Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients
Abstract
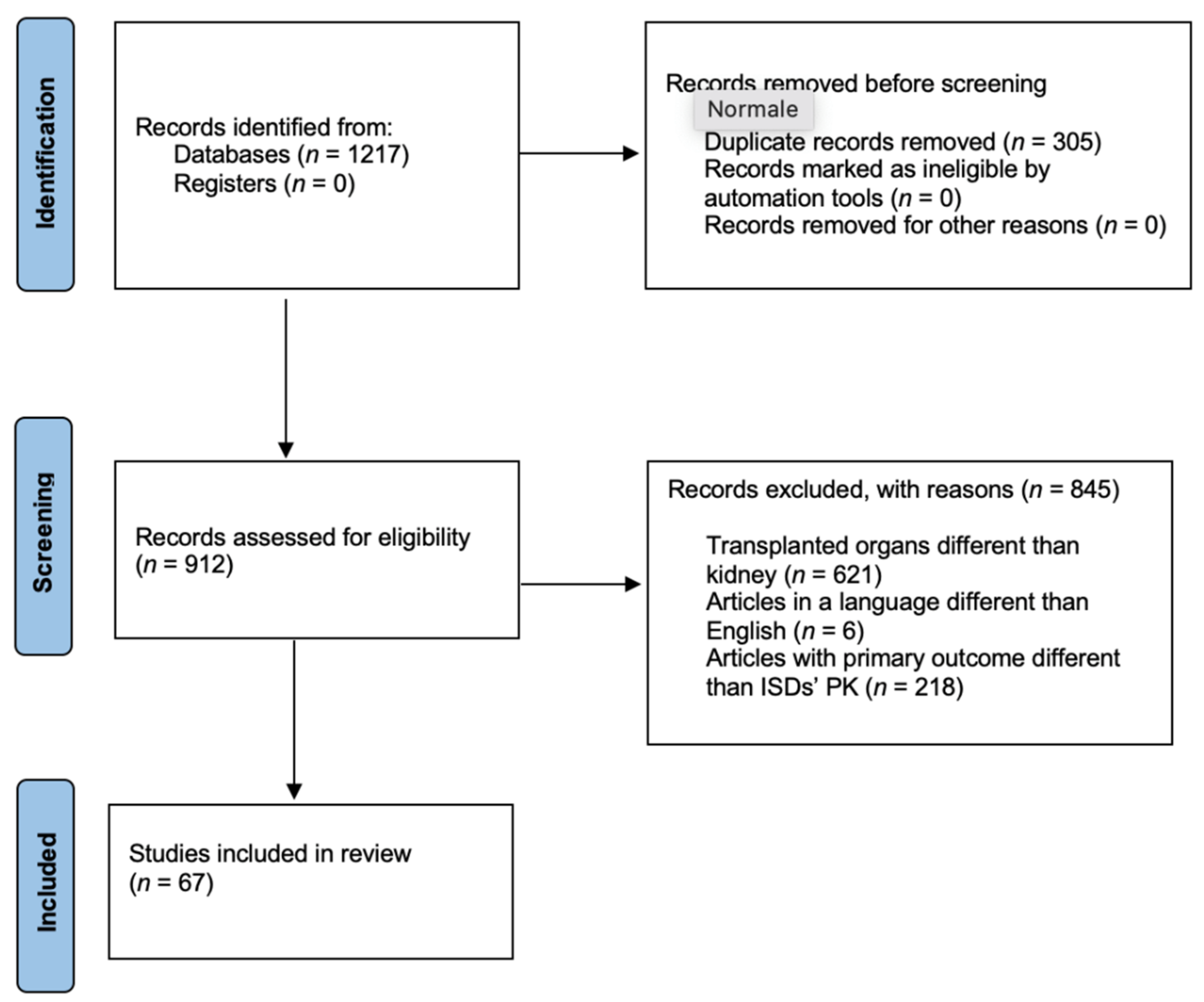
1. Introduction
2. Material and Methods
3. Results
3.1. Pharmacogenetics of Tacrolimus
3.2. Pharmacogenetics of Cyclosporine
3.3. Pharmacogenetics of Mycophenolic Acid
3.4. Pharmacogenetics of Everolimus and Sirolimus
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taddeo, A.; Prim, D.; Bojescu, E.D.; Segura, J.M.; Pfeifer, M.E. Point-of-Care Therapeutic Drug Monitoring for Precision Dosing of Immunosuppressive Drugs. J. Appl. Lab. Med. 2020, 54, 738–761. [Google Scholar] [CrossRef] [PubMed]
- Catić-Dordević, A.; Cvetković, T.; Stefanović, N.; Veličković-Radovanović, R. Current biochemical monitoring and risk management of immunosuppressive therapy after transplantation. J. Med. Biochem. 2017, 36, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jusko, W.J.; Piekoszewski, W.; Klintmalm, G.B.; Shaefer, M.S.; Hebert, M.F.; Piergies, A.A.; Lee, C.C.; Schechter, P.; Mekki, Q.A. Pharmacokinetics of tacrolimus in liver transplant patients. Clin. Pharmacol. Ther. 1995, 57, 281–290. [Google Scholar] [CrossRef]
- Faulds, D.; Goa, K.L.; Benfield, P. Cyclosporin. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic use in immunoregulatory disorders. Drugs 1993, 45, 953–1040. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C.; Eugui, E.M. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 2000, 47, 85–118. [Google Scholar] [CrossRef]
- Bullingham, R.; Nicholls, A.J.; Kamm, B.R. Clinical pharmacokinetics of mycophenolate mofetil. Clin. Pharmacokinet. 1998, 64, 429–455. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Peng, W.; Lin, Y.; Zhang, H.; Meng, K. Effect of ABCB1 3435C>T Genetic Polymorphism on Pharmacokinetic Variables of Tacrolimus in Adult Renal Transplant Recipients: A Systematic Review and Meta-analysis. Clin. Ther. 2020, 42, 2049–2065. [Google Scholar] [CrossRef]
- Staatz, C.E.; Tett, S.E. Clinical pharmacokinetics and pharmacodynamics of mycophenolate in solid organ transplant recipients. Clin. Pharm. 2007, 46, 13–58. [Google Scholar] [CrossRef]
- Barry, A.; Levine, M. A systematic review of the effect of CYP3A5 genotype on the apparent oral clearance of tacrolimus in renal transplant recipients. Ther. Drug Monit. 2010, 32, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, P.A.; Oetting, W.S.; Brearley, A.M.; Leduc, R.; Guan, W.; Schladt, D.; Matas, A.J.; Lamba, V.; Julian, B.A.; Mannon, R.B.; et al. Novel polymorphisms associated with tacrolims trough concentrations: Results from a multicenter kidney transplant consortium. Transplantation 2011, 91, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Hesselink, D.A.; Bouamar, R.; Elens, L.; van Schaik, R.H.; van Gelder, T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin. Pharm. 2014, 53, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Pallio, G.; Irrera, N.; Bitto, A.; Mannino, F.; Minutoli, L.; Rottura, M.; Pallio, S.; Altavilla, D.; Alibrandi, A.; Marciano, M.C.; et al. Failure of Achieving Tacrolimus Target Blood Concentration Might Be Avoided by a Wide Genotyping of Transplanted Patients: Evidence from a Retrospective Study. J. Pers. Med. 2020, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.L.; Tang, H.L.; Zhai, S.D. Effects of the CYP3A4*1B Genetic Polymorphism on the Pharmacokinetics of Tacrolimus in Adult Renal Transplant Recipients: A Meta-Analysis. PLoS ONE 2015, 10, e0127995. [Google Scholar] [CrossRef] [PubMed]
- Okubo, M.; Murayama, N.; Shimizu, M.; Shimada, T.; Guengerich, F.P.; Yamazaki, H. CYP3A4 intron 6 C>T polymorphism (CYP3A4*22) is associated with reduced CYP3A4 protein level and function in human liver microsomes. J. Toxicol. Sci. 2013, 38, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Masters, B.S. The journey from NADPH-cytochrome P450 oxidoreductase to nitric oxide synthases. Biochem. Biophys. Res. Commun. 2005, 338, 507–519. [Google Scholar] [CrossRef]
- De Jonge, H.; Metalidis, C.; Naesens, M.; Lambrechts, D.; Kuypers, D.R. The P450 oxidoreductase *28 SNP is associated with low initial tacrolimus exposure and increased dose requirements in CYP3A5-expressing renal recipients. Pharmacogenomics 2011, 12, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, S.; Chen, D.; Guo, F.; Zhong, L.; Fan, J.; Peng, Z. Influence of TLR4 rs1927907 locus polymorphisms on tacrolimus pharmacokinetics in the early stage after liver transplantation. Eur. J. Clin. Pharmacol. 2014, 70, 925–931. [Google Scholar] [CrossRef]
