Epitope Detection in Monocytes (EDIM) As a New Method of Liquid Biopsy in Pediatric Rhabdomyosarcoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Immunohistochemistry
2.3. Immunofluorescence Microscopy
2.4. Preparation of Tumor Material
2.5. RNA Isolation and Real Time PCR (qRT-PCR)
- TKTL1 fw: CGCCGAGCACTGCATAAA;
- TKTL1 rev: CCACATAAGTGTTCCACCCAA A;
- DNaseX/Apo10 fw: AGCTGGTGTCTGTGAAGAGG;
- DNaseX/Apo10 rev: CCGTGTAGACCTCAACCAAC;
- TBP fw: GCC CGA AAC GCC GAA TAT;
- TBP rev: CCG TGG TTC GTG GCT CTC.
2.6. Flow Cytometry—Spike Experiments
2.7. Patient’s Characteristics
2.8. Blood Samples
2.9. Flow Cytometry Measurements—EDIM Blood Test
2.10. Evaluation of Correlation between TKTL1 Expression and DNA-Hypomethylation in RMS Cells
2.11. In Silico Analysis of the TKTL1 Promoter for Identification of CpG Islands
2.12. Bisulfite Sequencing (BS) of the TKTL1 Promoter in Tumor Material from RMS Patients
- TKTL1 fw: TTGTGTAGAGAAAGAAGATTTTG;
- TKTL1 rev: ACCCCTTTAAAATCTAAAAACCC.
2.13. Statistics
3. Results
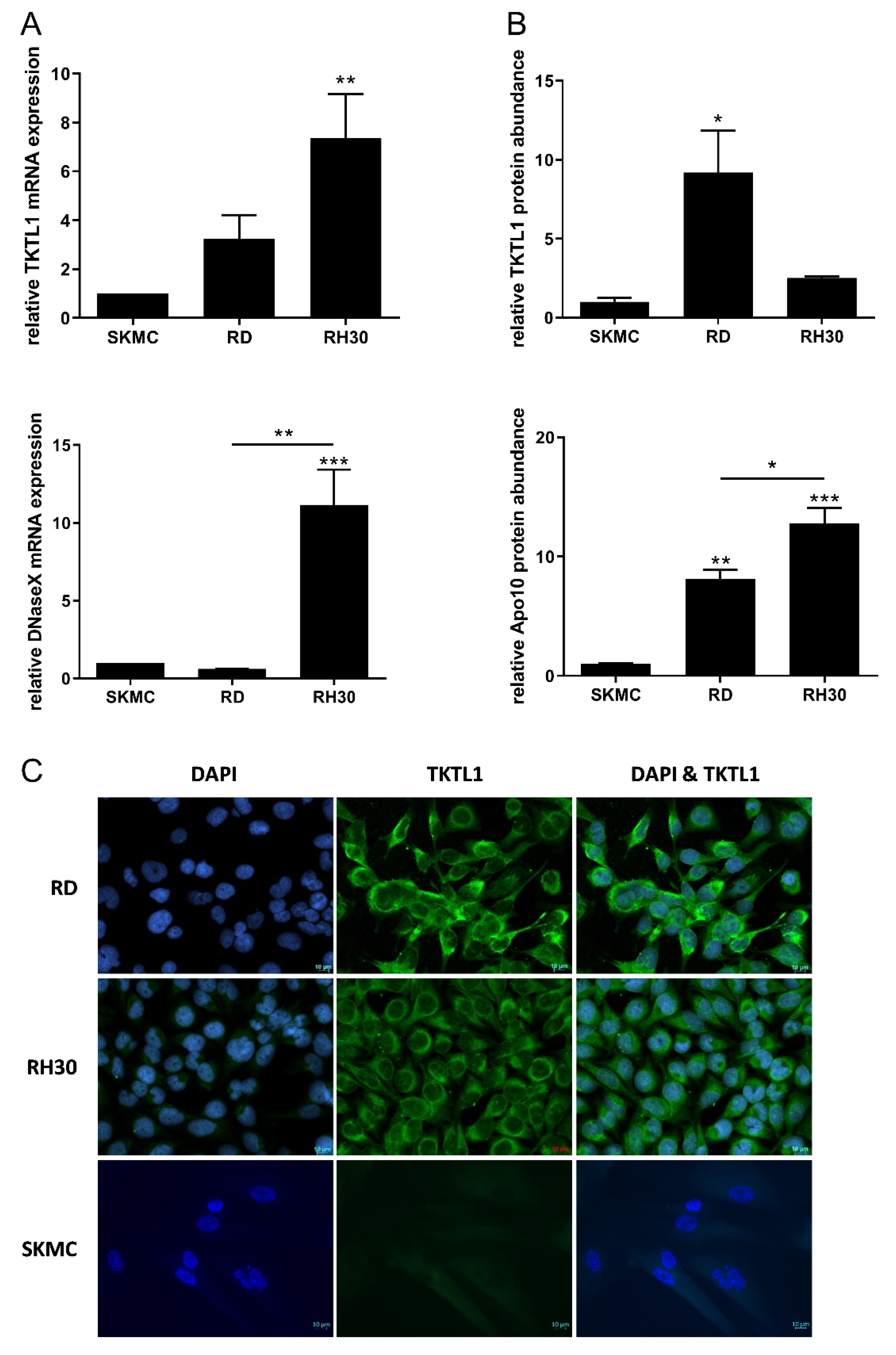
3.1. Expression of Biomarkers TKTL1 and Apo10 in RMS Cell Lines
3.2. Expression of Biomarkers TKTL1 and Apo10 in Tumor Material from Patients
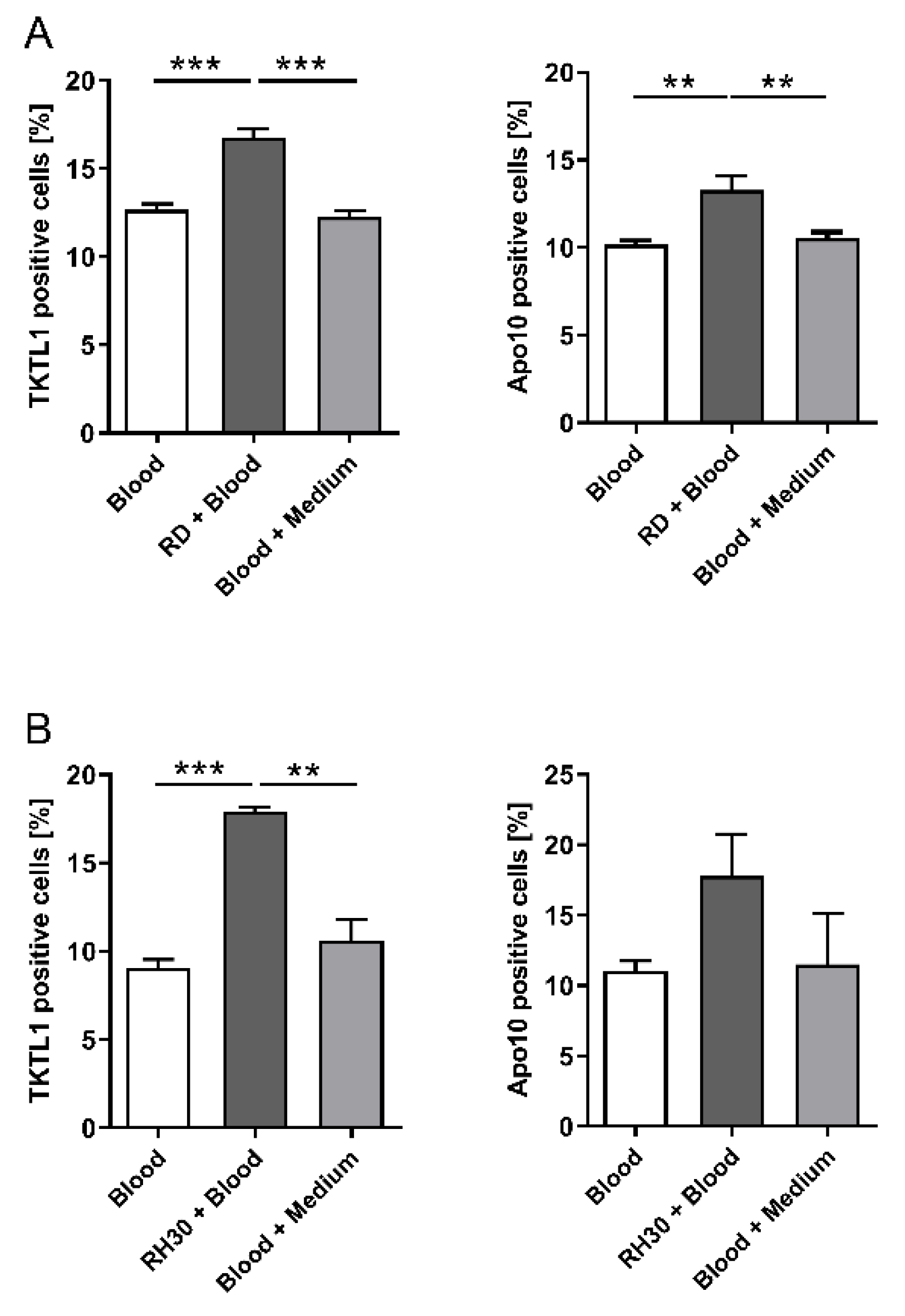
3.3. Co-Culture Experiments with Whole Blood and RMS Cell Lines
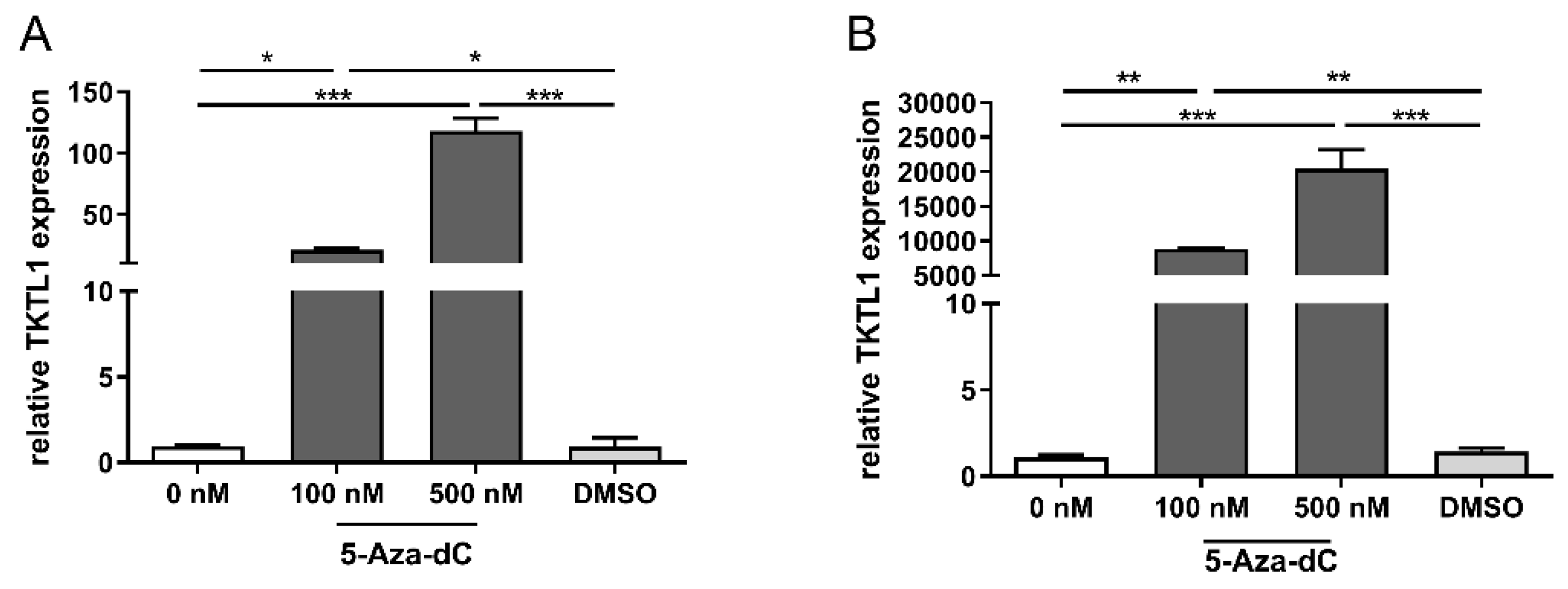
3.4. TKTL1 Expression and DNA-Hypomethylation in RMS Cells
3.5. TKTL1 Expression and DNA Hypomethylation in Tumor Material
3.6. EDIM Scores in Patients with RMS and Healthy Controls
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koscielniak, E.; Harms, D.; Henze, G.; Jurgens, H.; Gadner, H.; Herbst, M.; Klingebiel, T.; Schmidt, B.F.; Morgan, M.; Knietig, R.; et al. Results of treatment for soft tissue sarcoma in childhood and adolescence: A final report of the German Cooperative Soft Tissue Sarcoma Study CWS-86. J. Clin. Oncol. 1999, 17, 3706–3719. [Google Scholar] [CrossRef]
- Koscielniak, E.; Jurgens, H.; Winkler, K.; Burger, D.; Herbst, M.; Keim, M.; Bernhard, G.; Treuner, J. Treatment of soft tissue sarcoma in childhood and adolescence. A report of the German Cooperative Soft Tissue Sarcoma Study. Cancer 1992, 70, 2557–2567. [Google Scholar] [CrossRef]
