Effects of Extracellular Vesicles Secreted by TGFβ-Stimulated Umbilical Cord Mesenchymal Stem Cells on Skin Fibroblasts by Promoting Fibroblast Migration and ECM Protein Production
Abstract
:1. Introduction
2. Materials and Methods
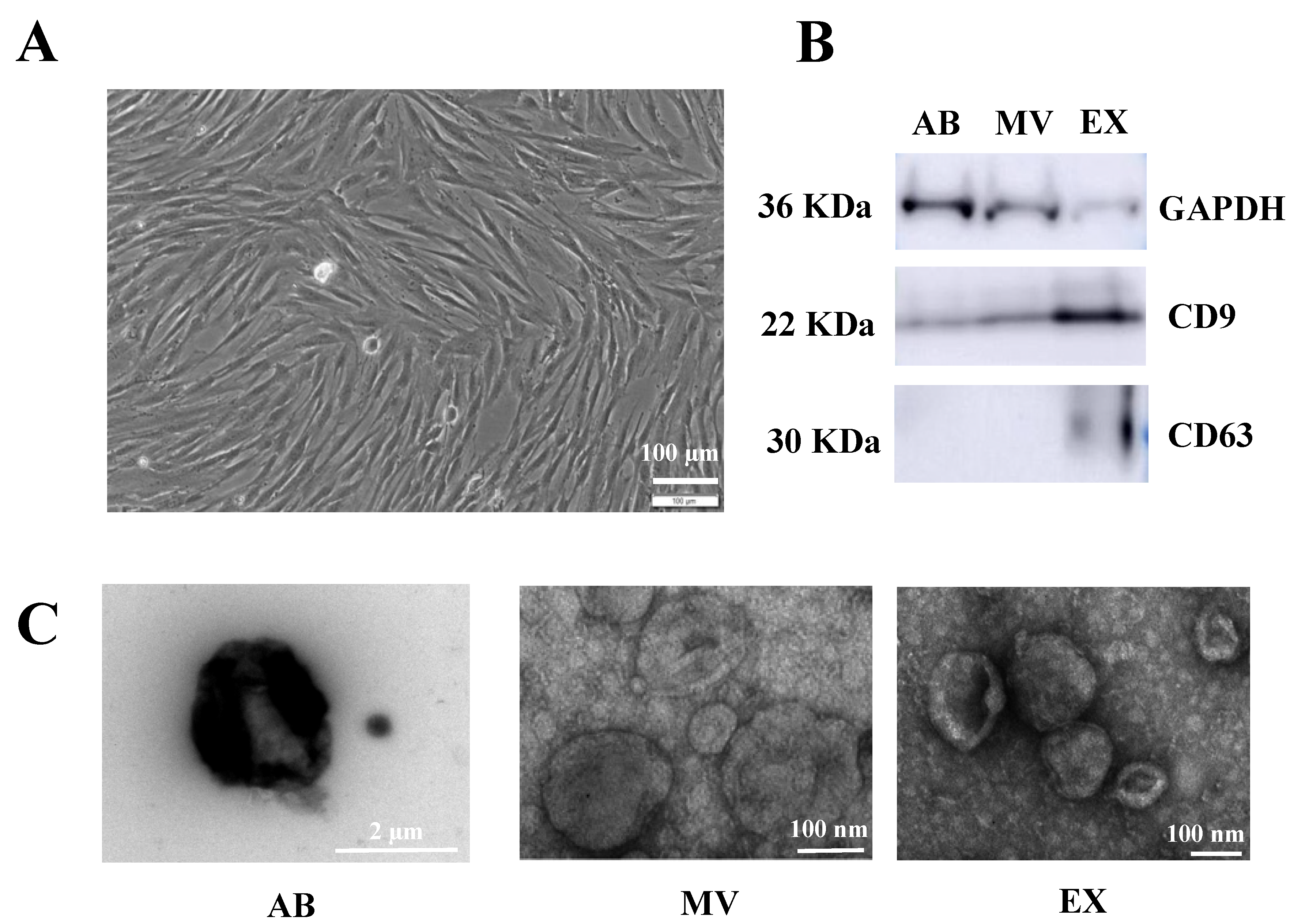
2.1. Umbilical Cord Mesenchymal Stem Cells Isolation and Culture
2.2. EV Isolation
2.3. Western Blot
2.4. Transmission Electron Microscopy (TEM)
2.5. Luminex Assay
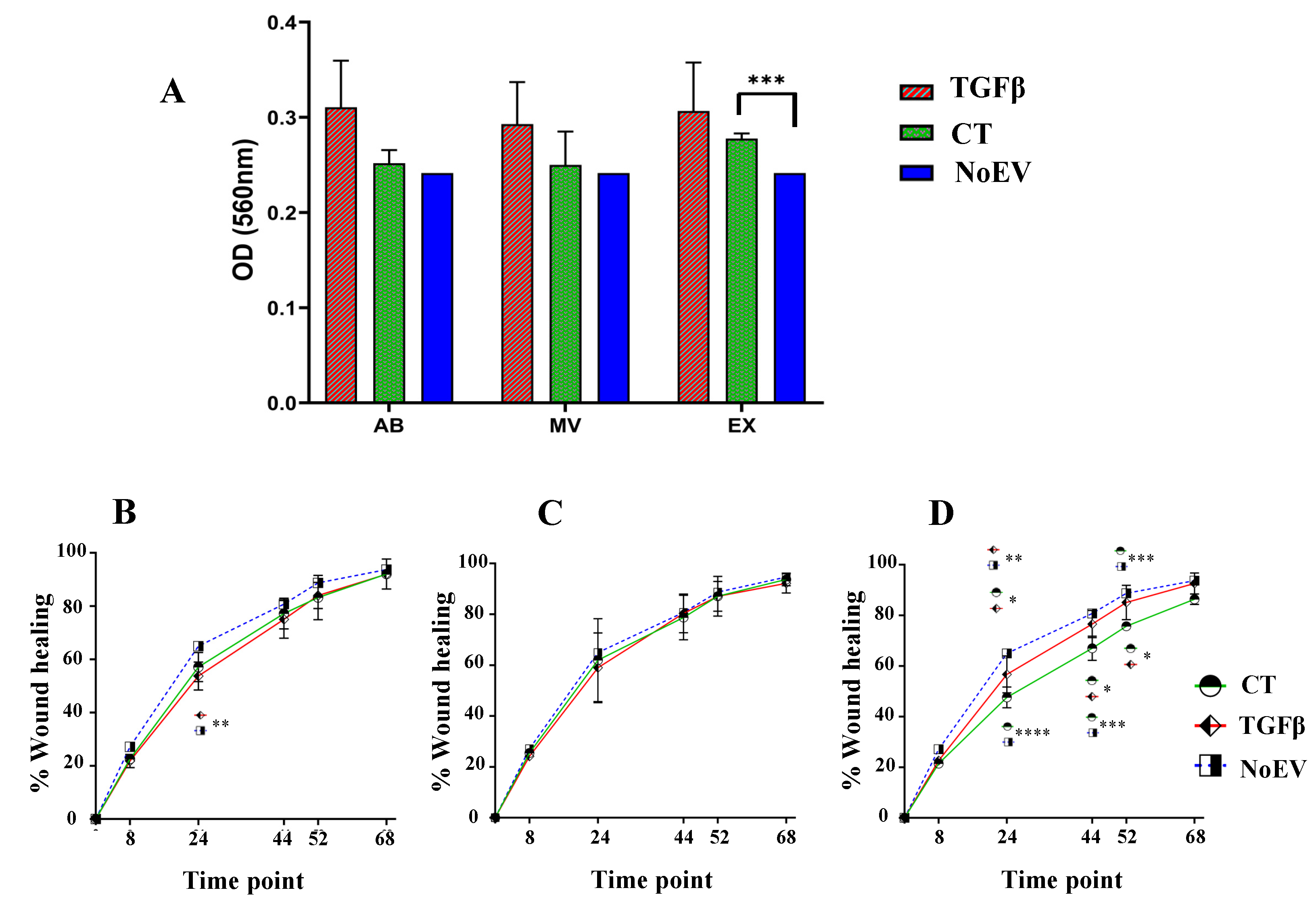
2.6. Proliferation Assay
2.7. Migration Assay
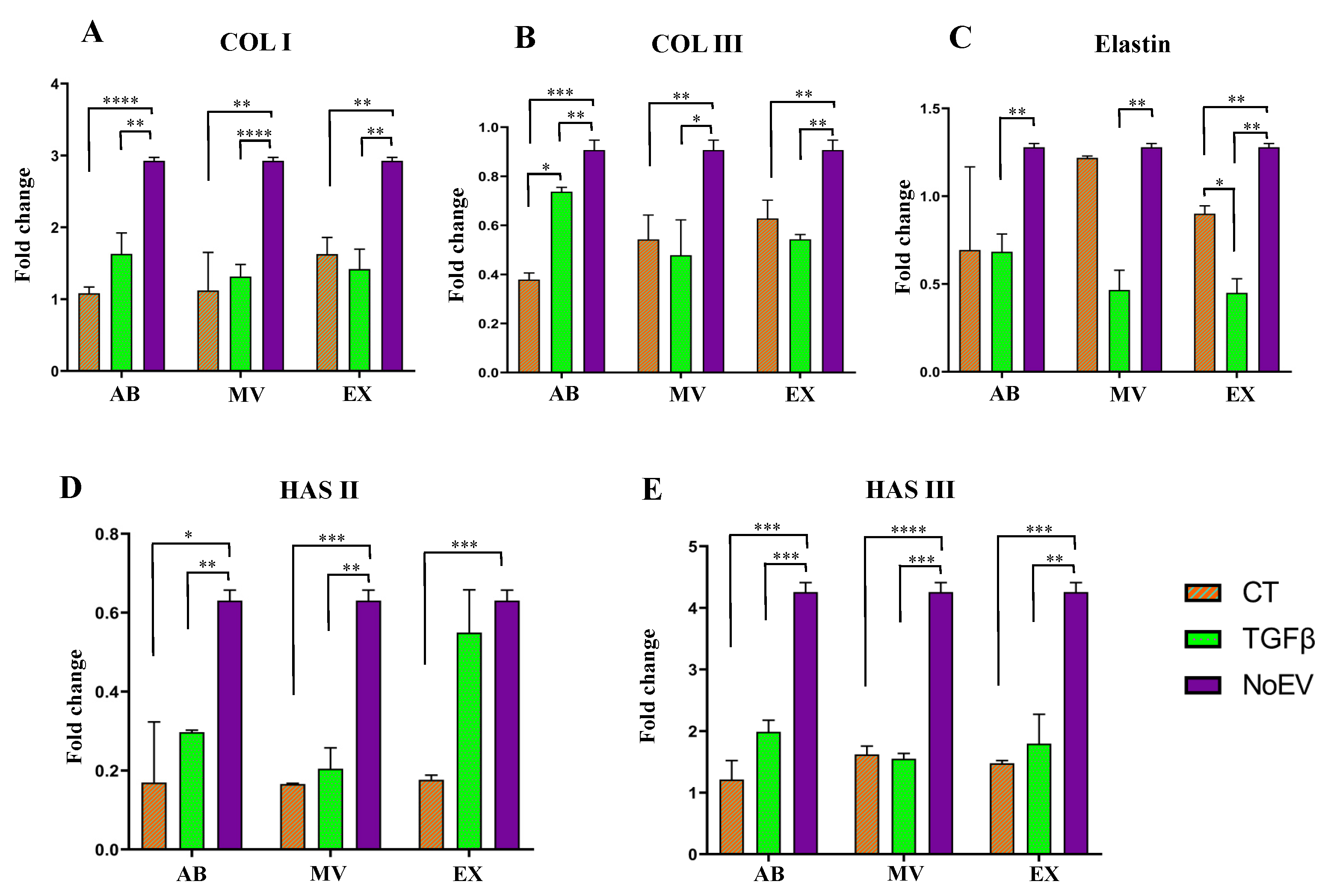
2.8. qRT-PCR to Detect Collagen Type I and Hyaluronic Acid
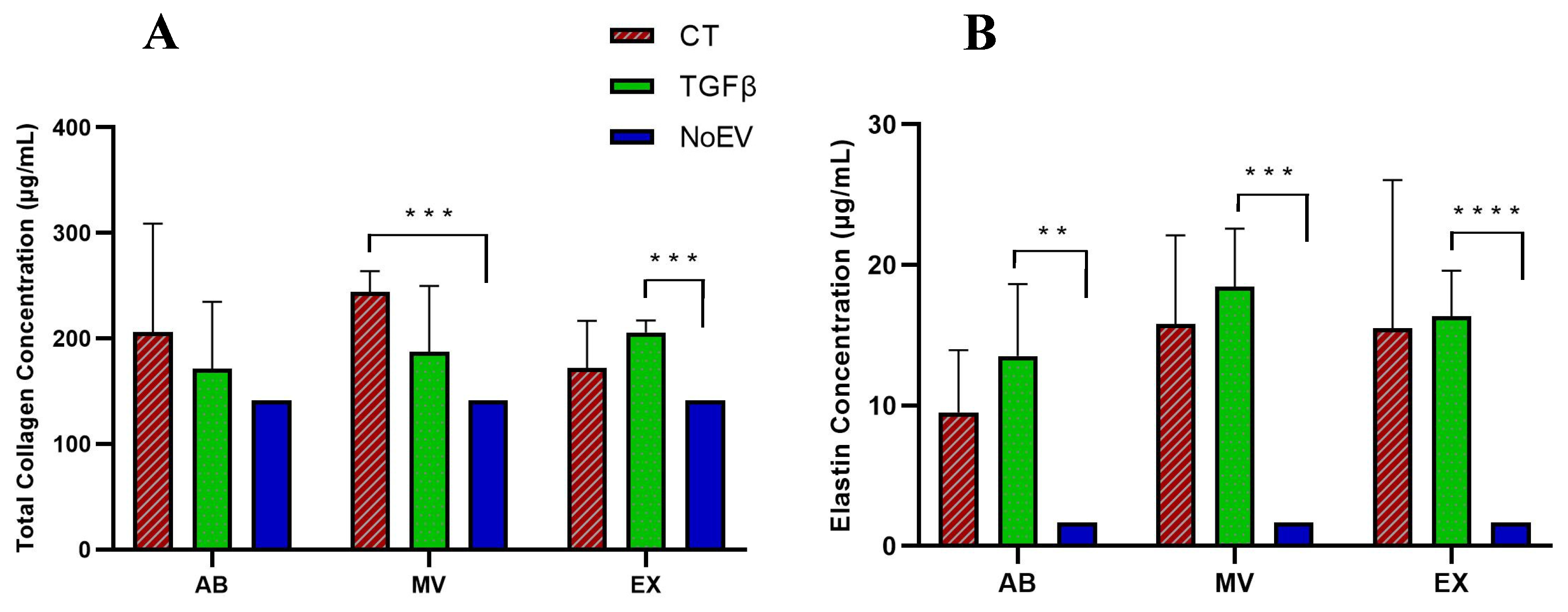
2.9. Elastin Detection Assay
2.10. Collagen Detection Assay
3. Results
3.1. Characteristics of EVs Originating from TGFβ-Stimulated UCMSCs
3.2. Growth Factor Expression in EVs Released by TGFβ-Stimulated UCMSCs
3.3. Capacity of EVs from TGFβ-Stimulated UCMSCs in Inducing Fibroblast Proliferation and Migration
3.4. Capacity of EVs from TGFβ-Stimulated UCMSCs to ECM Gene Expression by Dermal Fibroblasts
3.5. Capacity of EVs from TGFβ-Stimulated UCMSCs to Collagen and Elastin Protein Production by Dermal Fibroblasts
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cole, M.A.; Quan, T.; Voorhees, J.J.; Fisher, G.J. Extracellular matrix regulation of fibroblast function: Redefining our perspective on skin aging. J. Cell Commun. Signal. 