Downregulation of STAT3 in Epstein-Barr Virus-Positive Hodgkin Lymphoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Treatments
2.2. Polymerase Chain-Reaction (PCR) Analyses
2.3. Protein Analyses
2.4. Genomic Profiling Analysis
2.5. Gene Expression Profiling Analysis and Transcriptome Data
3. Results
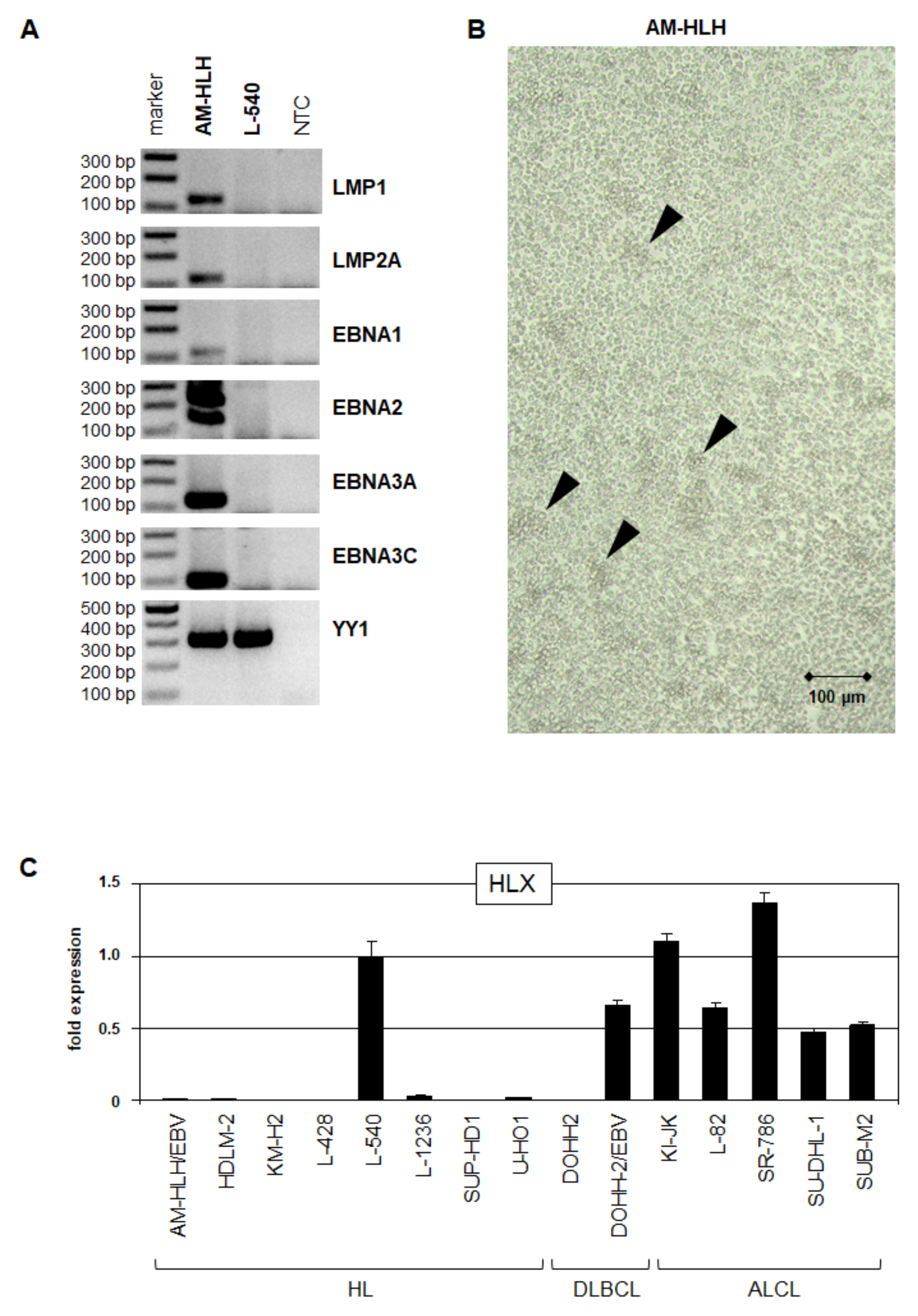
3.1. HL Cell Line AM-HLH Is EBV-Positive and HLX-Negative
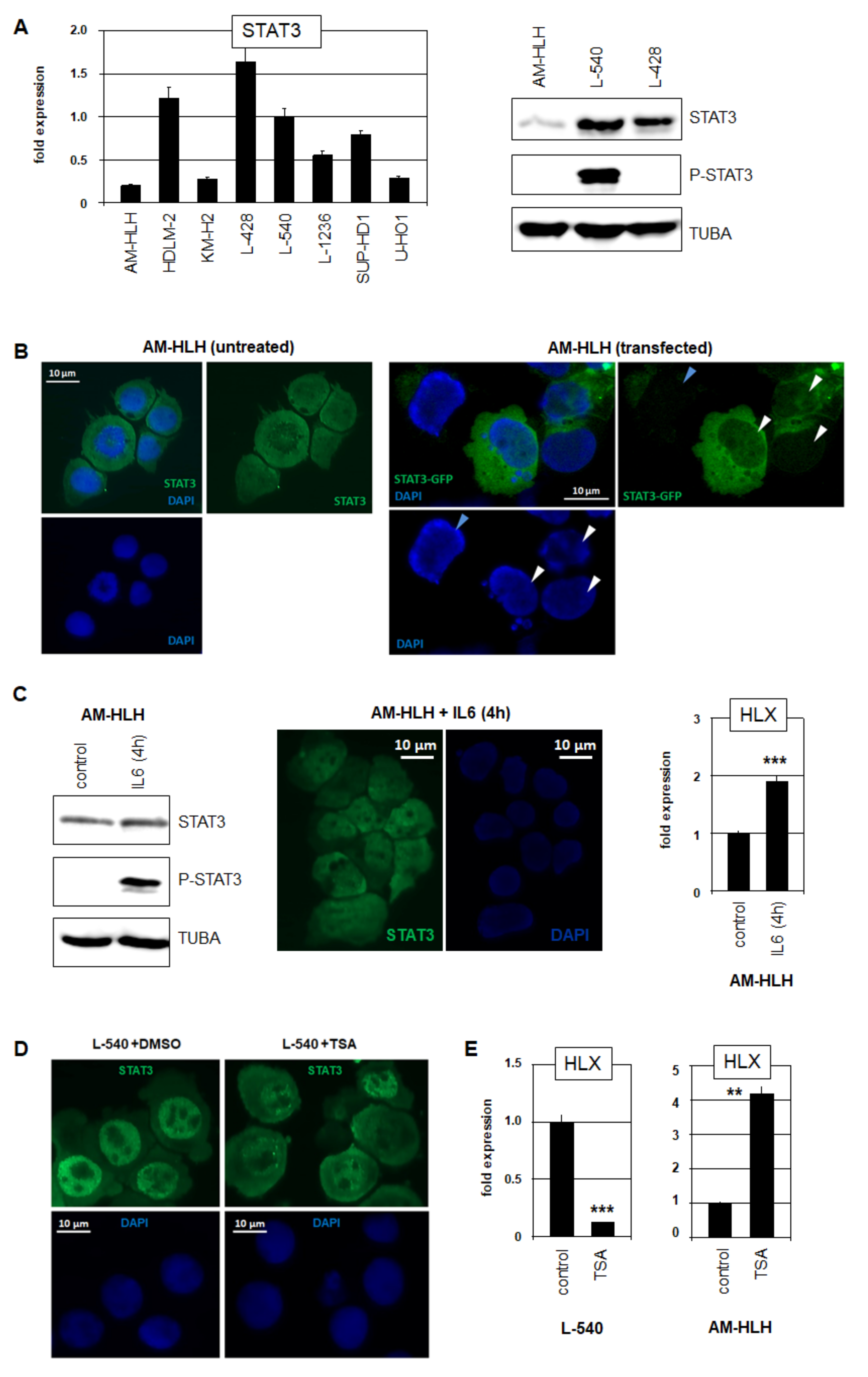
3.2. Analysis of STAT3 in HL Cell Lines AM-HLH and L-540
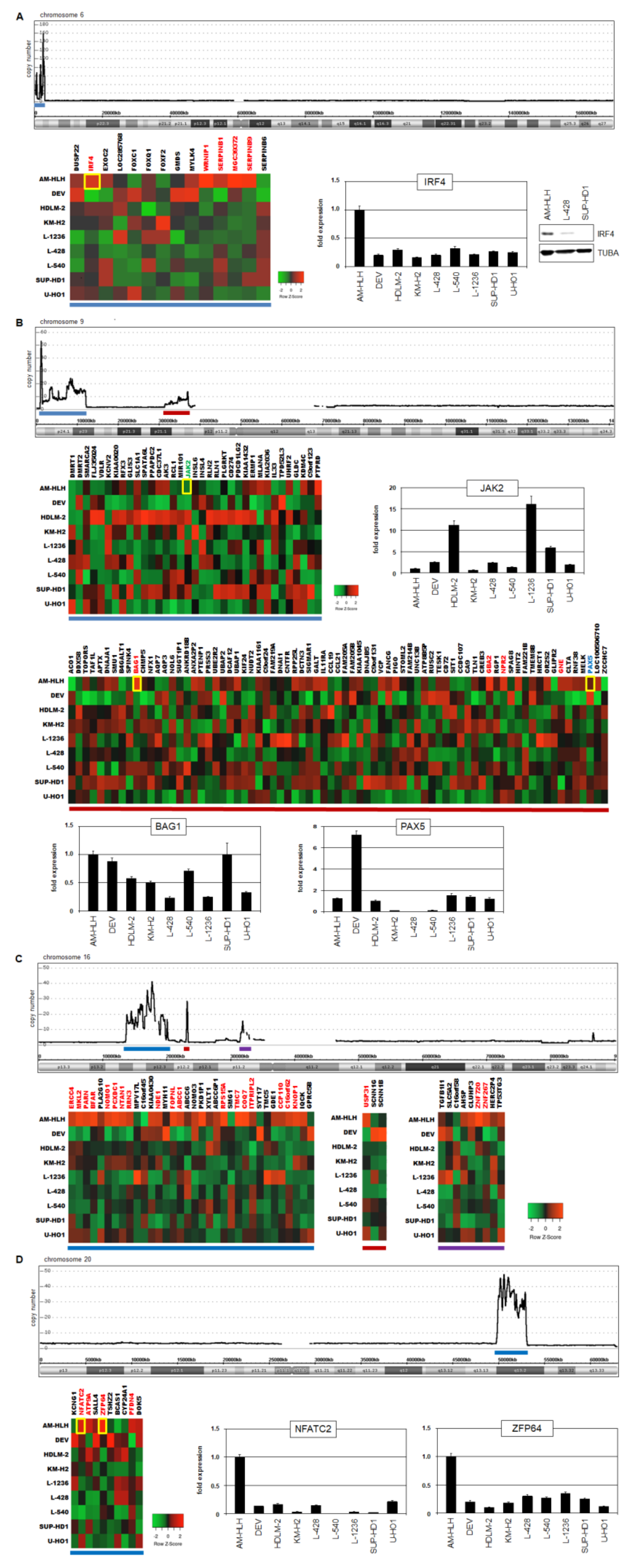
3.3. Analysis of Genomic Amplifications in AM-HLH
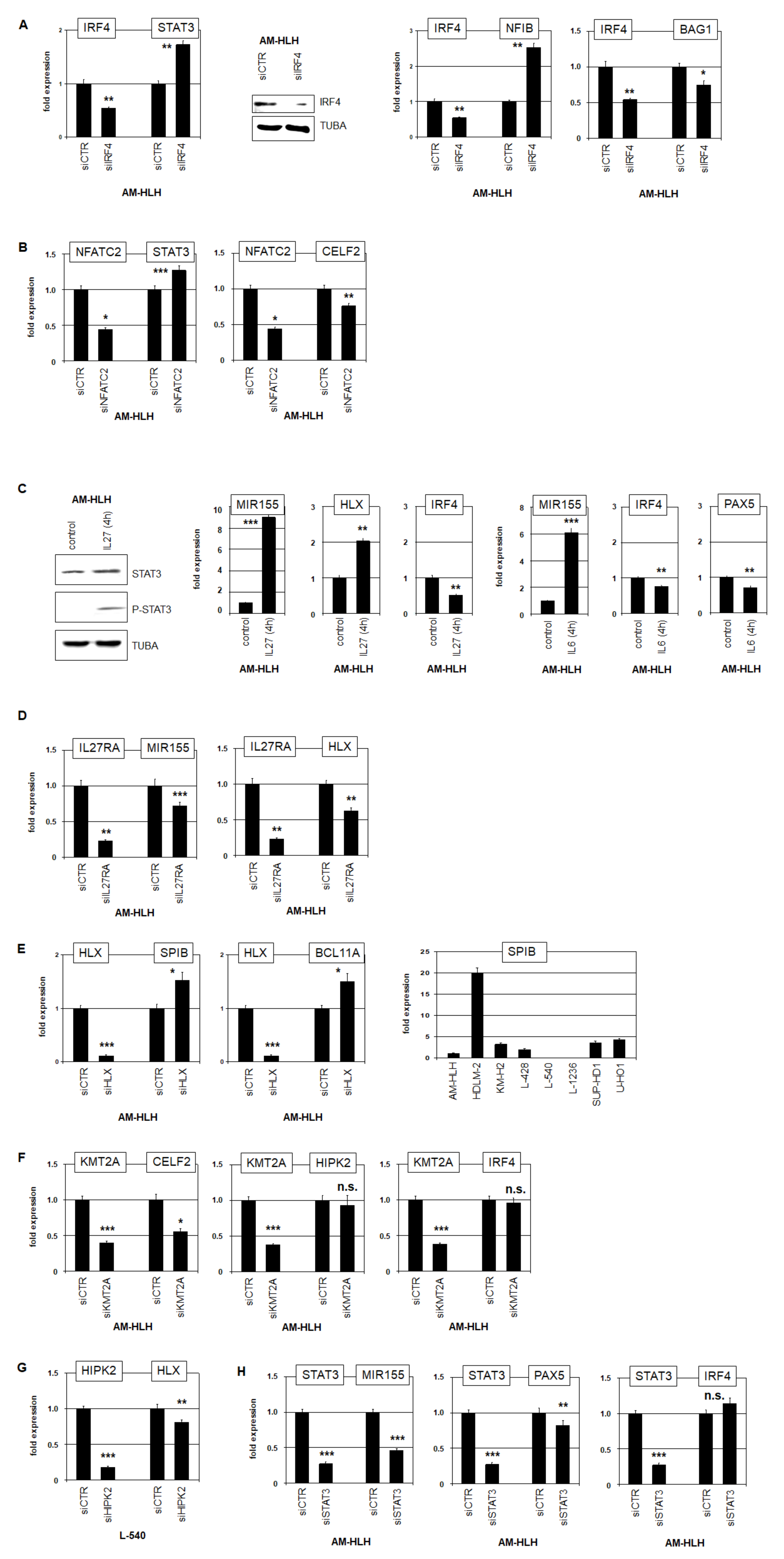
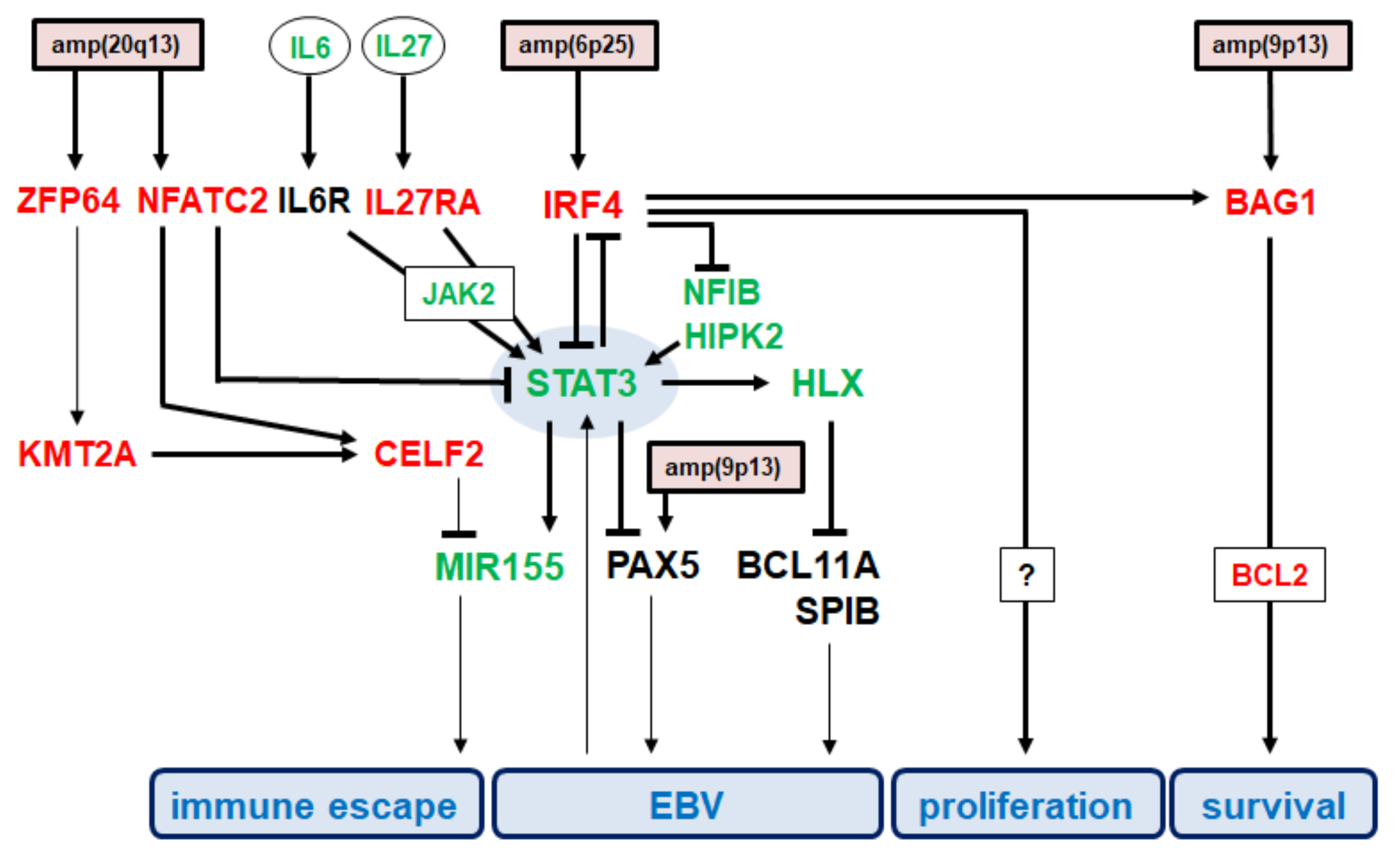
3.4. Impacts of Identified Gene Candidates in STAT3 Regulation
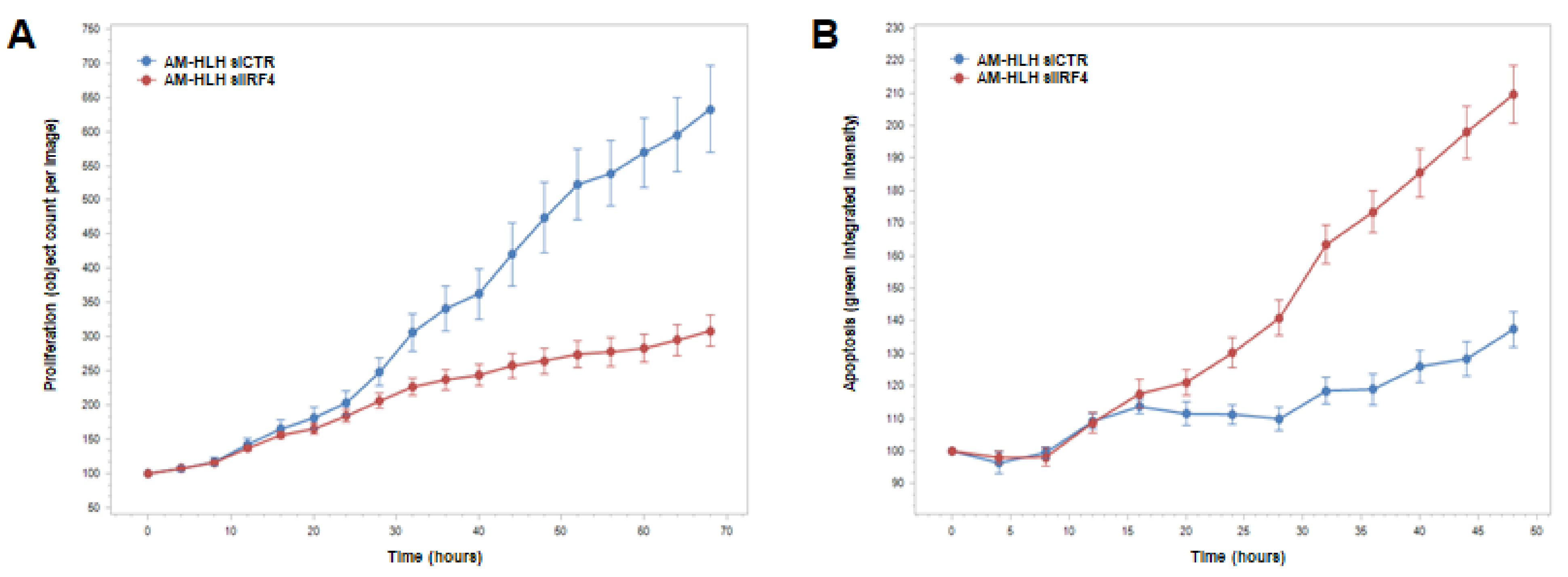
3.5. Functional Analysis of IRF4 in AM-HLH
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Küppers, R. The biology of Hodgkin’s lymphoma. Nat. Rev. Cancer 2009, 9, 15–27. [Google Scholar] [CrossRef]
- Küppers, R.; Engert, A.; Hansmann, M.L. Hodgkin lymphoma. J. Clin. Investig. 2012, 122, 3439–3447. [Google Scholar] [CrossRef] [PubMed]
- Drexler, H.G.; Pommerenke, C.; Eberth, S.; Nagel, S. Hodgkin lymphoma cell lines: To separate the wheat from the chaff. Biol. Chem. 2018, 399, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Küppers, R. Molecular biology of Hodgkin lymphoma. Leukemia 2021, 35, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Moss, W.N.; Yario, T.A.; Steitz, J.A. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 2015, 160, 607–618. [Google Scholar] [CrossRef] [Green Version]
- Kung, C.P.; Raab-Traub, N. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 2008, 82, 5486–5493. [Google Scholar] [CrossRef] [Green Version]