- Dasgupta, R.; Fuchs, J.; Rodeberg, D. Rhabdomyosarcoma. Semin. Pediatr. Surg. 2016, 25, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Dantonello, T.M.; Int-Veen, C.; Schuck, A.; Seitz, G.; Leuschner, I.; Nathrath, M.; Schlegel, P.G.; Kontny, U.; Behnisch, W.; Veit-Friedrich, I.; et al. Survival following disease recurrence of primary localized alveolar rhabdomyosarcoma. Pediatr. Blood Cancer 2013, 60, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Hayes-Jordan, A.; Doherty, D.K.; West, S.D.; Raney, R.B.; Blakely, M.L.; Cox, C.S., Jr.; Andrassy, R.J.; Lally, K.P. Outcome after surgical resection of recurrent rhabdomyosarcoma. J. Pediatr. Surg. 2006, 41, 633–638. [Google Scholar] [CrossRef]
- Grimm, M.; Kraut, W.; Hoefert, S.; Krimmel, M.; Biegner, T.; Teriete, P.; Cetindis, M.; Polligkeit, J.; Kluba, S.; Munz, A.; et al. Evaluation of a biomarker based blood test for monitoring surgical resection of oral squamous cell carcinomas. Clin. Oral Investig. 2016, 20, 329–338. [Google Scholar] [CrossRef]
- Jansen, N.; Coy, J.F. Diagnostic use of epitope detection in monocytes blood test for early detection of colon cancer metastasis. Future Oncol. 2013, 9, 605–609. [Google Scholar] [CrossRef] [Green Version]
- Japink, D.; Leers, M.P.; Sosef, M.N.; Nap, M. CEA in activated macrophages. New diagnostic possibilities for tumor markers in early colorectal cancer. Anticancer Res. 2009, 29, 3245–3251. [Google Scholar]
- Feyen, O.; Coy, J.F.; Prasad, V.; Schierl, R.; Saenger, J.; Baum, R.P. EDIM-TKTL1 blood test: A noninvasive method to detect upregulated glucose metabolism in patients with malignancies. Future Oncol. 2012, 8, 1349–1359. [Google Scholar] [CrossRef] [Green Version]
- Grimm, M.; Schmitt, S.; Teriete, P.; Biegner, T.; Stenzl, A.; Hennenlotter, J.; Muhs, H.J.; Munz, A.; Nadtotschi, T.; Konig, K.; et al. A biomarker based detection and characterization of carcinomas exploiting two fundamental biophysical mechanisms in mammalian cells. BMC Cancer 2013, 13, 569. [Google Scholar] [CrossRef]