2018, 12, 35–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavagnino, M.; Gardner, K.; Arnoczky, S.P. Age-related changes in the cellular, mechanical, and contractile properties of rat tail tendons. Connect. Tissue Res. 2013, 54, 70–75. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.C.; Hill, R.C.; Calderone, K.; Cui, Y.; Yan, Y.; Quan, T.; Fisher, G.J.; Hansen, K.C. Alterations in extracellular matrix composition during aging and photoaging of the skin. Matrix Biol. Plus 2020, 8, 100041. [Google Scholar] [CrossRef] [PubMed]
- Naylor, E.C.; Watson, R.E.; Sherratt, M.J. Molecular aspects of skin ageing. Maturitas 2011, 69, 249–256. [Google Scholar] [CrossRef]
- Baumann, L. Skin ageing and its treatment. J. Pathol. 2007, 211, 241–251. [Google Scholar] [CrossRef]
- Papakonstantinou, E.; Roth, M.; Karakiulakis, G. Hyaluronic acid: A key molecule in skin aging. Dermatoendocrinology 2012, 4, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-J.; Yoo, S.M.; Park, H.H.; Lim, H.J.; Kim, Y.-L.; Lee, S.; Seo, K.-W.; Kang, K.-S. Exosomes derived from human umbilical cord blood mesenchymal stem cells stimulates rejuvenation of human skin. Biochem. Biophys. Res. Commun. 2017, 493, 1102–1108. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Seo, D.H.; Lee, S.H.; Lee, S.-H.; An, G.-H.; Ahn, H.-J.; Kwon, D.; Seo, K.-W.; Kang, K.-S. Conditioned media from human umbilical cord blood-derived mesenchymal stem cells stimulate rejuvenation function in human skin. Biochem. Biophys. Rep. 2018, 16, 96–102. [Google Scholar] [CrossRef]
