TLR5 Variants Are Associated with the Risk for COPD and NSCLC Development, Better Overall Survival of the NSCLC Patients and Increased Chemosensitivity in the H1299 Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. SNP Selection Criterion
2.3. DNA Sample Preparation and Genotyping
2.4. Statistical Analysis for Association Studies
2.5. Cells Cultures
2.6. Generation of Adenoviral Vectors Containing TLR5WT and the TLR5N592S as a Transgenes
2.7. Reporter Gene Assays
2.8. Immunoblot Analysis
2.9. Proliferation Assay
2.10. Serum Concentration of the Cytokines
2.11. RNA-Seq Library Preparation and Sequencing
2.12. Bioinformatics Analysis
2.13. Statistical Analysis
3. Results
3.1. TLR5 Variant N592S Is Associated with an Increased Risk for COPD and NSCLC Development
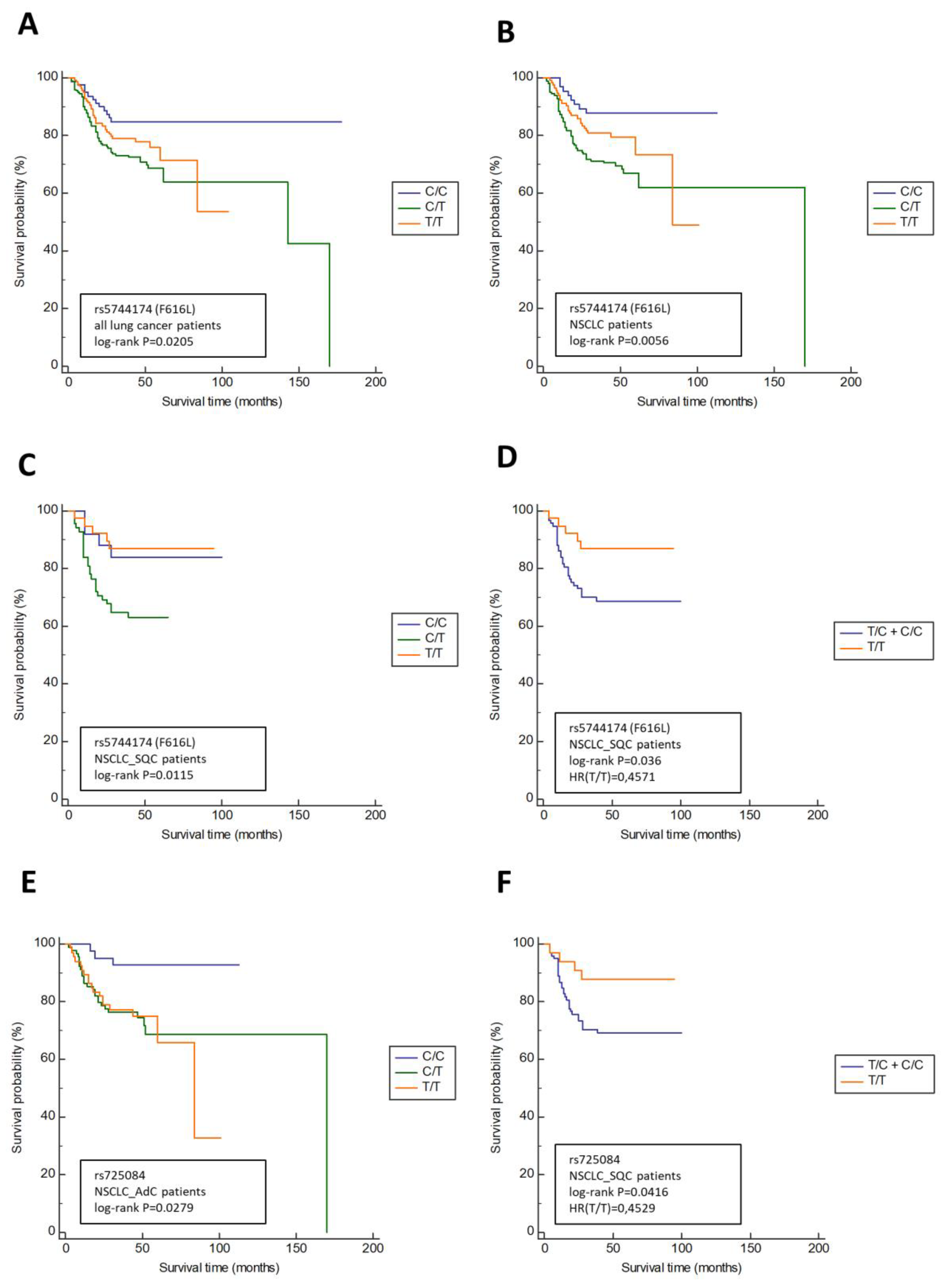
3.2. TLR5 Variants Are Associated with the Lymph Node Involment (N Status) and Overall Survival in NSCLC Patients
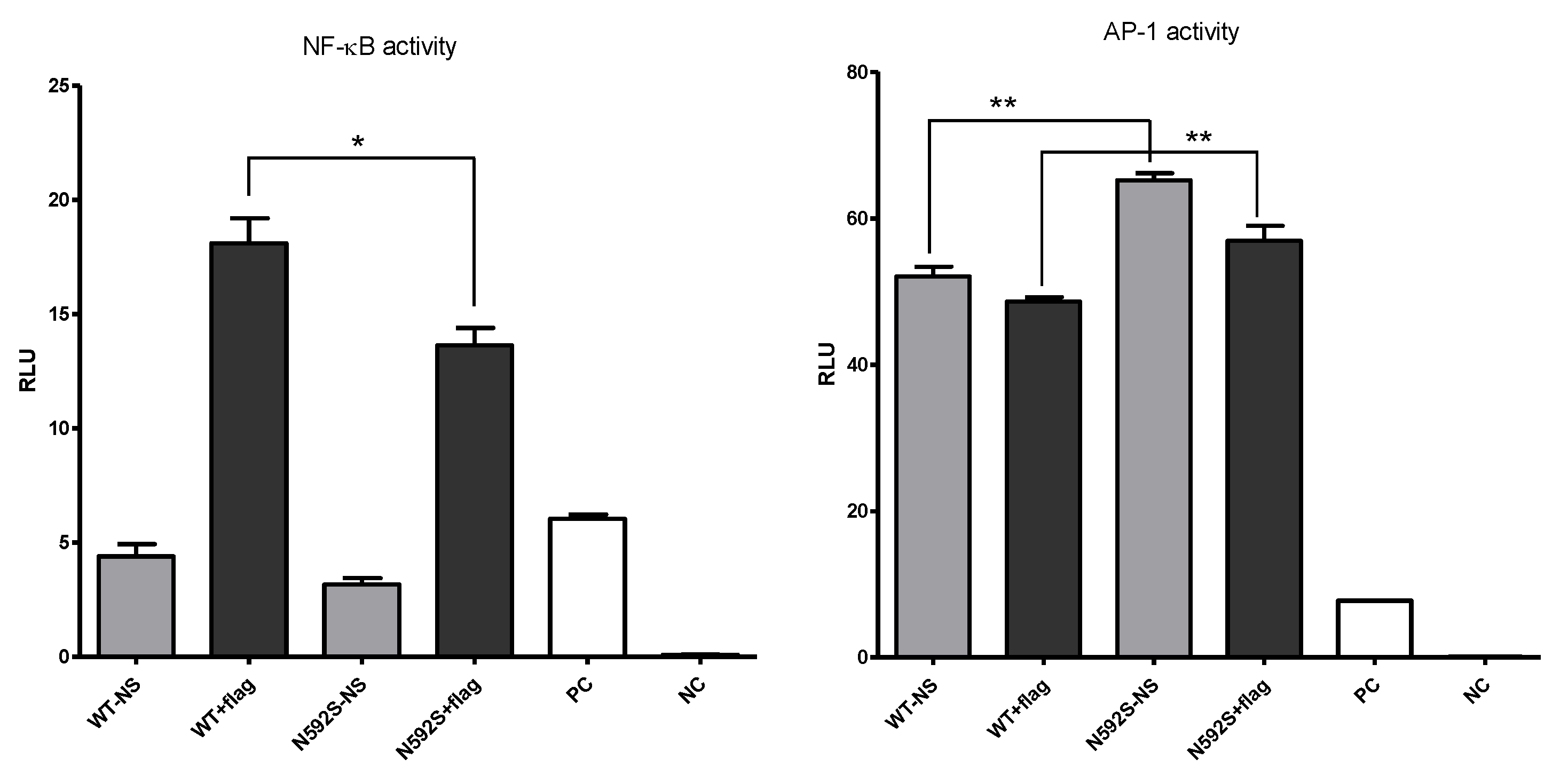
3.3. N592S Variant Affect the Activation of the NF-κB and AP-1 Transcription Factors
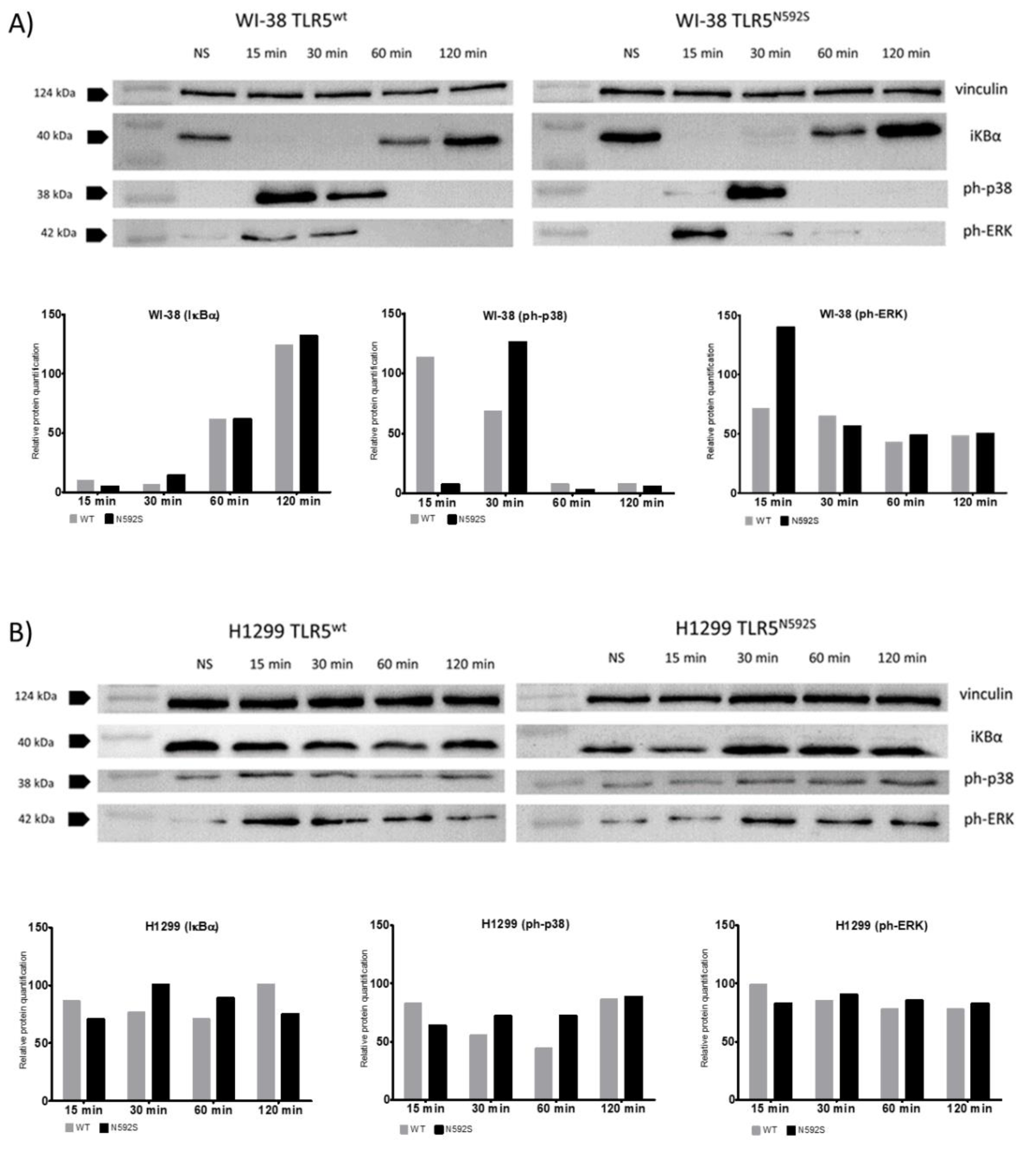
3.4. Activation of p38 and ERK Is Affected by the N592S TLR5 Coding Variant in the WI-38 Cell Line
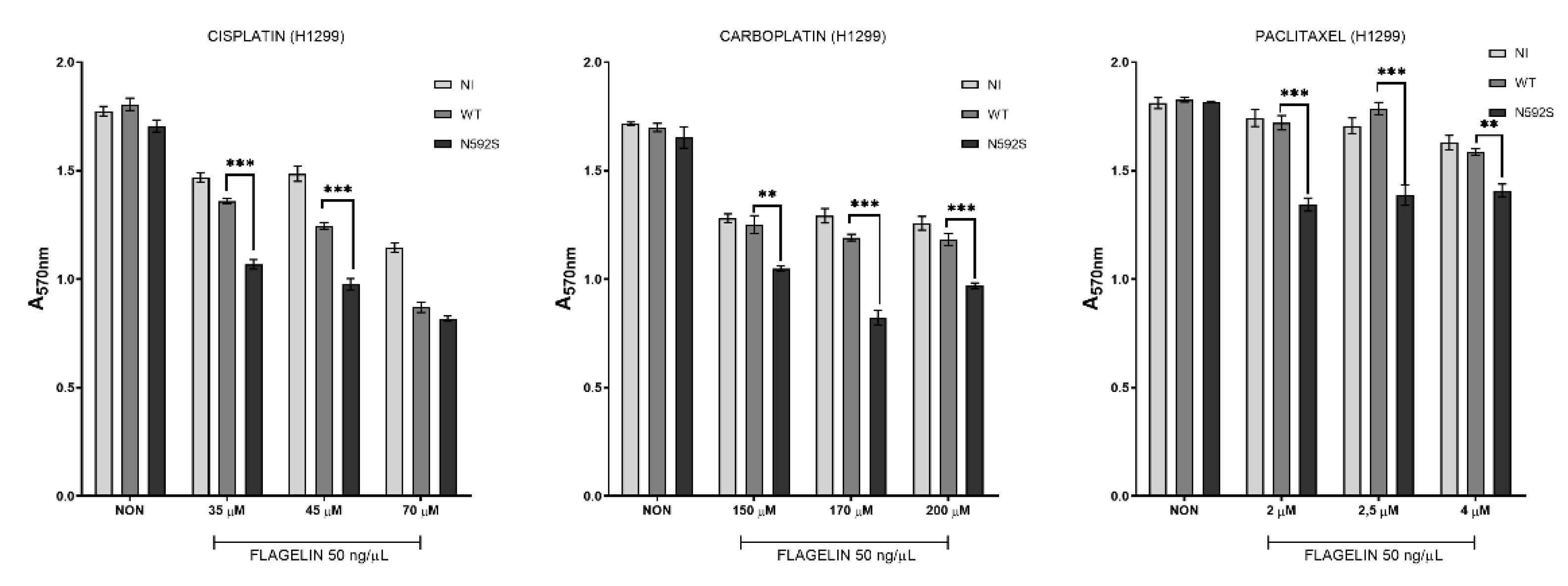
3.5. H1299 Cells Overexpressing N592S Variant Exhibit Increased Chemosensitivity
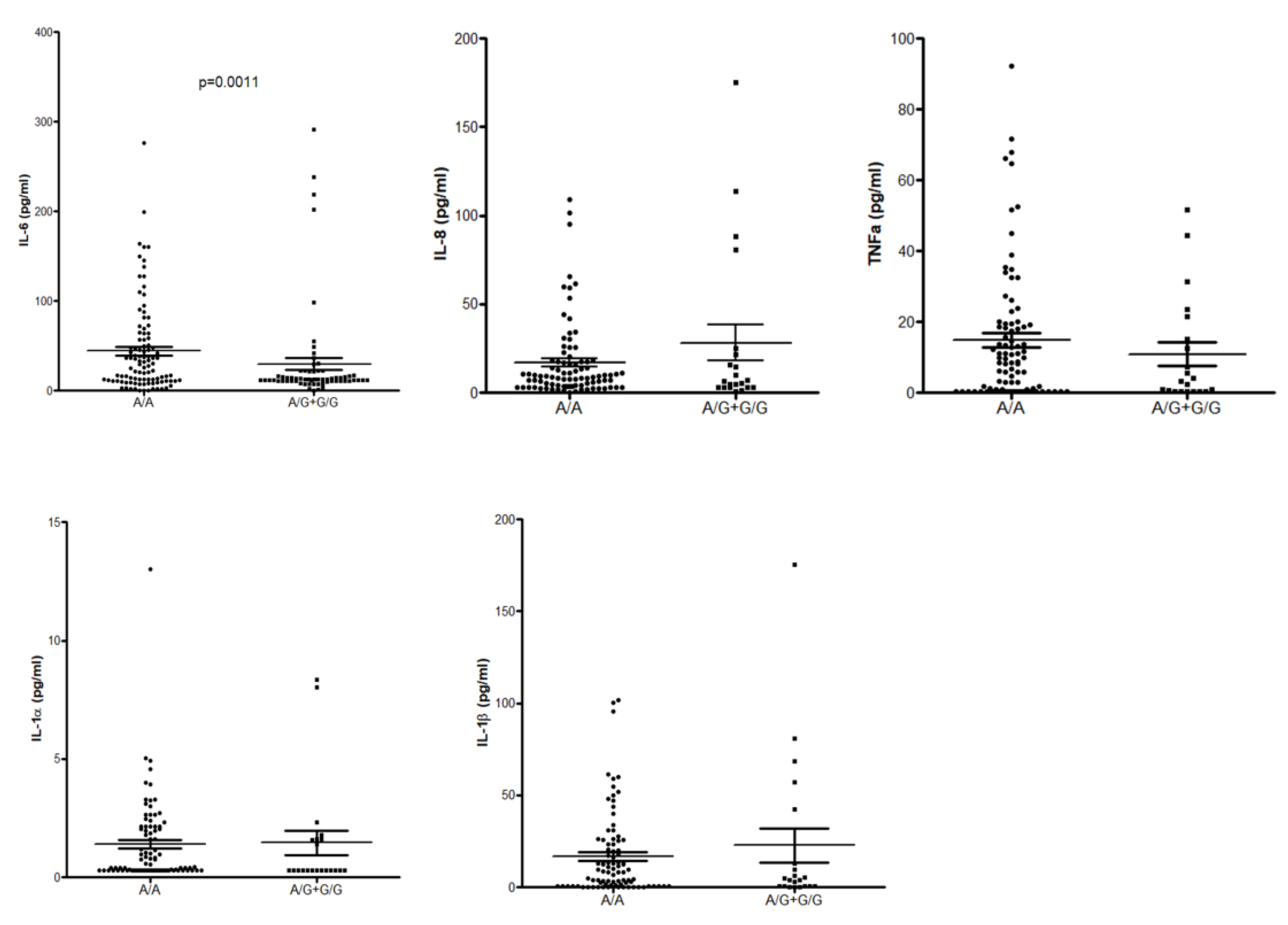
3.6. ELISA
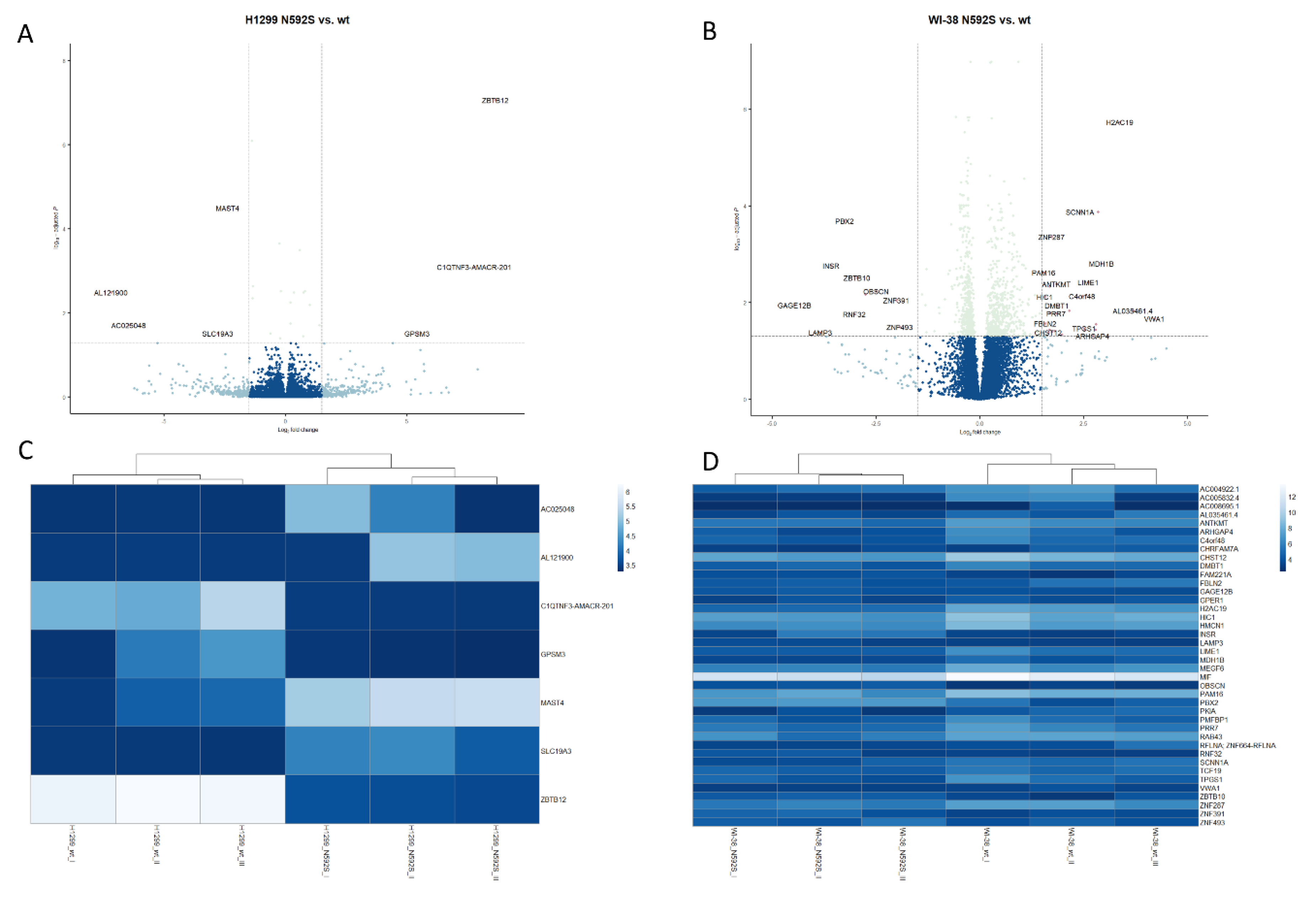
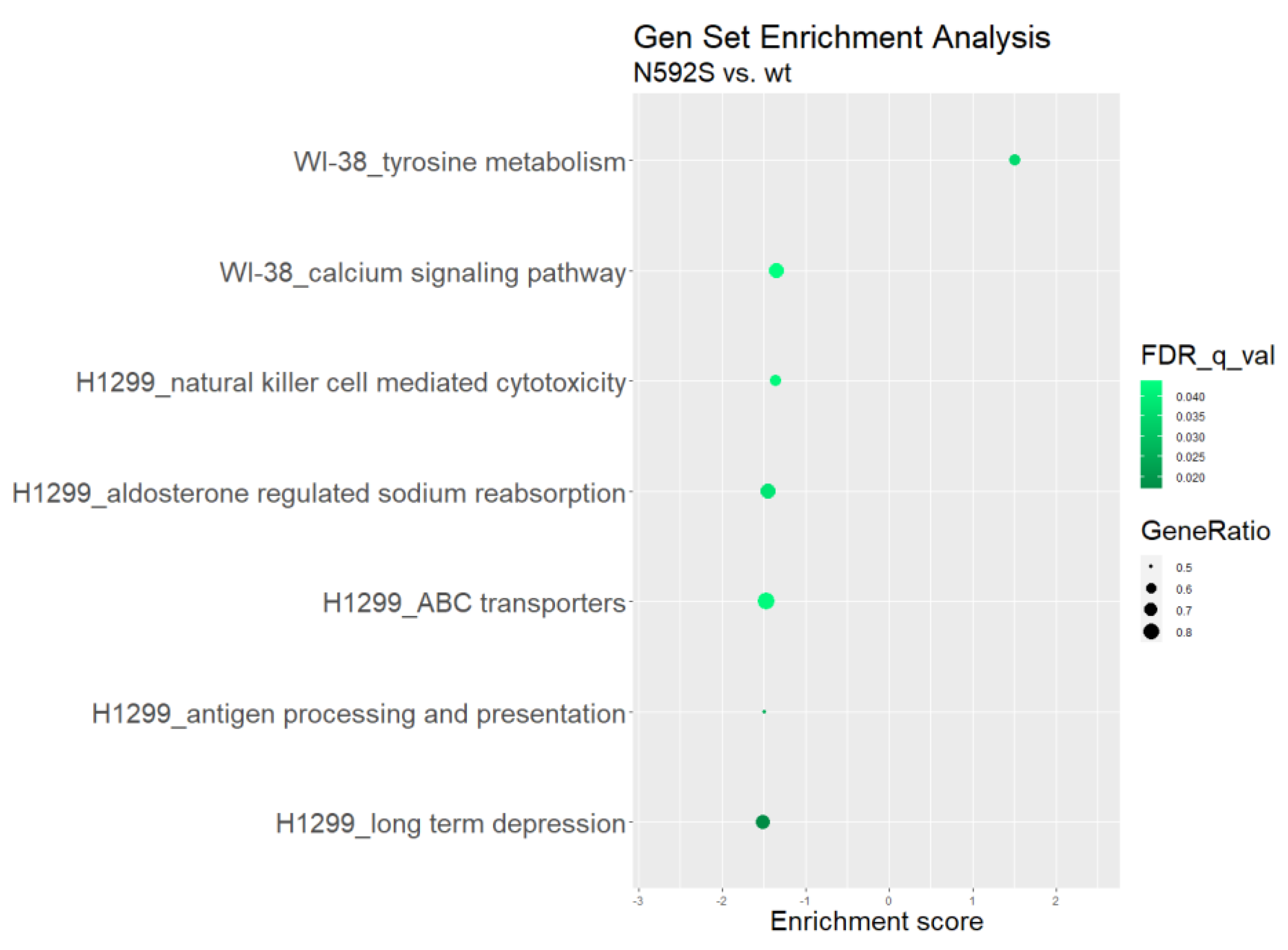
