Comprehensive Analysis of Disease Pathology in Immunocompetent and Immunocompromised Hosts following Pulmonary SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus and Cell Lines
2.2. SARS-CoV-2 Infectivity Titration
2.3. Golden Syrian Hamster Infection and Sample Collection
2.4. Histopathology Analysis
2.5. Immunohistochemistry Analysis
2.6. Virus Infectivity Assays on Tissues
2.7. Plaque Reduction Neutralization (PRNT) Assay
2.8. RNA Isolation from Hamster Lungs
2.9. RNAseq Analysis of Lung Transcriptome
2.10. Gene Network and Pathway Analysis
2.11. Determination of Total Viral Load by Quantitative PCR
2.12. Statistical Analysis
2.13. Ethics Statement
3. Results
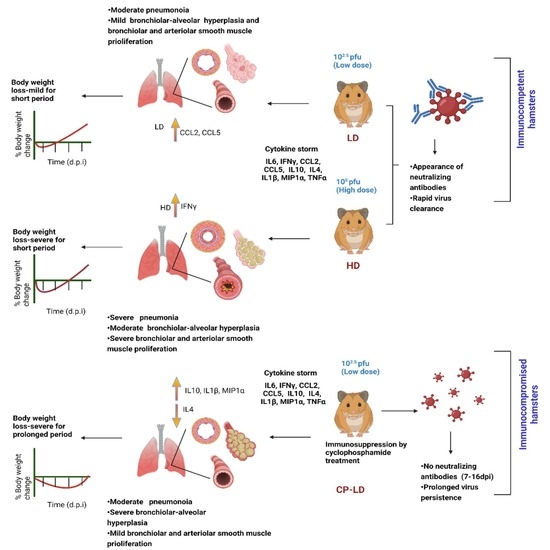
3.1. Distinct Disease Progression in SARS-CoV-2-Infected Immunocompetent and Immunocompromised Hamsters
3.2. Viral Burden in Immunocompetent Hamsters Infected with a Low or High Dose Inoculum