- Coy, J.F. EDIM-TKTL1/Apo10 Blood Test: An Innate Immune System Based Liquid Biopsy for the Early Detection, Characterization and Targeted Treatment of Cancer. Int. J. Mol. Sci. 2017, 18, 878. [Google Scholar] [CrossRef] [Green Version]
- Saman, S.; Stagno, M.J.; Warmann, S.W.; Malek, N.P.; Plentz, R.R.; Schmid, E. Biomarkers Apo10 and TKTL1: Epitope-detection in monocytes (EDIM) as a new diagnostic approach for cholangiocellular, pancreatic and colorectal carcinoma. Cancer Biomark. 2020, 27, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, I.M.; Glazer, C.A.; Mithani, S.K.; Ochs, M.F.; Sun, W.; Bhan, S.; Vostrov, A.; Abdullaev, Z.; Lobanenkov, V.; Gray, A.; et al. Coordinated activation of candidate proto-oncogenes and cancer testes antigens via promoter demethylation in head and neck cancer and lung cancer. PLoS ONE 2009, 4, e4961. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, A.; Lo, P.H.; Chueh, A.C.; Prithviraj, P.; Molania, R.; Davalos-Salas, M.; Anaka, M.; Walkiewicz, M.; Cebon, J.; Behren, A. Transketolase-like 1 ectopic expression is associated with DNA hypomethylation and induces the Warburg effect in melanoma cells. BMC Cancer 2016, 16, 134. [Google Scholar] [CrossRef] [Green Version]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Moralli, S.; Aguilar, E.; Marin, S.; Coy, J.F.; Dewerchin, M.; Antoniewicz, M.R.; Meca-Cortés, O.; Notebaert, L.; Ghesquière, B.; Eelen, G.; et al. A key role for transketolase-like 1 in tumor metabolic reprogramming. Oncotarget 2016, 7, 51875–51897. [Google Scholar] [CrossRef]
- Li, Y.; Yao, C.F.; Xu, F.J.; Qu, Y.Y.; Li, J.T.; Lin, Y.; Cao, Z.L.; Lin, P.C.; Xu, W.; Zhao, S.M.; et al. APC/C(CDH1) synchronizes ribose-5-phosphate levels and DNA synthesis to cell cycle progression. Nat. Commun. 2019, 10, 2502. [Google Scholar] [CrossRef]
- Langbein, S.; Zerilli, M.; Zur Hausen, A.; Staiger, W.; Rensch-Boschert, K.; Lukan, N.; Popa, J.; Ternullo, M.P.; Steidler, A.; Weiss, C.; et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br. J. Cancer 2006, 94, 578–585. [Google Scholar] [CrossRef] [Green Version]