3.7. Transcriptome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bozinovski, S.; Vlahos, R.; Anthony, D.; McQualter, J.; Anderson, G.; Irving, L.; Steinfort, D. COPD and squamous cell lung cancer: Aberrant inflammation and immunity is the common link. Br. J. Pharmacol. 2016, 173, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Houghton, A.M. Common Mechanisms Linking Chronic Obstructive Pulmonary Disease and Lung Cancer. Ann. Am. Thorac. Soc. 2018, 15, S273–S277. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.L.; Adcock, I.M. The relationship between COPD and lung cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Spyratos, D.; Papadaki, E.; Lampaki, S.; Kontakiotis, T. Chronic obstructive pulmonary disease in patients with lung cancer: Prevalence, impact and management challenges. Lung Cancer 2017, 8, 101–107. [Google Scholar] [CrossRef]
- Mattson, M.E.; Pollack, E.S.; Cullen, J.W. What are the odds that smoking will kill you? Am. J. Public Health 1987, 77, 425–431. [Google Scholar] [CrossRef]
- Šutić, M.; Vukić, A.; Baranašić, J.; Försti, A.; Džubur, F.; Samaržija, M.; Jakopović, M.; Brčić, L.; Knežević, J. Diagnostic, Predictive, and Prognostic Biomarkers in Non-Small Cell Lung Cancer (NSCLC) Management. J. Pers. Med. 2021, 11, 1102. [Google Scholar] [CrossRef]
- Zarember, K.A.; Godowski, P.J. Tissue Expression of Human Toll-Like Receptors and Differential Regulation of Toll-Like Receptor mRNAs in Leukocytes in Response to Microbes, Their Products, and Cytokines. J. Immunol. 2002, 168, 554LP–561LP. [Google Scholar] [CrossRef]
- Zhang, Z.; Schluesener, H.J. Mammalian toll-like receptors: From endogenous ligands to tissue regeneration. Cell. Mol. Life Sci. C 2006, 63, 2901–2907. [Google Scholar] [CrossRef]
