et al. Self-Collected Samples to Detect SARS-CoV-2: Direct Comparison of Saliva, Tongue Swab, Nasal Swab, Chewed Cotton Pads and Gargle Lavage. JCM 2021, 10, 5751. [Google Scholar] [CrossRef]
- WHO Technical Specifications for Selection of Essential in Vitro Diagnostics for SARS-CoV-2. Available online: https://www.who.int/publications/m/item/technical-specifications-for-selection-of-essential-in-vitro-diagnostics-for-sars-cov-2 (accessed on 12 April 2022).
- Zhou, Y.; O’Leary, T.J. Relative Sensitivity of Anterior Nares and Nasopharyngeal Swabs for Initial Detection of SARS-CoV-2 in Ambulatory Patients: Rapid Review and Meta-Analysis. PLoS ONE 2021, 16, e0254559. [Google Scholar] [CrossRef]
- Pan, Y.; Long, L.; Zhang, D.; Yuan, T.; Cui, S.; Yang, P.; Wang, Q.; Ren, S. Potential False-Negative Nucleic Acid Testing Results for Severe Acute Respiratory Syndrome Coronavirus 2 from Thermal Inactivation of Samples with Low Viral Loads. Clin. Chem. 2020, 66, 794–801. [Google Scholar] [CrossRef]
- Matuchansky Protection against SARS-CoV-2 after Vaccination and Previous Infection. N. Engl. J. Med. 2022, 386, 2534–2535. [CrossRef]
- Drain, P.K.; Ampajwala, M.; Chappel, C.; Gvozden, A.B.; Hoppers, M.; Wang, M.; Rosen, R.; Young, S.; Zissman, E.; Montano, M. A Rapid, High-Sensitivity SARS-CoV-2 Nucleocapsid Immunoassay to Aid Diagnosis of Acute COVID-19 at the Point of Care: A clinical performance study. Infect. Dis. Ther. 2021, 10, 753–761. [Google Scholar] [CrossRef]
- Stokes, W.; Berenger, B.M.; Scott, B.; Szelewicki, J.; Singh, T.; Portnoy, D.; Turnbull, L.; Pabbaraju, K.; Shokoples, S.; Wong, A.A.; et al. One Swab Fits All: Performance of a Rapid, Antigen-Based SARS-CoV-2 Test Using a Nasal Swab, Nasopharyngeal Swab for Nasal Collection, and RT–PCR Confirmation from Residual Extraction Buffer. J. Appl. Lab. Med. 2022, 7, 834–841. [Google Scholar] [CrossRef]
- Cubas-Atienzar, A.I.; Bell, F.; Byrne, R.L.; Buist, K.; Clark, D.J.; Cocozza, M.; Collins, A.M.; Cuevas, L.E.; Duvoix, A.; Easom, N.; et al. Accuracy of the Mologic COVID-19 Rapid Antigen Test: A Prospective Multi-Centre Analytical and Clinical Evaluation. Wellcome Open Res. 2021, 6, 132. [Google Scholar] [CrossRef]