3.3. Disparate Tissue Viral Burden between Immunocompetent and Immunocompromised Hamsters Infected with SARS-CoV-2
3.4. Heterogeneity in Neutralizing Antibody Titer in SARS-CoV-2-Infected Immunocompetent and Immunocompromised Hamsters
3.5. Cytokine Storm Marker Expression in SARS-CoV-2-Infected Immunocompetent and Immunocompromised Hamsters
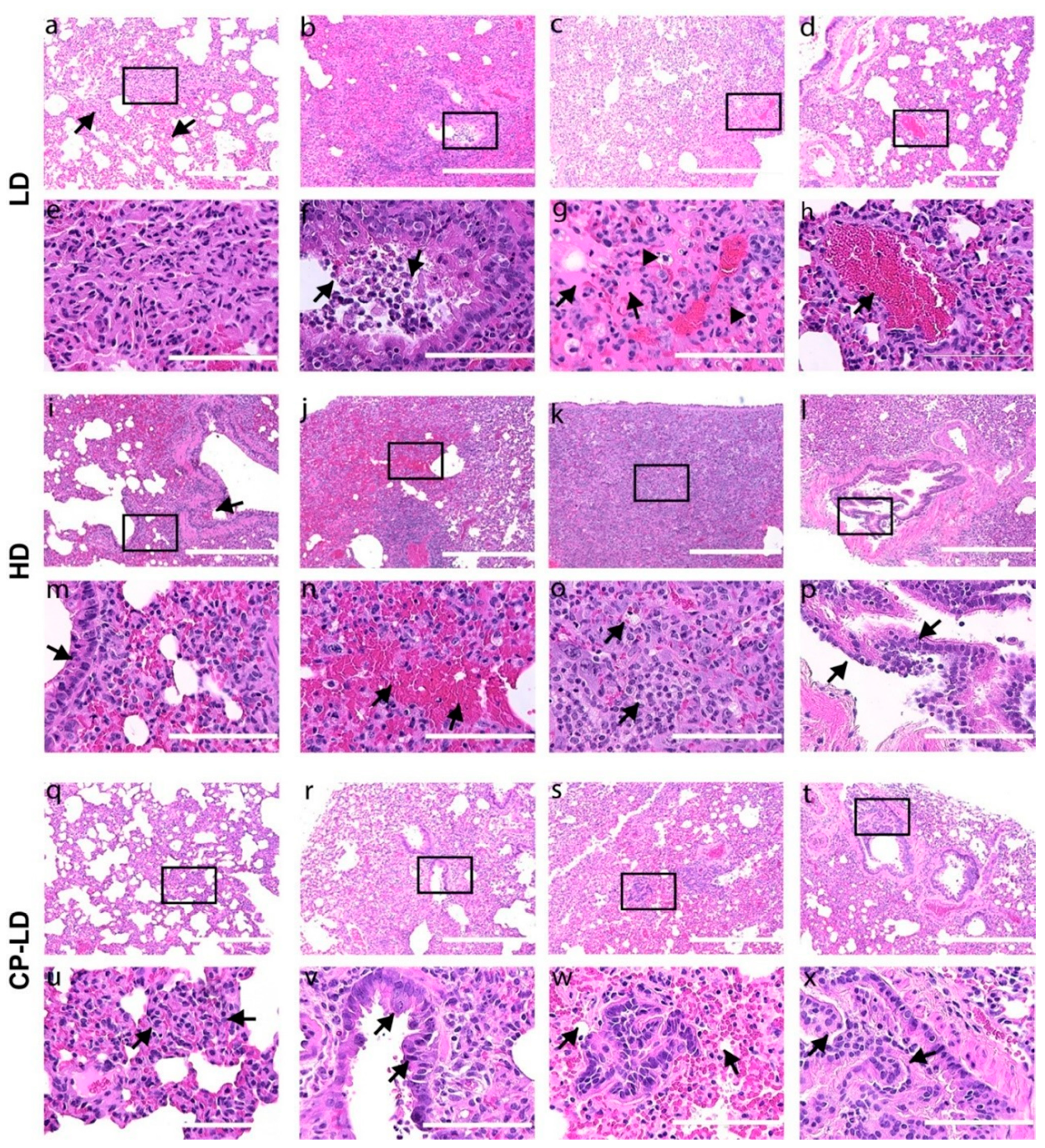
3.6. Severity of Pulmonary/Bronchiolar Pathology and Thrombosis in SARS-CoV-2-Infected Immunocompetent and Immunocompromised Hamsters