- Cascorbi, I. P-glycoprotein: Tissue distribution, substrates, and functional consequences of genetic variations. Handb. Exp. Pharmacol. 2011, 201, 261–283. [Google Scholar]
- Kravljaca, M.; Perovic, V.; Pravica, V.; Brkovic, V.; Milinkovic, M.; Lausevic, M.; Naumovic, R. The importance of MDR1 gene polymorphisms for tacrolimus dosage. Eur. J. Pharm. Sci. 2016, 83, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Tron, C.; Lemaitre, F.; Verstuyft, C.; Petitcollin, A.; Verdier, M.C.; Bellissant, E. Pharmacogenetics of Membrane Transporters of Tacrolimus in Solid Organ Transplantation. Clin. Pharm. 2019, 58, 593–613. [Google Scholar] [CrossRef] [PubMed]
- Kalliokoski, A.; Niemi, M. Impact of OATP transporters on pharmacokinetics. Br. J. Pharmacol. 2009, 158, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Gensburger, O.; Van Schaik, R.H.; Picard, N.; Le Meur, Y.; Rousseau, A.; Woillard, J.B.; Van Gelder, T.; Marquet, P. Polymorphisms in type I and II inosine monophosphate dehydrogenase genes and association with clinical outcome in patients on mycophenolate mofetil. Pharm. Genom. 2010, 20, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Picard, N.; Ratanasavanh, D.; Prémaud, A.; Le Meur, Y.; Marquet, P. Identification of the UDP-glucuronosyltransferase isoforms involved in mycophenolic acid phase II metabolism. Drug. Metab. Dispos. 2005, 33, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.Y.; Chan, K.M.; Wong, Y.T.; Chak, W.L.; Bekers, O.; van Hooff, J.P. Impact of CYP3A5 Genetic Polymorphism on Intrapatient Variability of Tacrolimus Exposure in Chinese Kidney Transplant Recipients. Transplant. Proc. 2019, 51, 1754–1757. [Google Scholar] [CrossRef] [PubMed]
- Allegri, L.; Baldan, F.; Vallone, C.; Tulissi, P.; Gropuzzo, M.; Canelles, M.F.; Righi, E.; Adani, G.L.; Baccarani, U.; Montanaro, D.; et al. Tacrolimus Therapeutic Drug Monitoring in Stable Kidney Transplantation and Individuation of CYP3A5 Genotype. Transpl. Proc. 2019, 51, 2917–2920. [Google Scholar] [CrossRef]
- Mourad, M.; Mourad, G.; Wallemacq, P.; Garrigue, V.; Van Bellingen, C.; Van Kerckhove, V.; De Meyer, M.; Malaise, J.; Eddour, D.C.; Lison, D.; et al. Sirolimus and tacrolimus trough concentrations and dose requirements after kidney transplantation in relation to CYP3A5 and MDR1 polymorphisms and steroids. Transplantation 2005, 80, 977–984. [Google Scholar] [CrossRef]
- Quteineh, L.; Verstuyft, C.; Furlan, V.; Durrbach, A.; Letierce, A.; Ferlicot, S.; Taburet, A.M.; Charpentier, B.; Becquemont, L. Influence of CYP3A5 genetic polymorphism on tacrolimus daily dose requirements and acute rejection in renal graft recipients. Basic Clin. Pharmacol. Toxicol. 2008, 103, 546–552. [Google Scholar] [CrossRef]
- Tada, H.; Tsuchiya, N.; Satoh, S.; Kagaya, H.; Li, Z.; Sato, K.; Miura, M.; Suzuki, T.; Kato, T.; Habuchi, T. Impact of CYP3A5 and MDR1(ABCB1) C3435T polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplant. Proc. 2005, 37, 1730–1732. [Google Scholar] [CrossRef]
- Tsuchiya, N.; Satoh, S.; Tada, H.; Li, Z.; Ohyama, C.; Sato, K.; Suzuki, T.; Habuchi, T.; Kato, T. Influence of CYP3A5 and MDR1 (ABCB1) polymorphisms on the pharmacokinetics of tacrolimus in renal transplant recipients. Transplantation 2004, 78, 1182–1187. [Google Scholar] [CrossRef]
- Muller, W.K.; Dandara, C.; Manning, K.; Mhandire, D.; Ensor, J.; Barday, Z.; Freercks, R. CYP3A5 polymorphisms and their effects on tacrolimus exposure in an ethnically diverse South African renal transplant population. S. Afr. Med. J. 2020, 110, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Gervasini, G.; Garcia, M.; Macias, R.M.; Cubero, J.J.; Caravaca, F.; Benitez, J. Impact of genetic polymorphisms on tacrolimus pharmacokinetics and the clinical outcome of renal transplantation. Transpl. Int. 2012, 25, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Thervet, E.; Anglicheau, D.; King, B.; Schlageter, M.H.; Cassinat, B.; Beaune, P.; Legendre, C.; Daly, A.K. Impact of cytochrome p450 3A5 genetic polymorphism on tacrolimus doses and concentration-to-dose ratio in renal transplant recipients. Transplantation 2003, 76, 1233–1235. [Google Scholar] [CrossRef]
- Yildirim, E.; Şahin, G.; Kaltuş, Z.; Çolak, E. Effect of CYP3A5 and ABCB1 Gene Polymorphisms on Tacrolimus Blood Concentration in Renal Transplant Recipients. Clin. Lab. 2019, 65, 11. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, J.; Deng, R.; Fu, Q.; Chen, J.; Huang, M.; Chen, X.; Wang, C. Dynamic effects of CYP3A5 polymorphism on dose requirement and trough concentration of tacrolimus in renal transplant recipients. J. Clin. Pharm. Ther. 2017, 42, 93–97. [Google Scholar] [CrossRef]