- Incrocci, R.; Barse, L.; Stone, A.; Vagvala, S.; Montesano, M.; Subramaniam, V.; Swanson-Mungerson, M. Epstein-Barr Virus Latent Membrane Protein 2A (LMP2A) enhances IL-10 production through the activation of Bruton’s tyrosine kinase and STAT3. Virology 2017, 500, 96–102. [Google Scholar] [CrossRef]
- Lu, F.; Weidmer, A.; Liu, C.G.; Volinia, S.; Croce, C.M.; Lieberman, P.M. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J. Virol. 2008, 82, 10436–10443. [Google Scholar] [CrossRef] [Green Version]
- Wood, C.D.; Carvell, T.; Gunnell, A.; Ojeniyi, O.O.; Osborne, C.; West, M.J. Enhancer Control of MicroRNA miR-155 Expression in Epstein-Barr Virus-Infected B Cells. J. Virol. 2018, 92, e00716-18. [Google Scholar] [CrossRef] [Green Version]
- Schlegelberger, B.; Weber-Matthiesen, K.; Himmler, A.; Bartels, H.; Sonnen, R.; Kuse, R.; Feller, A.C.; Grote, W. Cytogenetic findings and results of combined immunophenotyping and karyotyping in Hodgkin’s disease. Leukemia 1994, 8, 72–80. [Google Scholar]
- Nagel, S.; Meyer, C.; Quentmeier, H.; Kaufmann, M.; Drexler, H.G.; MacLeod, R.A. Chromothripsis in Hodgkin lymphoma. Genes Chromosomes Cancer 2013, 52, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Martin-Subero, J.I.; Gesk, S.; Harder, L.; Sonoki, T.; Tucker, P.W.; Schlegelberger, B.; Grote, W.; Novo, F.J.; Calasanz, M.J.; Hansmann, M.L.; et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood 2002, 99, 1474–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joos, S.; Küpper, M.; Ohl, S.; von Bonin, F.; Mechtersheimer, G.; Bentz, M.; Marynen, P.; Möller, P.; Pfreundschuh, M.; Trümper, L.; et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 2000, 60, 549–552. [Google Scholar] [PubMed]