- Hallman, M.; Rämet, M.; Ezekowitz, R.A. Toll-like Receptors as Sensors of Pathogens. Pediatr. Res. 2001, 50, 315–321. [Google Scholar] [CrossRef]
- Suzuki, T.; Chow, C.-W.; Downey, G.P. Role of innate immune cells and their products in lung immunopathology. Int. J. Biochem. Cell Biol. 2008, 40, 1348–1361. [Google Scholar] [CrossRef]
- Gu, J.; Liu, Y.; Xie, B.; Ye, P.; Huang, J.; Lu, Z. Roles of toll-like receptors: From inflammation to lung cancer progression. Biomed. Rep. 2018, 8, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, S.; Sajjan, U.S. Repair and Remodeling of airway epithelium after injury in Chronic Obstructive Pulmonary Disease. Curr. Respir. Care Rep. 2013, 2, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Scotland, R.S.; Whiteford, J.R. Toll-like receptors: Role in inflammation and therapeutic potential. BioFactors 2014, 40, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Kim, Y.-J. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 2007, 76, 447–480. [Google Scholar] [CrossRef] [PubMed]
- Rooney, C.; Sethi, T. The Epithelial Cell and Lung Cancer: The Link between Chronic Obstructive Pulmonary Disease and Lung Cancer. Respiration 2011, 81, 89–104. [Google Scholar] [CrossRef]
- Berg, J.; Halvorsen, A.R.; Bengtson, M.B.; Taskén, K.A.; Mælandsmo, G.M.; Yndestad, A.; Halvorsen, B.; Brustugun, O.T.; Aukrust, P.; Ueland, T.; et al. Levels and prognostic impact of circulating markers of inflammation, endothelial activation and extracellular matrix remodelling in patients with lung cancer and chronic obstructive pulmonary disease. BMC Cancer 2018, 18, 739. [Google Scholar] [CrossRef]