- Hesselink, D.A.; van Schaik, R.H.; van Agteren, M.; de Fijter, J.W.; Hartmann, A.; Zeier, M.; Budde, K.; Kuypers, D.R.; Pisarski, P.; Le Meur, Y.; et al. CYP3A5 genotype is not associated with a higher risk of acute rejection in tacrolimus-treated renal transplant recipients. Pharm. Genom. 2008, 18, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Z.H.; Zheng, J.M.; Chen, Z.H.; Tang, Z.; Chen, J.S.; Li, L.S. Influence of CYP3A5 and MDR1 polymorphisms on tacrolimus concentration in the early stage after renal transplantation. Clin. Transplant. 2005, 19, 638–643. [Google Scholar] [CrossRef]
- Ferraresso, M.; Tirelli, A.; Ghio, L.; Grillo, P.; Martina, V.; Torresani, E.; Edefonti, A. Influence of the CYP3A5 genotype on tacrolimus pharmacokinetics and pharmacodynamics in young kidney transplant recipients. Pediatric. Transplant. 2007, 11, 296–300. [Google Scholar] [CrossRef]
- Satoh, S.; Saito, M.; Inoue, T.; Kagaya, H.; Miura, M.; Inoue, K.; Komatsuda, A.; Tsuchiya, N.; Suzuki, T.; Habuchi, T. CYP3A5 *1 allele associated with tacrolimus trough concentrations but not subclinical acute rejection or chronic allograft nephropathy in Japanese renal transplant recipients. Eur. J. Clin. Pharmacol. 2009, 65, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Tirelli, S.; Ferraresso, M.; Ghio, L.; Meregalli, E.; Martina, V.; Belingheri, M.; Mattiello, C.; Torresani, E.; Edefonti, A. The effect of CYP3A5 polymorphisms on the pharmacokinetics of tacrolimus in adolescent kidney transplant recipients. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2008, 14, 251–254. [Google Scholar]
- Hu, R.; Barratt, D.T.; Coller, J.K.; Sallustio, B.C.; Somogyi, A.A. CYP3A5*3 and ABCB1 61A>G Significantly Influence Dose-adjusted Trough Blood Tacrolimus Concentrations in the First Three Months Post-Kidney Transplantation. Basic Clin. Pharmacol. Toxicol. 2018, 123, 320–326. [Google Scholar] [CrossRef]
- Li, Y.; Yan, L.; Shi, Y.; Bai, Y.; Tang, J.; Wang, L. CYP3A5 and ABCB1 genotype influence tacrolimus and sirolimus pharmacokinetics in renal transplant recipients. Springerplus 2015, 4, 637. [Google Scholar] [CrossRef]
- Roy, J.N.; Barama, A.; Poirier, C.; Vinet, B.; Roger, M. Cyp3A4, Cyp3A5, and MDR-1 genetic influences on tacrolimus pharmacokinetics in renal transplant recipients. Pharm. Genom. 2006, 16, 659–665. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Q.; Tian, X.; Yang, J.; Zhang, X. Tacrolimus Starting Dose Prediction Based on Genetic Polymorphisms and Clinical Factors in Chinese Renal Transplant Recipients. Genet. Test. Mol. Biomark. 2020, 24, 665–673. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, M.; Guan, D.; Bi, S.; Meng, J.; Li, Q.; Wang, W. Genetic polymorphisms of CYP3A5 genes and concentration of the cyclosporine and tacrolimus. Transplant. Proc. 2005, 37, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Teng, R.C.; Zhu, H.Z.; Fang, Y. CYP3A4/5 polymorphisms affect the blood level of cyclosporine and tacrolimus in Chinese renal transplant recipients. Int. J. Clin. Pharmacol. Ther. 2013, 51, 466–474. [Google Scholar] [CrossRef]
- Zhao, W.; Fakhoury, M.; Baudouin, V.; Storme, T.; Maisin, A.; Deschênes, G.; Jacqz-Aigrain, E. Population pharmacokinetics and pharmacogenetics of once daily prolonged-release formulation of tacrolimus in paediatric and adolescent kidney transplant recipients. Eur. J. Clin. Pharmacol. 2013, 69, 189–195. [Google Scholar] [CrossRef]
- Andrews, L.M.; Hesselink, D.A.; van Schaik, R.; van Gelder, T.; de Fijter, J.W.; Lloberas, N.; Elens, L.; Moes, D.; de Winter, B. A population pharmacokinetic model to predict the individual starting dose of tacrolimus in adult renal transplant recipients. Br. J. Clin. Pharmacol. 2019, 85, 601–615. [Google Scholar] [CrossRef]
- Zuo, X.C.; Ng, C.M.; Barrett, J.S. Effects of CYP3A4 and CYP3A5 polymorphisms on tacrolimus pharmacokinetics in Chinese adult renal transplant recipients: A population pharmacokinetic analysis. Pharm. Genom. 2013, 23, 251–261. [Google Scholar] [CrossRef]
- Hannachi, I.; Chadli, Z.; Kerkeni, E.; Kolsi, A.; Hammouda, M.; Chaabane, A.; Ben Fredj, N.; Touitou, Y.; Boughattas, N.A.; Aouam, K. Influence of CYP3A polymorphisms on tacrolimus pharmacokinetics in kidney transplant recipients. Pharm. J. 2021, 21, 69–77. [Google Scholar]