- Krenacs, L.; Himmelmann, A.W.; Quintanilla-Martinez, L.; Fest, T.; Riva, A.; Wellmann, A.; Bagdi, E.; Kehrl, J.H.; Jaffe, E.S.; Raffeld, M. Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood 1998, 92, 1308–1316. [Google Scholar] [CrossRef]
- Bohle, V.; Döring, C.; Hansmann, M.L.; Küppers, R. Role of early B-cell factor 1 (EBF1) in Hodgkin lymphoma. Leukemia 2013, 27, 671–679. [Google Scholar] [CrossRef] [Green Version]
- Dimitrova, L.; Seitz, V.; Hecht, J.; Lenze, D.; Hansen, P.; Szczepanowski, M.; Ma, L.; Oker, E.; Sommerfeld, A.; Jundt, F.; et al. PAX5 overexpression is not enough to reestablish the mature B-cell phenotype in classical Hodgkin lymphoma. Leukemia 2014, 28, 213–216. [Google Scholar] [CrossRef]
- Nagel, S.; MacLeod, R.A.F.; Meyer, C.; Kaufmann, M.; Drexler, H.G. NKL homeobox gene activities in B-cell development and lymphomas. PLoS ONE 2018, 13, e0205537. [Google Scholar] [CrossRef]
- Nagel, S.; Pommerenke, C.; Meyer, C.; Kaufmann, M.; MacLeod, R.A.F.; Drexler, H.G. Aberrant expression of NKL homeobox gene HLX in Hodgkin lymphoma. Oncotarget 2018, 9, 14338–14353. [Google Scholar] [CrossRef] [Green Version]
- Nagel, S.; Uphoff, C.C.; Dirks, W.G.; Pommerenke, C.; Meyer, C.; Drexler, H.G. Epstein-Barr virus (EBV) activates NKL homeobox gene HLX in DLBCL. PLoS ONE 2019, 14, e0216898. [Google Scholar] [CrossRef]
- Nagel, S.; Pommerenke, C.; MacLeod, R.A.F.; Meyer, C.; Kaufmann, M.; Drexler, H.G. The NKL-code for innate lymphoid cells reveals deregulated expression of NKL homeobox genes HHEX and HLX in anaplastic large cell lymphoma (ALCL). Oncotarget 2020, 11, 3208–3226. [Google Scholar] [CrossRef]