- Krüger, L.J.; Tanuri, A.; Lindner, A.K.; Gaeddert, M.; Köppel, L.; Tobian, F.; Brümmer, L.E.; Klein, J.A.F.; Lainati, F.; Schnitzler, P.; et al. Accuracy and Ease-of-Use of Seven Point-of-Care SARS-CoV-2 Antigen-Detecting Tests: A Multi-Centre Clinical Evaluation. eBioMedicine 2022, 75, 103774. [Google Scholar] [CrossRef] [PubMed]
- Galliez, R.M.; Bomfim, L.; Mariani, D.; Leitão, I.d.C.; Castiñeiras, A.C.P.; Gonçalves, C.C.A.; Ortiz da Silva, B.; Cardoso, P.H.; Arruda, M.B.; Alvarez, P.; et al. Evaluation of the Panbio COVID-19 Antigen Rapid Diagnostic Test in Subjects Infected with Omicron Using Different Specimens. Microbiol. Spectr. 2022, 10, e0125022. [Google Scholar] [CrossRef] [PubMed]
- Bayart, J.-L.; Degosserie, J.; Favresse, J.; Gillot, C.; Didembourg, M.; Djokoto, H.P.; Verbelen, V.; Roussel, G.; Maschietto, C.; Mullier, F.; et al. Analytical Sensitivity of Six SARS-CoV-2 Rapid Antigen Tests for Omicron versus Delta Variant. Viruses 2022, 14, 654. [Google Scholar] [CrossRef]
- Samsunder, N.; de Vos, M.; Ngcapu, S.; Giandhari, J.; Lewis, L.; Kharsany, A.B.M.; Cawood, C.; de Oliveira, T.; Karim, Q.A.; Karim, S.A.; et al. Clinical Evaluation of Severe Acute Respiratory Syndrome Coronavirus 2 Rapid Antigen Tests During the Omicron Wave in South Africa. J. Infect. Dis. 2022, jiac333. [Google Scholar] [CrossRef]
- Sitoe, N.; Sambo, J.; Nguenha, N.; Chilaule, J.; Chelene, I.; Loquiha, O.; Mudenyanga, C.; Viegas, S.; Cunningham, J.; Jani, I. Performance Evaluation of the STANDARDTM Q COVID-19 and PanbioTM COVID-19 Antigen Tests in Detecting SARS-CoV-2 during High Transmission Period in Mozambique. Diagnostics 2022, 12, 475. [Google Scholar] [CrossRef] [PubMed]
- Velavan, T.P.; Pallerla, S.R.; Kremsner, P.G. How to (Ab)Use a COVID-19 Antigen Rapid Test with Soft Drinks? Int. J. Infect. Dis. 2021, 111, 28–30. [Google Scholar] [CrossRef]
- Goggolidou, P.; Hodges-Mameletzis, I.; Purewal, S.; Karakoula, A.; Warr, T. Self-Testing as an Invaluable Tool in Fighting the COVID-19 Pandemic. J. Prim. Care Community Health 2021, 12, 215013272110477. [Google Scholar] [CrossRef]