- Chatterjee, S.; Crozet, L.; Damotte, D.; Iribarren, K.; Schramm, C.; Alifano, M.; Lupo, A.; Cherfils-Vicini, J.; Goc, J.; Katsahian, S.; et al. TLR7 promotes tumor progression, chemotherapy resistance, and poor clinical outcomes in non-small cell lung cancer. Cancer Res. 2014, 74, 5008–5018. [Google Scholar] [CrossRef]
- Kondo, Y.; Higa-Nakamine, S.; Noguchi, N.; Maeda, N.; Toku, S.; Isohama, Y.; Sugahara, K.; Kukita, I.; Yamamoto, H. Induction of epithelial-mesenchymal transition by flagellin in cultured lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2012, 303, 1057–1069. [Google Scholar] [CrossRef]
- Klimosch, S.N.; Foersti, A.; Eckert, J.; Knežević, J.; Bevier, M.; von Schoenfels, W.; Heits, N.; Walter, J.; Hinz, S.; Lascorz, J.; et al. Functional TLR5 Genetic Variants Affect Human Colorectal Cancer Survival. Cancer Res. 2013, 73, 7232LP–7242LP. [Google Scholar] [CrossRef]
- Curran, C.S.; Bolig, T.; Torabi-Parizi, P. Mechanisms and targeted therapies for pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 2018, 197, 708–727. [Google Scholar] [CrossRef]
- Ozretić, P.; da Silva Filho, M.I.; Catalano, C.; Sokolović, I.; Vukić-Dugac, A.; Šutić, M.; Kurtović, M.; Bubanović, G.; Popović-Grle, S.; Skrinjarić-Cincar, S.; et al. Association of NLRP1 Coding Polymorphism with Lung Function and Serum IL-1 β Concentration in Patients Diagnosed with Chronic Obstructive Pulmonary Disease (COPD). Genes 2019, 10, 783. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. Available online: http://www.jstor.org/stable/2346101 (accessed on 8 October 2020). [CrossRef]