- Villarino, A.V.; Kanno, Y.; Ferdinand, J.R.; O’Shea, J.J. Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 2015, 194, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Stumhofer, J.S.; Silver, J.S.; Laurence, A.; Porrett, P.M.; Harris, T.H.; Turka, L.A.; Ernst, M.; Saris, C.J.; O’Shea, J.J.; Hunter, C.A. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 2007, 8, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Tiacci, E.; Ladewig, E.; Schiavoni, G.; Penson, A.; Fortini, E.; Pettirossi, V.; Wang, Y.; Rosseto, A.; Venanzi, A.; Vlasevska, S.; et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 2018, 131, 2454–2465. [Google Scholar] [CrossRef] [Green Version]
- Escobar, T.; Yu, C.R.; Muljo, S.A.; Egwuagu, C.E. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest. Ophthalmol. Vis. Sci. 2013, 54, 4017–4025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozovski, U.; Calin, G.A.; Setoyama, T.; D’Abundo, L.; Harris, D.M.; Li, P.; Liu, Z.; Grgurevic, S.; Ferrajoli, A.; Faderl, S.; et al. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol. Cancer 2013, 12, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kube, D.; Holtick, U.; Vockerodt, M.; Ahmadi, T.; Haier, B.; Behrmann, I.; Heinrich, P.C.; Diehl, V.; Tesch, H. STAT3 is constitutively activated in Hodgkin cell lines. Blood 2001, 98, 762–770. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.B.; Yu, J.J.; Yu, R.Y.; Mendez, L.M.; Shaknovich, R.; Zhang, Y.; Cattoretti, G.; Ye, B.H. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 2008, 111, 1515–1523. [Google Scholar] [CrossRef] [Green Version]
- Lam, L.T.; Wright, G.; Davis, R.E.; Lenz, G.; Farinha, P.; Dang, L.; Chan, J.W.; Rosenwald, A.; Gascoyne, R.D.; Staudt, L.M. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 2008, 111, 3701–3713. [Google Scholar] [CrossRef] [Green Version]