- Lloberas, N.; Elens, L.; Llaudó, I.; Padullés, A.; van Gelder, T.; Hesselink, D.A.; Colom, H.; Andreu, F.; Torras, J.; Bestard, O.; et al. The combination of CYP3A4*22 and CYP3A5*3 single-nucleotide polymorphisms determines tacrolimus dose requirement after kidney transplantation. Pharm. Genom. 2017, 27, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Papaz, T.; Lafreniere-Roula, M.; Nalli, N.; Grasemann, H.; Schwartz, S.M.; Kamath, B.M.; Ng, V.; Parekh, R.S.; Manlhiot, C.; et al. A randomized clinical trial of age and genotype-guided tacrolimus dosing after pediatric solid organ transplantation. Pediatr. Transplant. 2018, 22, e13285. [Google Scholar] [CrossRef]
- Yanik, M.V.; Seifert, M.E.; Locke, J.E.; Hauptfeld-Dolejsek, V.; Crowley, M.R.; Cutter, G.R.; Mannon, R.B.; Feig, D.I.; Limdi, N.A. CYP3A5 genotype affects time to therapeutic tacrolimus level in paediatric kidney transplant recipients. Pediatr. Transplant. 2019, 23, e13494. [Google Scholar] [CrossRef] [PubMed]
- Trofe-Clark, J.; Brennan, D.C.; West-Thielke, P.; Milone, M.C.; Lim, M.A.; Neubauer, R.; Nigro, V.; Bloom, R.D. Results of ASERTAA, a Randomized Prospective Crossover Pharmacogenetic Study of Immediate-Release Versus Extended-Release Tacrolimus in African American Kidney Transplant Recipients. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2018, 73, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Tamashiro, E.Y.; Felipe, C.R.; Genvigir, F.; Rodrigues, A.C.; Campos, A.B.; Hirata, R.; Tedesco-Silva, H.; Medina-Pestana, J.O. Influence of CYP3A4 and CYP3A5 polymorphisms on tacrolimus and sirolimus exposure in stable kidney transplant recipients. Drug Metab. Pers. Ther. 2017, 32, 89–95. [Google Scholar] [CrossRef]
- Shuker, N.; Bouamar, R.; van Schaik, R.H.; Clahsen-van Groningen, M.C.; Damman, J.; Baan, C.C.; van de Wetering, J.; Rowshani, A.T.; Weimar, W.; van Gelder, T.; et al. A Randomized Controlled Trial Comparing the Efficacy of Cyp3a5 Genotype-Based With BodyWeight-Based Tacrolimus Dosing After Living Donor Kidney Transplantation. Am. J. Transplant. 2016, 16, 2085–2096. [Google Scholar] [CrossRef]
- Spierings, N.; Holt, D.W.; MacPhee, I.A.M. CYP3A5 Genotype Had no Impact on Intrapatient Variability of Tacrolimus Clearance in Renal Transplant Recipients. Drug Monit. 2013, 35, 328–331. [Google Scholar] [CrossRef]
- Sienkiewicz, B.; Hurkacz, M.; Kuriata-Kordek, M.; Augustyniak-Bartosik, H.; Wiela-Hojeńska, A.; Klinger, M. The impact of CYP3A5 on the metabolism of cyclosporine A and tacrolimus in the evaluation of efficiency and safety of immunosuppressive treatment in patients after kidney transplantation. Pharmazie 2016, 71, 562–565. [Google Scholar]
- Ogasawara, K.; Chitnis, S.D.; Gohh, R.Y.; Christians, U.; Akhlaghi, F. Multidrug resistance-associated protein 2 (MRP2/ABCC2) haplotypes significantly affect the pharmacokinetics of tacrolimus in kidney transplant recipients. Clin. Pharm. 2013, 52, 751–762. [Google Scholar] [CrossRef]
- Provenzani, A.; Notarbartolo, M.; Labbozzetta, M.; Poma, P.; Vizzini, G.; Salis, P.; Caccamo, C.; Bertani, T.; Palazzo, U.; Polidori, P.; et al. Influence of CYP3A5 and ABCB1 gene polymorphisms and other factors on tacrolimus dosing in Caucasian liver and kidney transplant patients. Int. J. Mol. Med. 2011, 28, 1093–1102. [Google Scholar] [CrossRef]
- Liu, S.; Chen, R.X.; Li, J.; Liu, X.M.; Huang, H.B.; Fu, Q.; Wang, C.X.; Huang, M.; Li, J.L. Associations of SLCO1B1 polymorphisms with tacrolimus concentrations in Chinese renal transplant recipients. Acta Pharm. Sin. 2016, 51, 1240–1244. [Google Scholar]
- Boivin, A.A.; Cardinal, H.; Barama, A.; Naud, J.; Pichette, V.; Hébert, M.J.; Roger, M. Influence of SLCO1B3 genetic variations on tacrolimus pharmacokinetics in renal transplant recipients. Drug Metab. Pharm. 2013, 28, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Anutrakulchai, S.; Pongskul, C.; Kritmetapak, K.; Limwattananon, C.; Vannaprasaht, S. Therapeutic concentration achievement and allograft survival comparing usage of conventional tacrolimus doses and CYP3A5 genotype-guided doses in renal transplantation patients. Br. J. Clin. Pharmacol. 2019, 85, 1964–1973. [Google Scholar] [CrossRef]