- Than, U.T.T.; Guanzon, D.; Leavesley, D.; Parker, T. Association of extracellular membrane vesicles with cutaneous wound healing. Int. J. Mol. Sci. 2017, 18, 956. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wang, M.; Gong, A.; Zhang, X.; Wu, X.; Zhu, Y.; Shi, H.; Wu, L.; Zhu, W.; Qian, H.; et al. HucMSC-exosome mediated-Wnt4 signaling is required for cutaneous wound healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, X.; Zhang, X.; Sun, Y.; Yan, Y.; Shi, H.; Zhu, Y.; Wu, L.; Pan, Z.; Zhu, W. Human umbilical cord mesenchymal stem cell exosomes enhance angiogenesis through the Wnt4/β-catenin pathway. Stem Cells Transl. Med. 2015, 4, 513–522. [Google Scholar] [CrossRef]
- Kim, W.-S.; Park, B.-S.; Park, S.-H.; Kim, H.-K.; Sung, J.-H. Antiwrinkle effect of adipose-derived stem cell: Activation of dermal fibroblast by secretory factors. J. Dermatol. Sci. 2009, 53, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.Y.; Liu, H.B.; Liu, H.W.; Zhu, Q.W. Mesenchymal Stem Cells Stimulated by Growth Factors Release Exosomes with Potent Proangiogenic Activity. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2018, 26, 1538–1542. (In Chinese) [Google Scholar] [CrossRef]
- Hu, L.; Wang, J.; Zhou, X.; Xiong, Z.; Zhao, J.; Yu, R.; Huang, F.; Zhang, H.; Chen, L. Exosomes derived from human adipose mensenchymal stem cells accelerates cutaneous wound healing via optimizing the characteristics of fibroblasts. Sci. Rep. 2016, 6, 32993. [Google Scholar] [CrossRef]