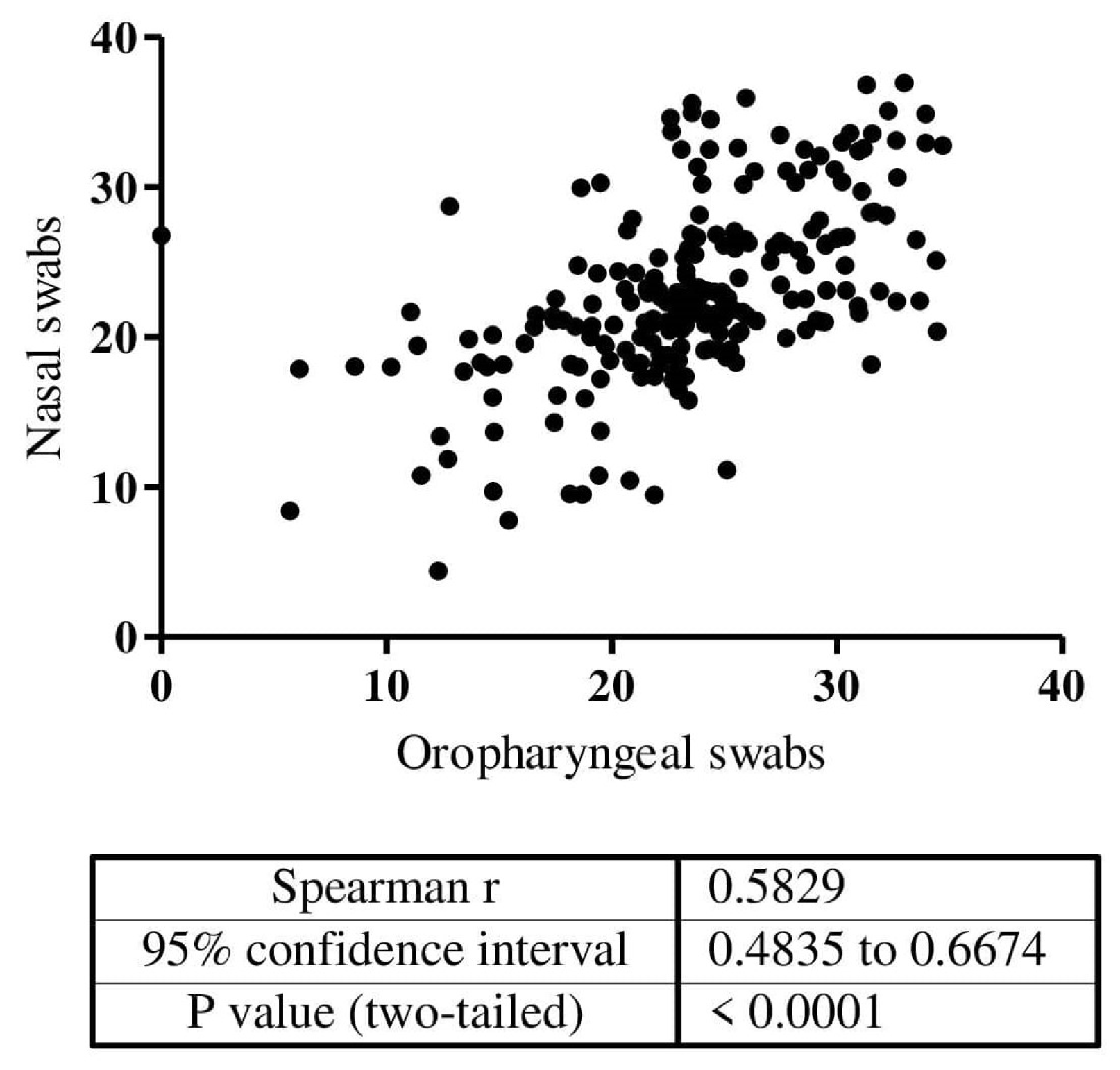
| Nasal Swabs Self-Collection 1 | Abbott Panbio 2 | Covios 3 | Lumira Dx 4 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N = 1498 | % | N = 400 | % | N = 1123 | % | N = 696 | % | ||
| Gold standard 5 (RT-PCR or OP 6 swabs) | Negative | 1229 | 82.04 | 186 | 46.5 | 586 | 53.0 | 310 | 44.54 |
| Positive | 269 | 17.96 | 214 | 53.5 | 520 | 47.0 | 386 | 55.46 | |
| Invalid | -- 7 | -- | -- | -- | -- | -- | -- | -- | |
| Indeterminate | -- | -- | -- | -- | -- | -- | -- | -- | |
| Technology under evaluation 8 | Negative | 1227 | 81.91 | 228 | 57.0 | 773 | 68.8 | 379 | 54.45 |
| Positive | 260 | 17.36 | 172 | 43.0 | 346 | 30.8 | 314 | 45.11 | |
| Invalid | -- | -- | -- | -- | 4 | 0.4 | 3 | 0.43 | |
| Indeterminate | 11 | 0.73 | -- | -- | -- | -- | -- | -- | |
| Study sites | CSM 9 | 200 | 13.35 | -- | -- | -- | -- | 256 | 36.78 |
| HGM 10 | 225 | 15.02 | -- | -- | 343 | 30.5 | 300 | 43.10 | |
| HGC 11 | 384 | 25.63 | 400 | 100 | 395 | 35.2 | -- | -- | |
| HPM 12 | 577 | 38.52 | -- | -- | 385 | 34.3 | 140 | 20.11 | |
| HCM 13 | 112 | 7.48 | -- | -- | -- | -- | -- | -- | |
| Age | Median in years | 35.0 | -- | 38.02 | -- | 33.0 | -- | 36.23 | -- |
| Sex | Female | 764 | 52.04 | 228 | 57.0 | 644 | 57.4 | 381 | 54.74 |
| Male | 734 | 48.9 | 172 | 43.0 | 479 | 42.6 | 315 | 45.26 | |
| Symptoms | Yes | 1356 | 90.52 | 397 | 99.25 | 1115 | 99.3 | 575 | 82.61 |
| No | 141 | 9.48 | 3 | 0.75 | 8 | 0.7 | 121 | 17.39 | |
| Onset symptoms | ≤7 days | 1168 | 86.13 | 369 | 92.95 | 1033 | 92.0 | 519 | 90.26 |
| >7 days | 188 | 13.87 | 28 | 7.05 | 82 | 7.3 | 56 | 9.74 | |
| Ct Value of PCR in OP samples, median (min-max) | 24.7 (5.7–36.5) | NA 14 | 26.7 (9.0–37.3) | NA | 25.2 (5.1–39.0) | NA | 25.1 (3.7–35.9) | NA | |
| Diagnostic Test | Patient Type | Sensitivity | Specificity | PPV | NPV | Observed Agreement | Cohen’s Kappa |
|---|---|---|---|---|---|---|---|
| Self-collected nasal swab | Overall N = 1498 | 80.6% [75.3–85.2%] | 96.4% [95.2–97.4%] | 83.1% [78.0–87.4%] | 95.8% [94.5–96.8%] | 93.5% [34.7–94.7%] | 0.78 [0.77–0.79] |