- Berndt, A.; Leme, A.S.; Shapiro, S.D. Emerging genetics of COPD. EMBO Mol. Med. 2012, 4, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Lyle, E.A.; Omueti, K.O.; Stepensky, V.A.; Yegin, O.; Alpsoy, E.; Hamann, L.; Schumann, R.R.; Tapping, R.I. Cutting Edge: A Common Polymorphism Impairs Cell Surface Trafficking and Functional Responses of TLR1 but Protects against Leprosy. J. Immunol. 2007, 178, 7520LP–7524LP. [Google Scholar] [CrossRef] [PubMed]
- Al-Daghri, N.M.; Clerici, M.; Al-Attas, O.; Forni, D.; Alokail, M.S.; Alkharfy, K.M.; Sabico, S.; Mohammed, A.K.; Cagliani, R.; Sironi, M. A Nonsense Polymorphism (R392X) in TLR5 Protects from Obesity but Predisposes to Diabetes. J. Immunol. 2013, 190, 3716–3720. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.S.; Kwan, C.K.; Yau, S.; So, P.P.F.; Poon, P.C.M.; Au, J.S.K. The role of inflammation in the pathogenesis of lung cancer. Expert Opin. Ther. Targets 2011, 15, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Ritter, M.; Mennerich, D.; Weith, A.; Seither, P. Characterization of Toll-like receptors in primary lung epithelial cells: Strong impact of the TLR3 ligand poly(I:C) on the regulation of Toll-like receptors, adaptor proteins and inflammatory response. J. Inflamm. 2005, 2, 16. [Google Scholar] [CrossRef]
- Waldner, M.J.; Foersch, S.; Neurath, M.F. Interleukin-6—A key regulator of colorectal cancer development. Int. J. Biol. Sci. 2012, 8, 1248–1253. [Google Scholar] [CrossRef]
- Kutikhin, A.G. Association of polymorphisms in TLR genes and in genes of the Toll-like receptor signaling pathway with cancer risk. Hum. Immunol. 2011, 72, 1095–1116. [Google Scholar] [CrossRef]