- Crescenzo, R.; Abate, F.; Lasorsa, E.; Tabbo’, F.; Gaudiano, M.; Chiesa, N.; Di Giacomo, F.; Spaccarotella, E.; Barbarossa, L.; Ercole, E.; et al. T-Cell Project: Prospective Collection of Data in Patients with Peripheral T-Cell Lymphoma and the AIRC 5xMille Consortium “Genetics-Driven Targeted Management of Lymphoid Malignancies”. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell 2015, 27, 516–532. [Google Scholar]
- Hayashida, M.; Daibata, M.; Tagami, E.; Taguchi, T.; Maekawa, F.; Takeoka, K.; Fukutsuka, K.; Shimomura, D.; Hayashi, T.; Iwatani, Y.; et al. Establishment and characterization of a novel Hodgkin lymphoma cell line, AM-HLH, carrying the Epstein-Barr virus genome integrated into the host chromosome. Hematol. Oncol. 2017, 35, 567–575. [Google Scholar] [CrossRef]
- Drexler, H.G.; MacLeod, R.A.; Nagel, S.; Dirks, W.G.; Uphoff, C.C.; Steube, K.G.; Quentmeier, H. Guide to Leukemia-Lymphoma Cell Lines on CD. Blood 2005, 116, 4340. [Google Scholar] [CrossRef]
- Nagel, S.; MacLeod, R.A.F.; Pommerenke, C.; Meyer, C.; Kaufmann, M.; Drexler, H.G. NKL homeobox gene NKX2-2 is aberrantly expressed in Hodgkin lymphoma. Oncotarget 2018, 9, 37480–37496. [Google Scholar] [CrossRef] [PubMed]
- Brune, V.; Tiacci, E.; Pfeil, I.; Döring, C.; Eckerle, S.; van Noesel, C.J.; Klapper, W.; Falini, B.; von Heydebreck, A.; Metzler, D.; et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J. Exp. Med. 2008, 205, 2251–2268. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Wang, D.; Liebowitz, D.; Wang, F.; Gregory, C.; Rickinson, A.; Larson, R.; Springer, T.; Kieff, E. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: Deletion of the amino terminus abolishes activity. J. Virol. 1988, 62, 4173–4184. [Google Scholar] [CrossRef] [Green Version]