| Symptomatic N = 1356 | 81.0% [75.5–85.7%] | 96.7% [95.5–97.7%] | 84.5% [79.2–88.9%] | 95.9% [94.5–97.0%] | 93.9% [34.7–95.1%] | 0.79 [0.78–0.79] | |
| Asymptomatic N = 141 | 90.5% [69.6–98.8%] | 93.1% [86.9–97.0%] | 70.4% [49.8–86.2%] | 98.2% [93.8–99.8%] | 92.7% [31.0–96.4%] | 0.75 [0.67–0.83] | |
| Symptoms ≤ 7 d 1 N = 1168 | 81.1% [75.2–86.2%] | 96.5% [95.1–97.6%] | 83.9% [78.1–88.7%] | 95.8% [94.3–97.0%] | 93.7% [34.6–95.0%] | 0.79 [0.78–0.80] | |
| Symptoms > 7 d N = 188 | 93.3% [68.1–99.8] | 98.1% [93.4–99.8%] | 87.5% [61.7–98.4%] | 99.0% [94.8–100.0%] | 97.5% [30.7–99.5%] | 0.89 [0.78–0.99] | |
| Abbott Panbio Nasal RDT | Overall N = 400 | 79.7% [73.3–85.1%] | 97.2% [93.5–99.1%] | 96.8% [92.8–99.0%] | 81.4% [75.5–86.4%] | 88.0% [41.9–91.2%] | 0.71 [0.70–0.72] |
| Symptomatic N = 397 | 79.9% [73.3–85.1%] | 97.1% [93.4–99.1%] | 96.8% [92.8–99.0%] | 81.2% [75.2–86.2%] | 87.9% [42.1–91.1%] | 0.71 [0.70–0.72] | |
| Asymptomatic N = 3 | NA | NA | NA | NA | NA | NA | |
| Symptoms ≤ 7 d N = 369 | 80.0% [73.3–85.7%] | 96.9% [93.0–99.0%] | 96.6% [92.1–98.9%] | 81.9% [75.7–87.0%] | 88.2% [41.7–91.4%] | 0.71 [0.70–0.72] | |
| Symptoms > 7 d N = 28 | 76.5% [50.1–93.1%] | 100.0% [69.2–NA] | 100.0% [75.3–NA] | 71.4% [41.9–91.6%] | 85.2% [34.1–95.8%] | 0.72 [0.59–0.85] | |
| Overall N= 1102 | 59.6% [55.2–63.8%] | 94.9% [92.8–96.5%] | 91.1% [87.6–93.9%] | 72.6% [69.3–75.8%] | 78.3% [37.0–80.7%] | 0.56 [0.551–0.560] | |
| Symptomatic N= 1094 | 59.7% [55.3–64.0%] | 94.8% [92.7–96.5%] | 91.1% [87.5–93.9%] | 72.7% [69.3–75.8%] | 78.3% [37.0–80.7%] | 0.56 [0.552–0.561] | |
| COVIOS Ag COVID-19 | Asymptomatic N= 8 | 33.3% [0.8–90.6%] | 100.0% [47.8–NA] | 100.0% [2.5–NA] | 71.4% [29.0–96.3%] | 75.0% [13.3–96.8%] | 0.38 [-0.628–1.397] |
| Symptoms ≤ 7 d N= 1013 | 60.9% [56.5–65.2%] | 94.6% [92.2–96.4%] | 91.6% [88.0–94.3%] | 71.4% [67.8–74.7%] | 78.0% [37.6–80.5%] | 0.56 [0.553–0.562] | |
| Symptoms > 7 d N= 81 | 20.0% [4.3–48.1%] | 97.0% [89.5–99.6%] | 60.0% [14.7–94.7%] | 84.2% [74.0–91.6%] | 82.7% [25.8–90.2%] | 0.23 [-0.156–0.613] | |
| LumiraDx SARS-CoV-2 Ag Test | Overall N = 696 | 78.0% [73.5–82.0%] | 95.8% [92.9–97.7%] | 95.9% [93.0–97.8%] | 77.6% [73.0–81.7%] | 85.9% [43.7–88.4%] | 0.72 [0.72–0.73] |