- Thervet, E.; Loriot, M.A.; Barbier, S.; Buchler, M.; Ficheux, M.; Choukroun, G.; Toupance, O.; Touchard, G.; Alberti, C.; Le Pogamp, P.; et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin. Pharmacol. Ther. 2010, 87, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Żochowska, D.; Wyzgał, J.; Pączek, L. Impact of CYP3A4*1B and CYP3A5*3 polymorphisms on the pharmacokinetics of cyclosporine and sirolimus in renal transplant recipients. Ann. Transplant. 2012, 17, 36–44. [Google Scholar]
- Meng, X.G.; Guo, C.X.; Feng, G.Q.; Zhao, Y.C.; Zhou, B.T.; Han, J.L.; Chen, X.; Shi, Y.; Shi, H.Y.; Yin, J.Y.; et al. Association of CYP3A polymorphisms with the pharmacokinetics of cyclosporine A in early post renal transplant recipients in China. Acta Pharmacol. Sin. 2012, 33, 1563–1570. [Google Scholar] [CrossRef]
- Qiu, X.Y.; Jiao, Z.; Zhang, M.; Zhong, L.J.; Liang, H.Q.; Ma, C.L.; Zhang, L.; Zhong, M.K. Association of MDR1, CYP3A4*18B, and CYP3A5*3 polymorphisms with cyclosporine pharmacokinetics in Chinese renal transplant recipients. Eur. J. Clin. Pharmacol. 2008, 64, 1069–1084. [Google Scholar] [CrossRef]
- Lunde, I.; Bremer, S.; Midtvedt, K.; Mohebi, B.; Dahl, M.; Bergan, S.; Åsberg, A.; Christensen, H. The influence of CYP3A, PPARA, and POR genetic variants on the pharmacokinetics of tacrolimus and cyclosporine in renal transplant recipients. Eur. J. Clin. Pharmacol. 2014, 70, 685–693. [Google Scholar] [CrossRef]
- Kotowski, M.J.; Bogacz, A.; Bartkowiak-Wieczorek, J. Effect of Multidrug-Resistant 1 (MDR1) and CYP3A4*1B Polymorphisms on Cyclosporine-Based Immunosuppressive Therapy in Renal Transplant Patients. Ann. Transplant. 2019, 24, 108–114. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.L.; Fu, Q.; Wang, X.D.; Liu, L.S.; Wang, C.X.; Xie, W.; Chen, Z.J.; Shu, W.Y.; Huang, M. Associations of ABCB1, NFKB1, CYP3A, and NR1I2 polymorphisms with cyclosporine trough concentrations in Chinese renal transplant recipients. Acta Pharmacol. Sin. 2013, 34, 555–560. [Google Scholar] [CrossRef][Green Version]
- Hu, Y.F.; Qiu, W.; Liu, Z.Q.; Zhu, L.J.; Liu, Z.Q.; Tu, J.H.; Wang, D.; Li, Z.; He, J.; Zhong, G.P.; et al. Effects of genetic polymorphisms of CYP3A4, CYP3A5 and MDR1 on cyclosporine pharmacokinetics after renal transplantation. Clin. Exp. Pharmacol. Physiol. 2006, 33, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Yates, C.R.; Zhang, W.; Song, P.; Li, S.; Gaber, A.O.; Kotb, M.; Honaker, M.R.; Alloway, R.R.; Meibohm, B. The effect of CYP3A5 and MDR1 polymorphic expression on cyclosporine oral disposition in renal transplant patients. J. Clin. Pharmacol. 2003, 43, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Anglicheau, D.; Thervet, E.; Etienne, I.; Hurault De Ligny, B.; Le Meur, Y.; Touchard, G.; Büchler, M.; Laurent-Puig, P.; Tregouet, D.; Beaune, P.; et al. CYP3A5 and MDR1 genetic polymorphisms and cyclosporine pharmacokinetics after renal transplantation. Clin. Pharmacol. Ther. 2004, 75, 422–433. [Google Scholar] [CrossRef]
- Bouamar, R.; Hesselink, D.A.; van Schaik, R.H.; Weimar, W.; Macphee, I.A.; de Fijter, J.W.; van Gelder, T. Polymorphisms in CYP3A5, CYP3A4, and ABCB1 are not associated with cyclosporine pharmacokinetics nor with cyclosporine clinical end points after renal transplantation. Ther. Drug Monit. 2011, 33, 178–184. [Google Scholar] [CrossRef]
- Fukuda, T.; Goebel, J.; Cox, S.; Maseck, D.; Zhang, K.; Sherbotie, J.R.; Ellis, E.N.; James, L.P.; Ward, R.M.; Vinks, A.A. UGT1A9, UGT2B7, and MRP2 genotypes can predict mycophenolic acid pharmacokinetic variability in pediatric kidney transplant recipients. Ther. Drug Monit. 2012, 34, 671–679. [Google Scholar] [CrossRef]
- Krall, P.; Yañez, D.; Rojo, A.; Delucchi, Á.; Córdova, M.; Morales, J.; Boza, P.; de la Rivera, A.; Espinoza, N.; Armijo, N.; et al. CCYP3A5 and UGT1A9 Polymorphisms Influence Immunosuppressive Therapy in Pediatric Kidney Transplant Recipients. Front. Pharmacol. 2021, 12, 653525. [Google Scholar] [CrossRef]
- Mazidi, T.; Rouini, M.R.; Ghahremani, M.H.; Dashti-Khavidaki, S.; Lessan-Pezeshki, M.; Ahmadi, F.L.; Salam-Zadeh, J.; Mandegary, A.; Gholami, K. Impact of UGT1A9 polymorphism on mycophenolic acid pharmacokinetic parameters in stable renal transplant patients. Iran. J. Pharm. Res. 2013, 12, 547–556. [Google Scholar]
- Xie, X.C.; Li, J.; Wang, H.Y.; Li, H.L.; Liu, J.; Fu, Q.; Huang, J.W.; Zhu, C.; Zhong, G.P.; Wang, X.D.; et al. Associations of UDP-glucuronosyltransferases polymorphisms with mycophenolate mofetil pharmacokinetics in Chinese renal transplant patients. Acta Pharmacol. Sin. 2015, 36, 644–650. [Google Scholar] [CrossRef]