- Beanes, S.R.; Dang, C.; Soo, C.; Ting, K. Skin repair and scar formation: The central role of TGF-β. Expert Rev. Mol. Med. 2004, 5, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhou, Y.; Ye, Z.; Tan, W.-S. Transforming growth factor-β1 stimulates mesenchymal stem cell proliferation by altering cell cycle through FAK-Akt-mTOR pathway. Connect. Tissue Res. 2019, 60, 406–417. [Google Scholar] [CrossRef] [PubMed]
- De Araújo Farias, V.; Carrillo-Gálvez, A.B.; Martín, F.; Anderson, P. TGF-β and mesenchymal stromal cells in regenerative medicine, autoimmunity and cancer. Cytokine Growth Factor Rev. 2018, 43, 25–37. [Google Scholar] [CrossRef]
- Hermansyah, D.; Putra, A.; Muhar, A.; Retnaningsih, R.; Wirastuti, K.; Dirja, B. Mesenchymal stem cells suppress TGF-β release to decrease α-SMA expression in ameliorating CCl4-induced liver fibrosis. Med. Arch. 2021, 75, 16–22. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Duong, C.M.; Nguyen, X.-H.; Than, U.T.T. Mesenchymal stem cell-derived extracellular vesicles for osteoarthritis treatment: Extracellular matrix protection, chondrocyte and osteocyte physiology, pain and inflammation management. Cells 2021, 10, 2887. [Google Scholar] [CrossRef]