- Cuomo, L.; Ramquist, T.; Trivedi, P.; Wang, F.; Klein, G.; Masucci, M.G. Expression of the Epstein-Barr virus (EBV)-encoded membrane protein LMP1 impairs the in vitro growth, clonability and tumorigenicity of an EBV-negative Burkitt lymphoma line. Int. J. Cancer 1992, 51, 949–955. [Google Scholar] [CrossRef]
- Bandini, C.; Pupuleku, A.; Spaccarotella, E.; Pellegrino, E.; Wang, R.; Vitale, N.; Duval, C.; Cantarella, D.; Rinaldi, A.; Provero, P.; et al. IRF4 Mediates the Oncogenic Effects of STAT3 in Anaplastic Large Cell Lymphomas. Cancers 2018, 10, 21. [Google Scholar] [CrossRef] [Green Version]
- Takayama, S.; Sato, T.; Krajewski, S.; Kochel, K.; Irie, S.; Millan, J.A.; Reed, J.C. Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell 1995, 80, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Nutt, S.L.; Heavey, B.; Rolink, A.G.; Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999, 401, 556–562. [Google Scholar] [CrossRef]
- Rafei, M.; Hsieh, J.; Fortier, S.; Li, M.; Yuan, S.; Birman, E.; Forner, K.; Boivin, M.N.; Doody, K.; Tremblay, M.; et al. Mesenchymal stromal cell-derived CCL2 suppresses plasma cell immunoglobulin production via STAT3 inactivation and PAX5 induction. Blood 2008, 112, 4991–4998. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Yang, Z.; Dai, C.; Wang, R.; Ke, X.; Zhang, S.; Xiang, X.; Chen, K.; Li, C.; Luo, J.; et al. STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis. Oncogenesis 2020, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B.; Klingbeil, O.; Tarumoto, Y.; Somerville, T.D.D.; Huang, Y.H.; Wei, Y.; Wai, D.C.; Low, J.K.K.; Milazzo, J.P.; Wu, X.S.; et