- Schwaab, J.; Horisberger, K.; Strobel, P.; Bohn, B.; Gencer, D.; Kahler, G.; Kienle, P.; Post, S.; Wenz, F.; Hofmann, W.K.; et al. Expression of Transketolase like gene 1 (TKTL1) predicts disease-free survival in patients with locally advanced rectal cancer receiving neoadjuvant chemoradiotherapy. BMC Cancer 2011, 11, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayser, G.; Sienel, W.; Kubitz, B.; Mattern, D.; Stickeler, E.; Passlick, B.; Werner, M.; Zur Hausen, A. Poor outcome in primary non-small cell lung cancers is predicted by transketolase TKTL1 expression. Pathology 2011, 43, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Ahopelto, K.; Böckelman, C.; Hagström, J.; Koskensalo, S.; Haglund, C. Transketolase-like protein 1 expression predicts poor prognosis in colorectal cancer. Cancer Biol. Ther. 2016, 17, 163–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Liu, D.; He, G. TKTL1 and p63 are biomarkers for the poor prognosis of gastric cancer patients. Cancer Biomark. 2015, 15, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.; Cetindis, M.; Lehmann, M.; Biegner, T.; Munz, A.; Teriete, P.; Reinert, S. Apoptosis resistance-related ABCB5 and DNaseX (Apo10) expression in oral carcinogenesis. Acta Odontol. Scand. 2015, 73, 336–342. [Google Scholar] [CrossRef]
- Stegmaier, S.; Sparber-Sauer, M.; Aakcha-Rudel, E.; Münch, P.; Reeh, T.; Feuchtgruber, S.; Hallmen, E.; Blattmann, C.; Bielack, S.; Klingebiel, T.; et al. Fusion transcripts as liquid biopsy markers in alveolar rhabdomyosarcoma and synovial sarcoma: A report of the Cooperative Weichteilsarkom Studiengruppe (CWS). Pediatr. Blood Cancer 2022, 69, e29652. [Google Scholar] [CrossRef] [PubMed]
- Regenbogen, S.; Stagno, M.J.; Schleicher, S.; Schilbach, K.; Bösmüller, H.; Fuchs, J.; Schmid, E.; Seitz, G. Cytotoxic drugs in combination with the CXCR4 antagonist AMD3100 as a potential treatment option for pediatric rhabdomyosarcoma. Int. J. Oncol. 2020, 57, 289–300. [Google Scholar] [CrossRef]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, A.L.; Chuang, A.; Jiang, W.W.; Lee, J.; Begum, S.; Poeta, L.; Zhao, M.; Jeronimo, C.; Henrique, R.; Nayak, C.S.; et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006, 66, 9401–9407. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Liu, Y.; Glazer, C.A.; Shao, C.; Bhan, S.; Demokan, S.; Zhao, M.; Rudek, M.A.; Ha, P.K.; Califano, J.A. TKTL1 is activated by promoter hypomethylation and contributes to head and neck squamous cell carcinoma carcinogenesis through increased aerobic glycolysis and HIF1alpha stabilization. Clin. Cancer Res. 2010, 16, 857–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herwig, R.; Horninger, W.; Rehder, P.; Klocker, H.; Ramoner, R.; Thurnher, M.; Pinggera, G.M.; Gozzi, C.; Konwalinka, G.; Bartsch, G. Ability of PSA-positive circulating macrophages to detect prostate cancer. Prostate 2005, 62, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.H.; Yang, J.H.; Zhang, D.T.; Zhang, S.; Wang, L.; Cai, P.C.; Zheng, J.F.; Huang, J.S. The TKTL1 gene influences total transketolase activity and cell proliferation in human colon cancer LoVo cells. Anticancer Drugs 2007, 18, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Foldi, M.; Stickeler, E.; Bau, L.; Kretz, O.; Watermann, D.; Gitsch, G.; Kayser, G.; Zur Hausen, A.; Coy, J.F. Transketolase protein TKTL1 overexpression: A potential biomarker and therapeutic target in breast cancer. Oncol. Rep. 2007, 17, 841–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volker, H.U.; Scheich, M.; Schmausser, B.; Kammerer, U.; Eck, M. Overexpression of transketolase TKTL1 is associated with shorter survival in laryngeal squamous cell carcinomas. Eur. Arch. Otorhinolaryngol. 2007, 264, 1431–1436. [Google Scholar] [CrossRef]
- Todenhofer, T.; Hennenlotter, J.; Keller, G.; Neumann, T.; Stenzl, A.; Bedke, J. Effect of radical prostatectomy on levels of cancer related epitopes in circulating macrophages of patients with clinically localized prostate cancer. Prostate 2017, 77, 1251–1258. [Google Scholar] [CrossRef]
- Hughes, P.; Marshall, D.; Reid, Y.; Parkes, H.; Gelber, C. The costs of using unauthenticated, over-passaged cell lines: How much more data do we need? Biotechniques 2007, 43, 575–586. [Google Scholar] [CrossRef]