- Ciftci, H.S.; Demir, E.; Karadeniz, M.S.; Tefik, T.; Nane, I.; Oguz, F.S.; Aydin, F.; Turkmen, A. Influence of uridine diphosphate-glucuronosyltransferases (1A9) polymorphisms on mycophenolic acid pharmacokinetics in patients with renal transplant. Ren. Fail. 2018, 40, 395–402. [Google Scholar] [CrossRef]
- Kuypers, D.R.; Naesens, M.; Vermeire, S.; Vanrenterghem, Y. The impact of uridine diphosphate-glucuronosyltransferase 1A9 (UGT1A9) gene promoter region single-nucleotide polymorphisms T-275A and C-2152T on early mycophenolic acid dose-interval exposure in de novo renal allograft recipients. Clin. Pharmacol. Ther. 2005, 78, 351–361. [Google Scholar] [CrossRef]
- Sánchez-Fructuoso, A.I.; Maestro, M.L.; Calvo, N.; Viudarreta, M.; Pérez-Flores, I.; Veganzone, S.; De la Orden, V.; Ortega, D.; Arroyo, M.; Barrientos, A. The prevalence of uridine diphosphate-glucuronosyltransferase 1A9 (UGT1A9) gene promoter region single-nucleotide polymorphisms T-275A and C-2152T and its influence on mycophenolic acid pharmacokinetics in stable renal transplant patients. Transplant. Proc. 2009, 41, 2313–2316. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Sheng, C.C.; Liao, G.Y.; Su, Y.; Feng, L.J.; Xia, Q.; Jiao, Z.; Xu, D.J. Genetic polymorphisms in metabolic enzymes and transporters have no impact on mycophenolic acid pharmacokinetics in adult kidney transplant patients co-treated with tacrolimus: A population analysis. J. Clin. Pharm. Ther. 2021, 46, 1564–1575. [Google Scholar] [CrossRef] [PubMed]
- Granata, S.; Dalla Gassa, A.; Carraro, A.; Brunelli, M.; Stallone, G.; Lupo, A.; Zaza, G. Sirolimus and Everolimus Pathway: Reviewing Candidate Genes Influencing Their Intracellular Effects. Int. J. Mol. Sci. 2016, 17, 735. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Jiménez, C.; García-Saiz, M.; Pérez-Tamajón, L.; Salido, E.; Torres, A. Influence of genetic polymorphisms of CYP3A5 and ABCB1 on sirolimus pharmacokinetics, patient and graft survival and other clinical outcomes in renal transplant. Drug Metab. Pers. Ther. 2017, 32, 49–58. [Google Scholar] [CrossRef]
- Lolita, L.; Zheng, M.; Zhang, X.; Han, Z.; Tao, J.; Fei, S.; Wang, Z.; Guo, M.; Yang, H.; Ju, X.; et al. The Genetic Polymorphism of CYP3A4 rs2242480 is Associated with Sirolimus Trough Concentrations Among Adult Renal Transplant Recipients. Curr. Drug Metab. 2020, 21, 1052–1059. [Google Scholar] [CrossRef]
- Lee, J.; Huang, H.; Chen, Y.; Lu, X. ABCB1 haplotype influences the sirolimus dose requirements in Chinese renal transplant recipients. Biopharm. Drug Dispos. 2014, 35, 164–172. [Google Scholar] [CrossRef]
- Miao, L.Y.; Huang, C.R.; Hou, J.Q.; Qian, M.Y. Association study of ABCB1 and CYP3A5 gene polymorphisms with sirolimus trough concentration and dose requirements in Chinese renal transplant recipients. Biopharm. Drug Dispos. 2008, 29, 1–5. [Google Scholar] [CrossRef]
- Moes, D.J.A.R.; Press, R.R.; den Hartigh, J. Population Pharmacokinetics and Pharmacogenetics of Everolimus in Renal Transplant Patients. Clin. Pharm. 2012, 51, 467–480. [Google Scholar] [CrossRef]
- Azarfar, A.; Ravanshad, Y.; Mehrad-Majd, H.; Esmaeeli, M.; Aval, S.B.; Emadzadeh, M.; Salehi, M.; Moradi, A.; Golsorkhi, M.; Khazaei, M.R. Comparison of tacrolimus and cyclosporine for immunosuppression after renal transplantation: An updated systematic review and meta-analysis. Saudi J. Kidney Dis. Transpl. 2018, 29, 1376–1385. [Google Scholar] [CrossRef]
- Cheung, C.Y.; Tang, S.C.W. Personalized immunosuppression after kidney transplantation. Nephrology 2022, 27, 475–483. [Google Scholar] [CrossRef]
- Birdwell, K.A.; Decker, B.; Barbarino, J.M.; Peterson, J.F.; Stein, C.M.; Sadee, W.; Wang, D.; Vinks, A.A.; He, Y.; Swen, J.J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Provenzani, A.; Santeusanio, A.; Mathis, E.; Notarbartolo, M.; Labbozzetta, M.; Poma, P.; Provenzani, A.; Polidori, C.; Vizzini, G.; Polidori, P.; et al. Pharmacogenetic considerations for optimizing tacrolimus dosing in liver and kidney transplant patients. World J. Gastroenterol. 2013, 19, 9156–9173. [Google Scholar] [CrossRef] [PubMed]
| Study | Number of Patients | Drug | Gene | RefSNP | Clinical Effects |
|---|---|---|---|---|---|
| Cheung et al., 2019 | 86 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5 expressers |
| Allegri et al., 2019 | 20 | Tacrolimus | CYP3A5 | rs776746 | Higher doses in CYP3A5*1/*1 and *1/*3 carriers |
| Mourad et al., 2005 | 85 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1 carrier |