- Hoang, D.H.; Nguyen, T.D.; Nguyen, H.-P.; Nguyen, X.-H.; Do, P.T.X.; Dang, V.D.; Dam, P.T.M.; Bui, H.T.H.; Trinh, M.Q.; Vu, D.M.; et al. Differential wound healing capacity of mesenchymal stem cell-derived exosomes originated from bone marrow, adipose tissue and umbilical cord under serum- and xeno-free condition. Front. Mol. Biosci. 2020, 7, 119. [Google Scholar] [CrossRef]
- Than, U.T.; Guanzon, D.; Broadbent, J.A.; Leavesley, D.I.; Salomon, C.; Parker, T.J. Differential expression of keratinocyte-derived extracellular vesicle mirnas discriminate exosomes from apoptotic bodies and microvesicles. Front. Endocrinol. 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, H.; Chen, M.; Greening, D.; He, W.; Rai, A.; Zhang, W.; Simpson, R.J. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS ONE 2014, 9, e110314. [Google Scholar] [CrossRef] [Green Version]
- Grafe, I.; Alexander, S.; Peterson, J.R.; Snider, T.N.; Levi, B.; Lee, B.; Mishina, Y. TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb. Perspect. Biol. 2018, 10, a022202. [Google Scholar] [CrossRef] [PubMed]
- Than, U.T.T.; Guanzon, D.; Broadbent, J.A.; Parker, T.J.; Leavesley, D.I. Deep sequencing microRNAs from extracellular membrane vesicles revealed the association of the vesicle cargo with cellular origin. Int. J. Mol. Sci. 2020, 21, 1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Kou, X.; Chen, C.; Liu, S.; Liu, Y.; Yu, W.; Yu, T.; Yang, R.; Wang, R.; Zhou, Y.; et al. Circulating apoptotic bodies maintain mesenchymal stem cell homeostasis and ameliorate osteopenia via transferring multiple cellular factors. Cell Res. 2018, 28, 918–933. [Google Scholar] [CrossRef] [Green Version]