- Mathieu, E.; Escribano-Vazquez, U.; Descamps, D.; Cherbuy, C.; Langella, P.; Riffault, S.; Remot, A.; Thomas, M. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front. Physiol. 2018, 9, 1168. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Lim, S.H. Bidirectional interaction between intestinal microbiome and cancer: Opportunities for therapeutic interventions. Biomark. Res. 2020, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, T.; Liang, J.; Huang, Q.; Huang, J.D.; Ke, Y.; Sun, H. Current status of intratumour microbiome in cancer and engineered exogenous microbiota as a promising therapeutic strategy. Biomed. Pharmacother. 2022, 145, 112443. [Google Scholar] [CrossRef]
- Sfondrini, L.; Rossini, A.; Besusso, D.; Merlo, A.; Tagliabue, E.; Mènard, S.; Balsari, A. Antitumor Activity of the TLR-5 Ligand Flagellin in Mouse Models of Cancer. J. Immunol. 2006, 176, 6624–6630. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, M.R.; Stephen, T.L.; Svoronos, N.; Allegrezza, M.J.; Tesone, A.J.; Perales-Puchalt, A.; Brencicova, E.; Escovar-Fadul, X.; Nguyen, J.M.; Cadungog, M.G.; et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell 2015, 27, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Groysman, L.; Carlsen, L.; Huntington, K.E.; Shen, W.H.; Zhou, L.; El-Deiry, W.S. Chemotherapy-induced cytokines and prognostic gene signatures vary across breast and colorectal cancer. Am. J. Cancer Res. 2021, 11, 6086–6106. Available online: https://pubmed.ncbi.nlm.nih.gov/35018244/0A (accessed on 19 January 2022).