al. A Transcription Factor Addiction in Leukemia Imposed by the MLL Promoter Sequence. Cancer Cell 2018, 34, 970–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, K.L.; Jaganathan, A.; Hu, Y.; Xu, F.; Lejeune, A.; Sharma, R.; Caescu, C.I.; Meslamani, J.; Vincek, A.; Zhang, F.; et al. HIPK2 directs cell type-specific regulation of STAT3 transcriptional activity in Th17 cell differentiation. Proc. Natl. Acad. Sci. USA 2022, 119, e2117112119. [Google Scholar] [CrossRef]
- Wu, C.; Zhu, X.; Liu, W.; Ruan, T.; Wan, W.; Tao, K. NFIB promotes cell growth, aggressiveness, metastasis and EMT of gastric cancer through the Akt/Stat3 signaling pathway. Oncol. Rep. 2018, 40, 1565–1573. [Google Scholar] [CrossRef]
- Gatto, G.; Rossi, A.; Rossi, D.; Kroening, S.; Bonatti, S.; Mallardo, M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008, 36, 6608–6619. [Google Scholar] [CrossRef]
- Yoon, J.S.J.; Wu, M.K.; Zhu, T.H.; Zhao, H.; Cheung, S.T.; Chamberlain, T.C.; Mui, A.L. Interleukin-10 control of pre-miR155 maturation involves CELF2. PLoS ONE 2020, 15, e0231639. [Google Scholar] [CrossRef]
- Su, G.H.; Ip, H.S.; Cobb, B.S.; Lu, M.M.; Chen, H.M.; Simon, M.C. The Ets protein Spi-B is expressed exclusively in B cells and T cells during development. J. Exp. Med. 1996, 184, 203–214. [Google Scholar] [CrossRef]
- Laux, G.; Adam, B.; Strobl, L.J.; Moreau-Gachelin, F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994, 13, 5624–5632. [Google Scholar] [CrossRef]
- Zhao, B.; Sample, C.E. Epstein-barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 2000, 74, 5151–5160. [Google Scholar] [CrossRef]