- Bruno, S.; Grange, C.; Deregibus, M.C.; Calogero, R.A.; Saviozzi, S.; Collino, F.; Morando, L.; Busca, A.; Falda, M.; Bussolati, B.; et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 2009, 20, 1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, Q.; Qi, L.; Dai, X.; Liu, H.; Wang, Y. Low levels of TGF-β1 enhance human umbilical cord-derived mesenchymal stem cell fibronectin production and extend survival time in a rat model of lipopolysaccharide-induced acute lung injury. Mol. Med. Rep. 2016, 14, 1681–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene name | Primer | Sequence | Size (bp) |
|---|---|---|---|
| COL I | Forward | CCTCAAGGGCTCCAACGAG | 117 |
| Reverse | TCAATCACTGTCTTGCCCCA | ||
| COL III | Forward | GAAGGGCAGGGAACAACTTG | 243 |
| Reverse | TTTGGCATGGTTCTGGCTTC | ||
| Elastin | Forward | GGCCATTCCTGGTGGAGTTCC | 106 |
| Reverse | AACTGGCTTAAGAGGTTTGCCTCCA | ||
| HAS II | Forward | ATGGGCAGAGACAAAT CAGC | 249 |
| Reverse | GGCTGGGTCAAGCATAGTGT | ||
| HAS III | Forward | AGCACCTTCTCGTGCATCAT | 159 |
| Reverse | CTCCAGGACTCGAAGCATCT | ||
| GAPDH | Forward | GGTGTGAACCATGAGAAGTATGA | 123 |