| Subjects (N, %) | Cases (974) | COPD (500; 51.3%) | COPD + LC (280; 28.7%) | LC (194; 19.9%) | Controls (1283) |
|---|---|---|---|---|---|
| Age, year (SD 1) | 60 (9.58) | 59 (10.4) | 63 (7.38) | 62 (9.8) | 38 (12) |
| Sex (N, %) | |||||
| male | 671 (68.9) | 355 (71) | 205 (73) | 111 (57.3) | 1120 (87) |
| female | 303 (31.1) | 145 (29) | 75 (26) | 83 (42.7) | 163 (12.7) |
| Smoking status (N, %) | |||||
| active | 523 (50.8) | 139 (25.8) | 148 (52.5) | 53 (26.3) | 325 (25.3) |
| ex-smoker | 347 (33.6) | 317 (57.4) | 124 (43.9) | 90 (43.8) | 4 (0.2) |
| non | 104 (9.8) | 44 (8) | 8 (2.5) | 51 (25.3) | 954 (74.4) |
| CHARACTERISTICS | COPD (N = 500) | COPD + LC and LConly (N = 474) |
|---|---|---|
| FEV1 % (N) 1 | ||
| 80–120% | 55 | 235 |
| 50–80% | 207 | 185 |
| 30–50% | 148 | 42 |
| ≤30% | 90 | 12 |
| TYPE OF CANCER | ||
| Lung cancer | / | 474 |
| NSCLC/AdC | / | 225 |
| NSCLC/SQC | / | 168 |
| other | / | 81 |
| TNM-stage (N) | ||
| 1 | / | 118 |
| 2 | / | 100 |
| 3 | / | 102 |
| 4 | / | 77 |
| nonclasified | / | 77 |
| T stage (N) | ||
| 1 | / | 60 |
| 2 | / | 149 |
| 3 | / | 58 |
| 4 | / | 25 |
| nonclasified | / | 182 |
| N stage (N) | ||
| 0 | / | 153 |
| 1 | / | 55 |
| 2 | / | 109 |
| 3 | / | 8 |
| nonclasified | / | 149 |
| M stage (N) | ||
| 0 | / | 258 |
| 1 | / | 60 |
| nonclasified | / | 156 |
| SNP | Chr. | Position | Allele | Location | MAF 1 | Aa Change | Sift 2 | PolyPhen 3 |
|---|---|---|---|---|---|---|---|---|
| rs5744174 | 1 | 223284528 | C/T | Exon | 0.45 (T) | F616L | Tolerated | Benign |
| rs2072493 | 1 | 223284599 | A/G | Exon | 0.12 (G) | N592S | Tolerated | Benign |
| rs725084 | 1 | 223323951 | T/C | promotor | 0.44 (T) | / | / | / |
| COPD vs. Healthy | Lung Cancer vs. Healthy | NSCLC vs. Healthy | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | Cases (N, %) | Controls | OR (95% CI) | p Value | Cases (N, %) | Controls (N, %) | OR (95% CI) | p Value | Cases (N, %) | Controls (N, %) | OR (95% CI) | p Value |
| rs5744174_TC | T/T | 216 (33.4) | 381 (30.8) | 1 | 145 (33.2) | 381 (30.8) | 1 | 132 (32.5) | 381 (30.8) | 1 | |||
| (coding F616L) | T/C | 296 (45.7) | 606 (49) | 0.64 (0.32–1.28) | 0.206 | 210 (48) | 606 (49) | 1.09 (0.54–2.18) | 0.8073 | 199 (49) | 606 (49) | 1.33 (0.64–2.74) | 0.4384 |
| C/C | 135 (20.9) | 249 (20.2) | 0.97 (0.47–2.01) | 0.9382 | 82 (18.8) | 249 (20.2) | 0.38 (0.13–1.14) | 0.0851 | 75 (18.5) | 249 (20.2) | 0.46 (0.15–1.43) | 0.1803 | |
| T/C + C/C | 431 (66.6) | 855 (69.2) | 0.87 (0.51–1.51) | 0.6314 | 292 (66.8) | 855 (69.2) | 0.88 (0.45–1.71) | 0.6987 | 274 (67.5) | 855 (69.2) | 0.96 (0.56–1.64) | 0.8854 | |
| rs725084_TC | T/T | 208 (32.4) | 325 (26.6) | 1 | 124 (28.8) | 325 (26.6) | 1 | 111 (28.1) | 325 (26.6) | 1 | |||
| (promoter) | T/C | 301 (46.9) | 628 (51.4) | 0.85 (0.62–1.16) | 0.3126 | 217 (50) | 628 (51.4) | 1.12 (0.78–1.59) | 0.5431 | 201 (50.7) | 628 (51.4) | 1.07 (0.72–1.57) | 0.744 |
| C/C | 133 (20.7) | 269 (22) | 0.83 (0.57–1.22) | 0.3502 | 91 (21.1) | 269 (22) | 0.81 (0.52–1.26) | 0.3496 | 84 (21.2) | 269 (22) | 0.82 (0.51–1.33) | 0.4311 | |
| T/C + C/C | 434 (67.6) | 897 (73.4) | 0.85 (0.63–1.13) | 0.2635 | 308 (71.1) | 897 (73.4) | 1.02 (0.72–1.43) | 0.9213 | 285 (71.9) | 897 (73.4) | 0.99 (0.68–1.43) | 0.95 | |
| rs2072493_AG | A/A | 413 (64.5) | 967 (80) | 1 | 248 (57.7) | 967 (80) | 1 | 226 (57.1) | 967 (80) | 1 | |||