| Symptomatic N = 575 | 79.9% [75.4–83.9%] | 95.7% [92.0–98.0%] | 97.0% [94.4–98.6%] | 73.4% [67.7–78.5%] | 85.7% [47.2–88.5%] | 0.71 [0.70–0.72] | |
| Asymptomatic N = 121 | 47.8% [26.8–69.4%] | 95.9% [89.8–98.9%] | 73.3% [44.9–92.2%] | 88.6% [80.9–94.0%] | 86.7% [28.7–92.2%] | 0.50 [0.36–0.65] | |
| Symptoms ≤ 7 d N = 519 | 81.6% [77.0–85.6%] | 96.2% [92.4–98.5%] | 97.5% [94.9–99.0%] | 74.6% [68.6–80.0%] | 86.8% [47.9–89.6%] | 0.73 [0.72–0.74] | |
| Symptoms > 7 d N = 56 | 62.5% [43.7–78.9%] | 91.7% [73.0–99.0%] | 90.9% [70.8–98.9%] | 64.7% [46.5–80.3%] | 75.0% [32.2–85.6%] | 0.52 [0.44–0.59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitoe, N.; Sambo, J.; Mabunda, N.; Nguenha, N.; Chilaúle, J.; Rafael, J.; Macicame, A.; Chelene, I.; Mudenyanga, C.; Sacks, J.; et al. Clinical Performance of Self-Collected Nasal Swabs and Antigen Rapid Tests for SARS-CoV-2 Detection in Resource-Poor Settings. Biomedicines 2022, 10, 2327. https://doi.org/10.3390/biomedicines10092327
Sitoe N, Sambo J, Mabunda N, Nguenha N, Chilaúle J, Rafael J, Macicame A, Chelene I, Mudenyanga C, Sacks J, et al. Clinical Performance of Self-Collected Nasal Swabs and Antigen Rapid Tests for SARS-CoV-2 Detection in Resource-Poor Settings. Biomedicines. 2022; 10(9):2327. https://doi.org/10.3390/biomedicines10092327
Chicago/Turabian StyleSitoe, Nádia, Júlia Sambo, Nédio Mabunda, Neuza Nguenha, Jorfélia Chilaúle, Júlio Rafael, Anésio Macicame, Imelda Chelene, Chishamiso Mudenyanga, Jillian Sacks, and et al. 2022. "Clinical Performance of Self-Collected Nasal Swabs and Antigen Rapid Tests for SARS-CoV-2 Detection in Resource-Poor Settings" Biomedicines 10, no. 9: 2327. https://doi.org/10.3390/biomedicines10092327
APA StyleSitoe, N., Sambo, J., Mabunda, N., Nguenha, N., Chilaúle, J., Rafael, J., Macicame, A., Chelene, I., Mudenyanga, C., Sacks, J., Viegas, S., Loquiha, O., & Jani, I. (2022). Clinical Performance of Self-Collected Nasal Swabs and Antigen Rapid Tests for SARS-CoV-2 Detection in Resource-Poor Settings. Biomedicines, 10(9), 2327. https://doi.org/10.3390/biomedicines10092327