| Reverse | GAGTCCTTCCACGATACC AAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, D.M.; Nguyen, V.-T.; Nguyen, T.H.; Do, P.T.X.; Dao, H.H.; Hai, D.X.; Le, N.T.; Nguyen, X.-H.; Than, U.T.T. Effects of Extracellular Vesicles Secreted by TGFβ-Stimulated Umbilical Cord Mesenchymal Stem Cells on Skin Fibroblasts by Promoting Fibroblast Migration and ECM Protein Production. Biomedicines 2022, 10, 1810. https://doi.org/10.3390/biomedicines10081810
Vu DM, Nguyen V-T, Nguyen TH, Do PTX, Dao HH, Hai DX, Le NT, Nguyen X-H, Than UTT. Effects of Extracellular Vesicles Secreted by TGFβ-Stimulated Umbilical Cord Mesenchymal Stem Cells on Skin Fibroblasts by Promoting Fibroblast Migration and ECM Protein Production. Biomedicines. 2022; 10(8):1810. https://doi.org/10.3390/biomedicines10081810
Chicago/Turabian StyleVu, Duc Minh, Van-Tinh Nguyen, Thu Huyen Nguyen, Phuong Thi Xuan Do, Huy Hoang Dao, Do Xuan Hai, Nhi Thi Le, Xuan-Hung Nguyen, and Uyen Thi Trang Than. 2022. "Effects of Extracellular Vesicles Secreted by TGFβ-Stimulated Umbilical Cord Mesenchymal Stem Cells on Skin Fibroblasts by Promoting Fibroblast Migration and ECM Protein Production" Biomedicines 10, no. 8: 1810. https://doi.org/10.3390/biomedicines10081810
APA StyleVu, D. M., Nguyen, V.-T., Nguyen, T. H., Do, P. T. X., Dao, H. H., Hai, D. X., Le, N. T., Nguyen, X.-H., & Than, U. T. T. (2022). Effects of Extracellular Vesicles Secreted by TGFβ-Stimulated Umbilical Cord Mesenchymal Stem Cells on Skin Fibroblasts by Promoting Fibroblast Migration and ECM Protein Production. Biomedicines, 10(8), 1810. https://doi.org/10.3390/biomedicines10081810