| (coding N592S) | A/G | 196 (30.6) | 226 (18.7) | 4.48 (2.34–8.57) | <0.0001 | 150 (34.9) | 226 (18.7) | 4.63 (2.06–10.39) | 0.0002 | 142 (35.8) | 226 (18.7) | 4.93 (2.12–11.49) | 0.0002 |
| G/G | 31 (4.9) | 15 (1.3) | 4.08 (0.72–23.13) | 0.1122 | 32 (7.4) | 15 (1.3) | 3.94 (0.77–20.14) | 0.0995 | 28 (7.1) | 15 (1.3) | 8.44 (1.25–56.85) | 0.0284 | |
| A/G + G/G | 227 (35.5) | 241 (20) | 4.41 (2.36–8.24) | <0.0001 | 182 (42.9) | 241 (20) | 4.61 (2.15–9.87) | 0.0001 | 170 (42.9) | 241 (20) | 5.17 (2.37–11.31) | <0.0001 | |
| NSCLC + COPD (Cases) vs. COPD (Controls) | |||||
|---|---|---|---|---|---|
| SNP | Genotype | Cases (N, %) | Controls (N, %) | OR (95% CI) | p Value |
| rs5744174_TC | T/T | 43 (17.7) | 88 (22.6) | 1 | |
| (coding F616L) | C/T | 118 (48.6) | 173 (44.3) | 1.41 (0.87–2.28) | 0.1558 |
| C/C | 82 (33.7) | 129 (33.1) | 1.43 (0.86–2.36) | 0.1594 | |
| C/T + C/C | 200 (82.3) | 302 (77.4) | 1.41 (0.90–2.19) | 0.1312 | |
| rs725084_TC | T/T | 45 (18.7) | 84 (21.6) | 1 | |
| (promoter) | C/T | 121 (50.4) | 174 (44.8) | 1.18 (0.73–1.91) | 0.4912 |
| C/C | 74 (30.9) | 130 (33.6) | 1.06 (0.64–1.73) | 0.8156 | |
| C/T + T/T | 195 (81.3) | 304 (78.4) | 1.13 (0.73–1.76) | 0.5683 | |
| rs2072493_AG | A/A | 144 (59.5) | 264 (68.6) | 1 | |
| (coding N592S) | A/G | 80 (33.1) | 110 (28.6) | 1.52 (1.02–2.25) | 0.0365 |
| G/G | 18 (7.4) | 11 (2.8) | 4.49 (1.9–10.61) | 0.0006 | |
| A/G + G/G | 98 (40.5) | 121 (31.4) | 1.75 (1.21–2.54) | 0.0031 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baranašić, J.; Šutić, M.; Catalano, C.; Drpa, G.; Huhn, S.; Majhen, D.; Nestić, D.; Kurtović, M.; Rumora, L.; Bosnar, M.; et al. TLR5 Variants Are Associated with the Risk for COPD and NSCLC Development, Better Overall Survival of the NSCLC Patients and Increased Chemosensitivity in the H1299 Cell Line. Biomedicines 2022, 10, 2240. https://doi.org/10.3390/biomedicines10092240
Baranašić J, Šutić M, Catalano C, Drpa G, Huhn S, Majhen D, Nestić D, Kurtović M, Rumora L, Bosnar M, et al. TLR5 Variants Are Associated with the Risk for COPD and NSCLC Development, Better Overall Survival of the NSCLC Patients and Increased Chemosensitivity in the H1299 Cell Line. Biomedicines. 2022; 10(9):2240. https://doi.org/10.3390/biomedicines10092240
Chicago/Turabian StyleBaranašić, Jurica, Maja Šutić, Calogerina Catalano, Gordana Drpa, Stefanie Huhn, Dragomira Majhen, Davor Nestić, Matea Kurtović, Lada Rumora, Martina Bosnar, and et al. 2022. "TLR5 Variants Are Associated with the Risk for COPD and NSCLC Development, Better Overall Survival of the NSCLC Patients and Increased Chemosensitivity in the H1299 Cell Line" Biomedicines 10, no. 9: 2240. https://doi.org/10.3390/biomedicines10092240
APA StyleBaranašić, J., Šutić, M., Catalano, C., Drpa, G., Huhn, S., Majhen, D., Nestić, D., Kurtović, M., Rumora, L., Bosnar, M., Vukić Dugac, A., Sokolović, I., Popovic-Grle, S., Oršolić, N., Škrinjarić-Cincar, S., Jakopović, M., Samaržija, M., Weber, A. N. R., Försti, A., & Knežević, J. (2022). TLR5 Variants Are Associated with the Risk for COPD and NSCLC Development, Better Overall Survival of the NSCLC Patients and Increased Chemosensitivity in the H1299 Cell Line. Biomedicines, 10(9), 2240. https://doi.org/10.3390/biomedicines10092240