- Wagner, J.R.; Busche, S.; Ge, B.; Kwan, T.; Pastinen, T.; Blanchette, M. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014, 15, R37. [Google Scholar] [CrossRef] [Green Version]
- Feinberg, A.P.; Tycko, B. The history of cancer epigenetics. Nat. Rev. Cancer 2004, 4, 143–153. [Google Scholar] [CrossRef]
- Hanada, M.; Delia, D.; Aiello, A.; Stadtmauer, E.; Reed, J.C. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood 1993, 82, 1820–1828. [Google Scholar] [CrossRef]
- Nishigaki, M.; Aoyagi, K.; Danjoh, I.; Fukaya, M.; Yanagihara, K.; Sakamoto, H.; Yoshida, T.; Sasaki, H. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 2005, 65, 2115–2124. [Google Scholar] [CrossRef] [Green Version]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Los, M.; Neubuser, D.; Coy, J.F.; Mozoluk, M.; Poustka, A.; Schulze-Osthoff, K. Functional characterization of DNase X, a novel endonuclease expressed in muscle cells. Biochemistry 2000, 39, 7365–7373. [Google Scholar] [CrossRef] [Green Version]
- Grimm, M.; Hoefert, S.; Krimmel, M.; Biegner, T.; Feyen, O.; Teriete, P.; Reinert, S. Monitoring carcinogenesis in a case of oral squamous cell carcinoma using a panel of new metabolic blood biomarkers as liquid biopsies. Oral Maxillofac. Surg. 2016, 20, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xia, W.; Lv, Z.; Ni, C.; Xin, Y.; Yang, L. Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cell. Physiol. Biochem. 2017, 41, 755–768. [Google Scholar] [CrossRef]
- Weiser, D.A.; West-Szymanski, D.C.; Fraint, E.; Weiner, S.; Rivas, M.A.; Zhao, C.W.T.; He, C.; Applebaum, M.A. Progress toward liquid biopsies in pediatric solid tumors. Cancer Metastasis Rev. 2019, 38, 553–571. [Google Scholar] [CrossRef]



| Characteristics | Number of Patients n = 29 (%) |
|---|---|
| Age | |
| ≥10 years | 5 (17%) |
| <10 years | 24 (83%) |
| Sex | |
| Female | 12 (41%) |
| Male | 17 (59%) |
| Tumor localization | |
| Favorable | 3 (10%) |
| Unfavorable | 26 (90%) |
| Tumor size | |
| ≥5 cm | 18 (62%) |
| <5 cm | 11(38%) |
| IRS Group | |
| I | 0 (0%) |
| II | 1 (3%) |
| III | 21 (72%) |
| IV | 7 (25%) |
| Histology | |
| ARMS | 5 (17%) |
| ERMS | 24 (83%) |