- Klein, B.; Tarte, K.; Jourdan, M.; Mathouk, K.; Moreaux, J.; Jourdan, E.; Legouffe, E.; De Vos, J.; Rossi, J.F. Survival and proliferation factors of normal and malignant plasma cells. Int. J. Hematol. 2003, 78, 106–113. [Google Scholar] [CrossRef]
- Busslinger, M.; Urbánek, P. The role of BSAP (Pax-5) in B-cell development. Curr. Opin. Genet. Dev. 1995, 5, 595–601. [Google Scholar] [CrossRef]
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.J.; van den Berg, A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005, 207, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, R.; Zhao, H.J.; Fu, D.; Zhong, H.J.; Weng, X.Q.; Qu, B.; Zhao, Y.; Wang, L.; Zhao, W.L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer 2019, 18, 54. [Google Scholar] [CrossRef]
- Peng, L.; Chen, Z.; Chen, Y.; Wang, X.; Tang, N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019, 8, 7161–7173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagel, S.; Meyer, C.; Eberth, S.; Haake, J.; Pommerenke, C. Downregulation of STAT3 in Epstein-Barr Virus-Positive Hodgkin Lymphoma. Biomedicines 2022, 10, 1608. https://doi.org/10.3390/biomedicines10071608
Nagel S, Meyer C, Eberth S, Haake J, Pommerenke C. Downregulation of STAT3 in Epstein-Barr Virus-Positive Hodgkin Lymphoma. Biomedicines. 2022; 10(7):1608. https://doi.org/10.3390/biomedicines10071608
Chicago/Turabian StyleNagel, Stefan, Corinna Meyer, Sonja Eberth, Josephine Haake, and Claudia Pommerenke. 2022. "Downregulation of STAT3 in Epstein-Barr Virus-Positive Hodgkin Lymphoma" Biomedicines 10, no. 7: 1608. https://doi.org/10.3390/biomedicines10071608
APA StyleNagel, S., Meyer, C., Eberth, S., Haake, J., & Pommerenke, C. (2022). Downregulation of STAT3 in Epstein-Barr Virus-Positive Hodgkin Lymphoma. Biomedicines, 10(7), 1608. https://doi.org/10.3390/biomedicines10071608