| Quteineh et al., 2008 | 136 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1 carrier |
| Tada et al., 2005 | 28 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1 carrier |
| Tsuchiya et al., 2004 | 30 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1 carrier |
| Muller, 2020 | 43 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1/*1 and *1/*3 carriers |
| Gervasini et al., 2012 | 103 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1 carrier |
| Thervet et al., 2003 | 80 | Tacrolimus | CYP3A5 | rs776746 | Lower dose in CYP3A5*3/*3 carrier |
| Yildrim et al., 2019 | 67 | Tacrolimus | CYP3A5 | rs776746 | Lower dose in CYP3A5*3/*3 carrier |
| Chen et al., 2017 | 194 | Tacrolimus | CYP3A5 | rs776746 | Lower C0 in CYP3A5 expressers |
| Hesselink et al., 2008 | 136 | Tacrolimus | CYP3A5 | rs776746 | Lower C0 in carrier of at least one CYP3A5*1 allele |
| Zhang et al., 2005 | 118 | Tacrolimus | CYP3A5 | rs776746 | Lower C0 in CYP3A5 expressers |
| Ferraresso et al., 2007 | 30 | Tacrolimus | CYP3A5 | rs776746 | Lower C0 in CYP3A5 expressers |
| Satoh et al., 2009 | 41 | Tacrolimus | CYP3A5 | rs776746 | Lower C0 in CYP3A5 expressers |
| Tirelli et al., 2008 | 26 | Tacrolimus | CYP3A5 | rs776746 | Lower C0 in CYP3A5 expressers |
| Hu et al., 2018 | 165 | Tacrolimus | CYP3A5 | rs776746 | Lower C0/D in CYP3A5 expressers |
| Li et al., 2015 | 112 | Tacrolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5*3/*3 carrier |
| Roy et al., 2006 | 44 | Tacrolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5*3/*3 carrier and lower C0/D in patients with less than three copies of MDR-1 polymorphisms. |
| ABCB1 | rs1045642 | ||||
| ABCB1 | rs2032582 | ||||
| ABCB1 | rs3213619 | ||||
| Wang et al., 2020 | 406 | Tacrolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5*3/*3 carrier |
| Zhao et al., 2005 | 30 | Tacrolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5*3/*3 carrier |
| Li et al., 2013 | 83 | Tacrolimus | CYP3A5 | rs776746 | Higher C0/D in carrier of haplotype GG |
| CYP3A4 | rs28371759 | ||||
| Zhao et al., 2013 | 22 | Tacrolimus | CYP3A5 | rs776746 | Lower clearance in CYP3A5*3/*3 carrier |
| Andrews et al., 2019 | 337 | Tacrolimus | CYP3A5 | rs776746 | Higher clearance in CYP3A5 expressers and lower clearance in CYP3A4*22 carrier |
| CYP3A4 | rs35599367 | ||||
| Zuo et al., 2013 | 161 | Tacrolimus | CYP3A5 | rs776746 | Higher clearance in CYP3A5*1 |
| Hannachi et al., 2021 | 80 | Tacrolimus | CYP3A5 | rs776746 | Decreased C0/D in CYP3A4*1B and CYP3A5*1 carrier. Increased C0/D in CYP3A4*22 carrier |
| CYP3A4 | rs2740574 | ||||
| CYP3A4 | rs35599367 | ||||
| Yanik et al., 2019 | 98 | Tacrolimus | CYP3A5 | rs776746 | Longer time to achieve a steady therapeutic concentration in CYP3A5*1 expresser. Higher incidence of early allograft rejection in CYP3A5*1 expressers |
| Spierings et al., 2013 | 118 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5 expressers |
| Ogasawara et al., 2013 | 102 | Tacrolimus | ABCC2 | rs3740066 | Lower C0/D in ABCC2 3972T allele carrier |
| Kravljaca et al., 2016 | 91 | Tacrolimus | ABCB1 | rs1045642 | Lower C0/D in CTT/TTT carrier |
| ABCB1 | rs1128503 | ||||
| ABCB1 | rs2032582 | ||||
| Provenzani et al., 2011 | 50 | Tacrolimus | CYP3A5 | rs776746 | Lower C0/D in patients with one copy of the CYP3A5*1 allele |
| Liu et al., 2016 | 89 | Tacrolimus | SLCO1B1 | rs2306283 | Higher C0 in CC carrier |
| Boivin et al., 2013 | 38 | Tacrolimus | SLCO1B3 | rs4149117 | Higher risk of over-exposure in SLCO1B3 334G and 699A homozygous haplotype |
| SLCO1B3 | rs7311358 | ||||
| Anutrakulcha et al., 2019 | 63 | Tacrolimus | CYP3A5 | rs776746 | More patients with achieved therapeutic range and lower proportion of over-therapeutic concentration in the genotype-guided group |
| Thervet et al., 2010 | 280 | Tacrolimus | CYP3A5 | rs776746 | C0 above the target range in CY3A5*3/*3 carriers and below the target in CYP3A5*1/*1 carrier |
| Quteineh et al., 2008 | 136 | Tacrolimus | CYP3A5 | rs776746 | Higher dose in CYP3A5*1/*1 carrier. Increased risk of acute rejection in CYP3A5*1/*1 |
| Żochowska et al., 2012 | 100 | Cyclosporine | CYP3A5 | rs776746 | Higher dose in CYP3A5*1 or CYP3A4*1B carrier |
| CYP3A4 | rs2740574 | ||||
| Meng et al., 2012 | 126 | Cyclosporine | CYP3A5 | rs776746 | Higher C0 and C0/D in CYP3A5*3 G/G carrier |
| Lunde et al., 2014 | 177 | Cyclosporine | CYP3A4 | rs35599367 | Higher C2/D in CYP3A4*22 carrier |
| Kotowski et al., 2019 | 184 | Cyclosporine | CYP3A4 | rs2740574 | Lower dose in CYP3A4*1/*1 |