| Variable | N | EDIM-TKTL1 | EDIM-Apo10 | EDIM-TKTL1/Apo10 | |||
|---|---|---|---|---|---|---|---|
| Mean ± SE | p | Mean ± SE | p | Mean ± SE | p | ||
| Histology | |||||||
| ARMS | 5 | 135 ± 7 | 0.431 | 163.8 ± 10 | 0.952 | 299 ± 12.3 | 0.829 |
| ERMS | 24 | 141.6 ± 4.2 | 157.5 ± 4.3 | 299.1 ± 6.3 | |||
| Age | |||||||
| <10 years | 24 | 140 ± 4 | 0.887 | 159.4 ± 4.2 | 0.318 | 299.5 ± 5.8 | 0.172 |
| ≥10 years | 5 | 141.8 ± 8.2 | 155 ± 11.6 | 296.8 ± 17.9 | |||
| Size | |||||||
| <5 cm | 11 | 141.8 ± 4.8 | 0.521 | 155.5 ± 6.5 | 0.659 | 297.36 ± 7.9 | 0.715 |
| ≥5 cm | 18 | 139.6 ± 5.1 | 160.5 ± 4.9 | 300.1 ± 7.7 | |||
| Localization | |||||||
| Favorable | 3 | 128 ± 2.3 | 0.076 | 157.3 ± 15.3 | 0.772 | 285.3 ± 13 | 0.532 |
| Unfavorable | 26 | 141.8 ± 3.9 | 158.7 ± 4.1 | 300.6 ± 6 | |||
| IRS Group | |||||||
| I–III | 22 | 138.8 ± 3.4 | 0.089 | 157 ± 4.7 | 0.417 | 295.8 ± 6 | 0.622 |
| IV | 7 | 145.6 ± 11 | 163 ± 6.9 | 309.3 ± 13.5 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urla, C.; Stagno, M.J.; Schmidt, A.; Handgretinger, R.; Fuchs, J.; Warmann, S.W.; Schmid, E. Epitope Detection in Monocytes (EDIM) As a New Method of Liquid Biopsy in Pediatric Rhabdomyosarcoma. Biomedicines 2022, 10, 1812. https://doi.org/10.3390/biomedicines10081812
Urla C, Stagno MJ, Schmidt A, Handgretinger R, Fuchs J, Warmann SW, Schmid E. Epitope Detection in Monocytes (EDIM) As a New Method of Liquid Biopsy in Pediatric Rhabdomyosarcoma. Biomedicines. 2022; 10(8):1812. https://doi.org/10.3390/biomedicines10081812
Chicago/Turabian StyleUrla, Cristian, Matias Julian Stagno, Andreas Schmidt, Rupert Handgretinger, Jörg Fuchs, Steven W. Warmann, and Evi Schmid. 2022. "Epitope Detection in Monocytes (EDIM) As a New Method of Liquid Biopsy in Pediatric Rhabdomyosarcoma" Biomedicines 10, no. 8: 1812. https://doi.org/10.3390/biomedicines10081812
APA StyleUrla, C., Stagno, M. J., Schmidt, A., Handgretinger, R., Fuchs, J., Warmann, S. W., & Schmid, E. (2022). Epitope Detection in Monocytes (EDIM) As a New Method of Liquid Biopsy in Pediatric Rhabdomyosarcoma. Biomedicines, 10(8), 1812. https://doi.org/10.3390/biomedicines10081812