| Zhang et al., 2013 | 101 | Cyclosporine | ABCB1 | rs1045642 | Higher C0/D in ABCB1 2677 T/T carrier. Higher C0/D in ABCB1 3435 T/T carrier. Higher C0/D in ABCB1 1236TT-2677TT-3435TT haplotype compared to other genotypes |
| ABCB1 | rs1128503 | ||||
| ABCB1 | rs2032582 | ||||
| Hu et al., 2006 | 106 | Cyclosporine | CYP3A5 | rs776746 | Lower C0/D in CYP3A5*1/*1 carrier. Lower C0/D in wild-type homozygotes for ABCB1 C3435T |
| ABCB1 | rs1045642 | ||||
| Yates et al., 2003 | 19 | Cyclosporine | ABCB1 | rs1045642 | Patients with at least one ABCB1 3435T allele had a significantly higher CsA clearance than homozygous wild-type |
| Fukuda et al., 2012 | 32 | Mycophenolic acid | MRP2 | rs717620 | Higher dose in MRP2-24T>C heterozygous with also UGT1A9-440C>T or UGT2B7-900A>G and in MRP2-24T>C wild-type with both UGT1A9-440C>T and UGT2B7-900A>G |
| UGT1A9 | rs2741045 | ||||
| UGT2B7 | rs7438135 | ||||
| Krall et al., 2021 | 104 | Mycophenolic acid | UGT1A9 | rs6714486 | Lower AUC0–12/D in UGT1A9-275A carrier |
| Mazidi et al., 2013 | 40 | Mycophenolic acid | UGT1A9 | rs6714486 | Lower AUC0–12 and Cmax in UGT1A9 275A carrier |
| Xie et al., 2015 | 127 | Mycophenolic acid | UGT2B7 | rs62298861 | Higher AUC0–12 in UGT2B7 IVS1+985AG, UGT1A9-1818CT, UGT1A9-440C>T and -331T>C. UGT1A8*2 allele is related to lower AUC0–12 as well as the UGT1A7 622TT genotype |
| UGT1A9 | rs13418420 | ||||
| UGT1A9 | rs2741045 | ||||
| UGT1A9 | rs2741046 | ||||
| UGT1A8 | rs1042597 | ||||
| UGT1A7 | rs11692021 | ||||
| Ciftci et al., 2018 | 125 | Mycophenolic acid | UGT1A9 | rs2741049 | Higher C0 and lower doses in UGT1A9 1399 T/T carrier |
| Kuypers et al., 2005 | 95 | Mycophenolic acid | UGT1A9 | rs6714486 | Lower exposure in T275A and C2152T carrier |
| UGT1A9 | rs17868320 | ||||
| Sánchez-Fructuoso et al., 2009 | 133 | Mycophenolic acid | UGT1A9 | rs6714486 | Lower exposure in UGT1A9 T-275A and C-2152T carrier |
| UGT1A9 | rs17868320 | ||||
| Rodríguez-Jiménez et al., 2017 | 48 | Sirolimus | CYP3A5 | rs776746 | Lower C0 in CYP3A5*1/*3 carrier. Higher C0 in ABCB1 3435 C/T carrier |
| ABCB1 | rs1045642 | ||||
| Lolita et al., 2020 | 69 | Sirolimus | CYP3A4 | rs2242480 | Higher C0 in C/C carrier |
| Lee et al., 2014 | 85 | Sirolimus | CYP3A5 | rs776746 | Lower C0/D in CYP3A5*1 carrier |
| Li et al., 2015 | 43 | Sirolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5*3/*3. No correlation between SRL trough concentrations or dose requirements with CYP3A4 and ABCB1 SNPs |
| ABCB1 | rs1045642 | ||||
| ABCB1 | rs1128503 | ||||
| ABCB1 | rs2032582 | ||||
| Miao et al., 2008 | 50 | Sirolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5*3/*3 carrier. No differences between C0/D and ABCB1 SNPs |
| Tamashiro et al., 2017 | 46 | Sirolimus | CYP3A5 | rs776746 | Higher C0/D in CYP3A5 TT carrier. |
| RefSNP | Drug | Clinical Effects |
|---|---|---|
| rs776746 | Tacrolimus | Higher dose, lower C0, lower C0/D, higher clearance, higher risk of allograft rejection |
| rs776746 | Cyclosporine | Higher dose, lower C0, lower C0/D |
| rs776746 | Sirolimus | Lower C0, lower C0/D |
| rs2740574 | Tacrolimus | Higher dose, lower C0/D |
| rs2740574 | Cyclosporine | Higher dose |
| rs35599367 | Tacrolimus | Higher C0/D, lower clearance |
| rs35599367 | Cyclosporine | Higher C2/D |
| rs1045642 | Cyclosporine | Higher C0/D |
| rs2032582 | Cyclosporine | Higher C0/D |
| rs6714486 | Mycophenolic acid | Lower exposure |
| rs17868320 | Mycophenolic acid | Lower exposure |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urzì Brancati, V.; Scarpignato, C.; Minutoli, L.; Pallio, G. Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients. Biomedicines 2022, 10, 1798. https://doi.org/10.3390/biomedicines10081798
Urzì Brancati V, Scarpignato C, Minutoli L, Pallio G. Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients. Biomedicines. 2022; 10(8):1798. https://doi.org/10.3390/biomedicines10081798
Chicago/Turabian StyleUrzì Brancati, Valentina, Carmelo Scarpignato, Letteria Minutoli, and Giovanni Pallio. 2022. "Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients" Biomedicines 10, no. 8: 1798. https://doi.org/10.3390/biomedicines10081798
APA StyleUrzì Brancati, V., Scarpignato, C., Minutoli, L., & Pallio, G. (2022). Use of Pharmacogenetics to Optimize Immunosuppressant Therapy in Kidney-Transplanted Patients. Biomedicines, 10(8), 1798. https://doi.org/10.3390/biomedicines10081798