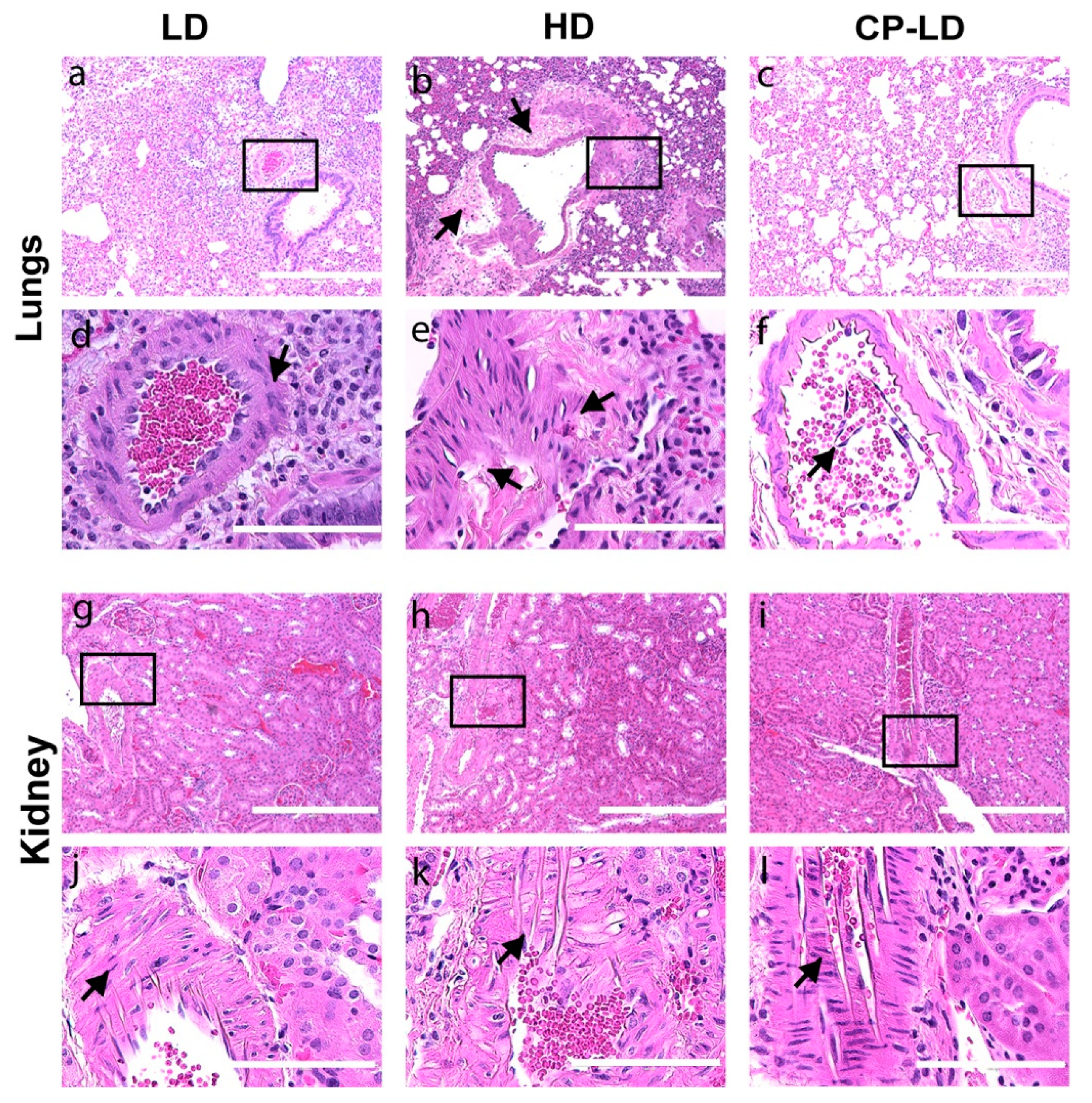
3.7. Hypertension-like Vascular Smooth Muscle Hyperplasia/Hypertrophy in the Lungs and Kidneys of SARS-CoV-2-Infected Hamsters
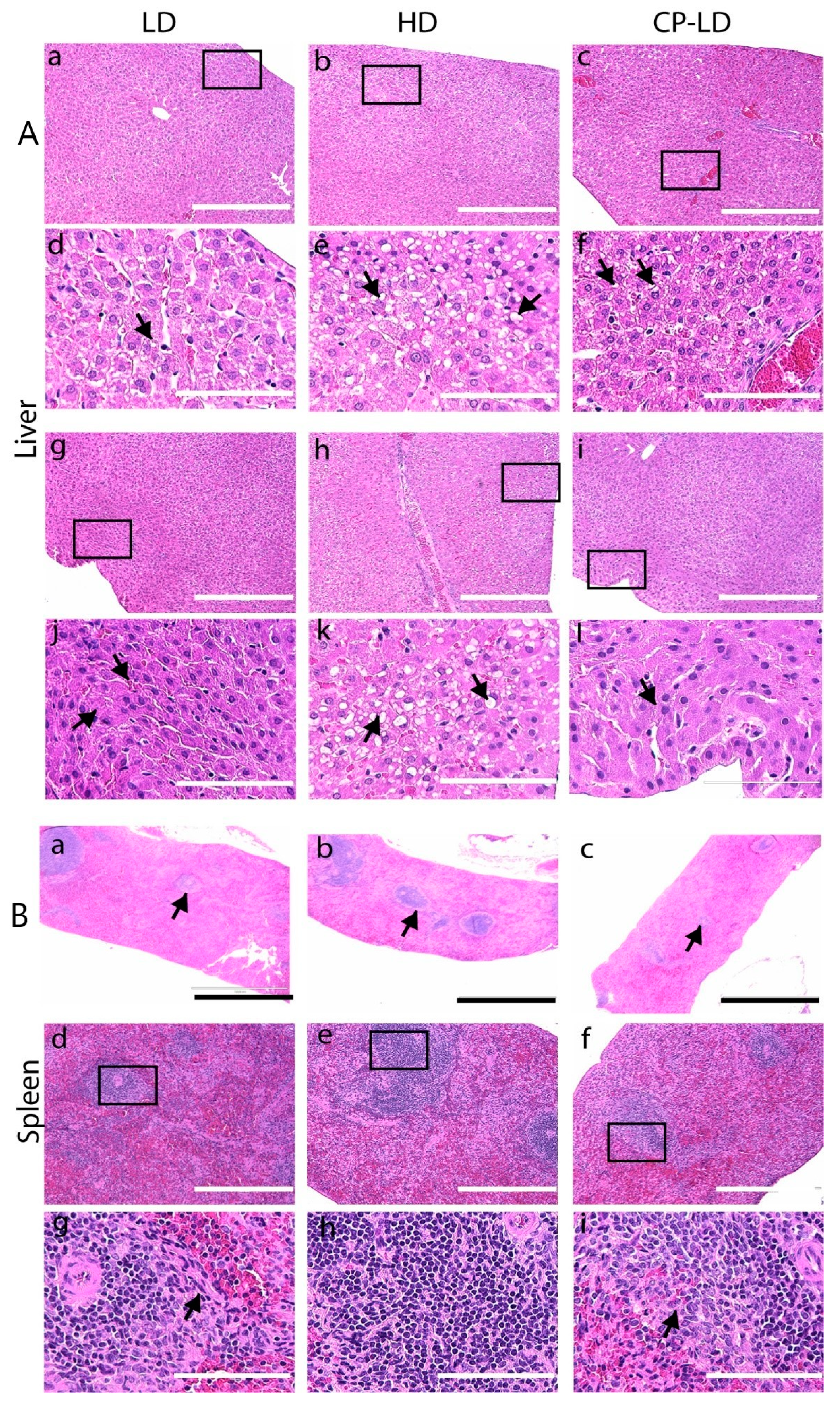
3.8. Multi-Organ Pathology Induced by SARS-CoV-2 Infection in Immunocompetent and Immunocompromised Hamsters
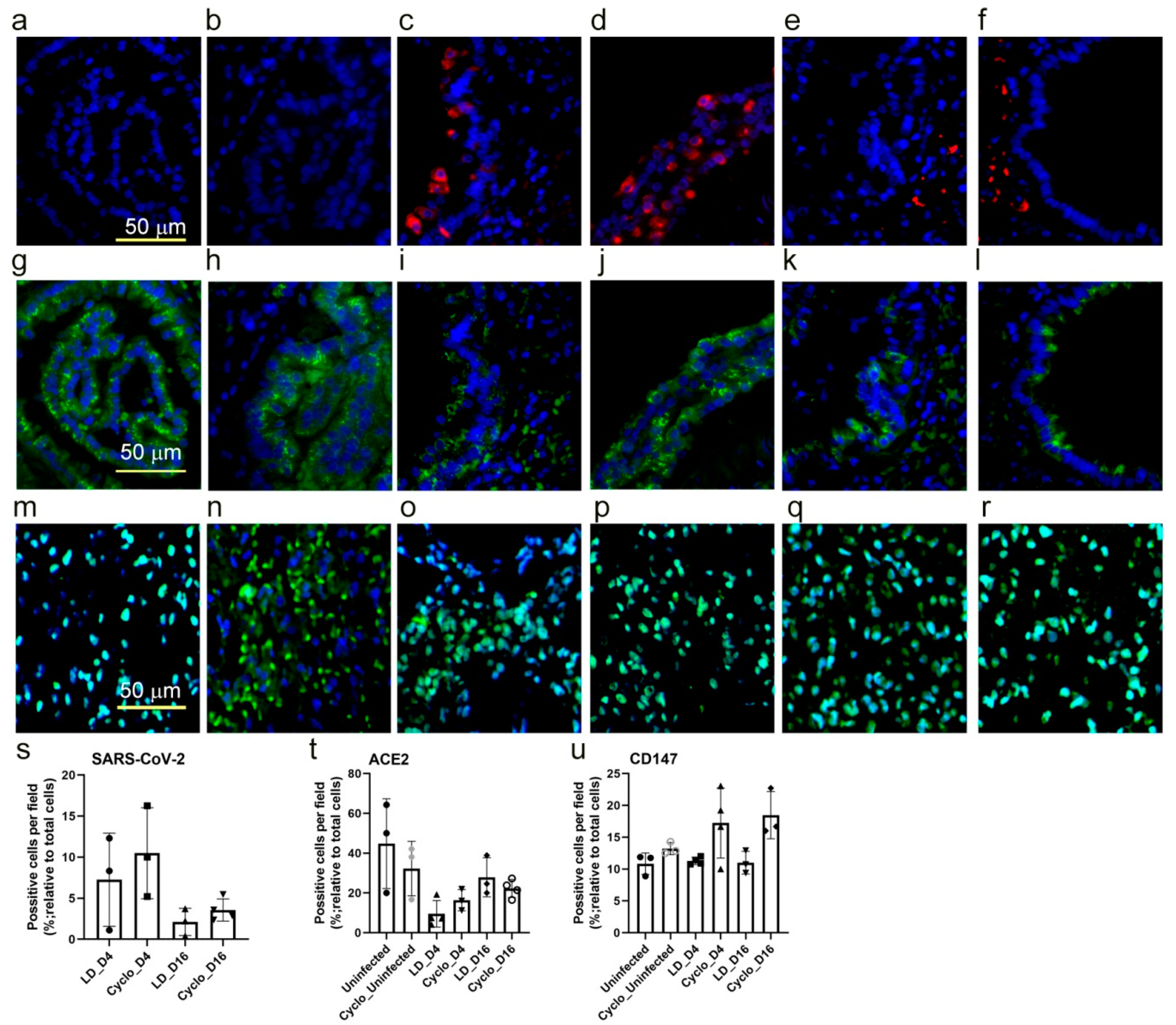
3.9. SARS-CoV-2 Host Cell Entry Receptor Expression in Immunocompetent and Immunocompromised Hamster Lungs
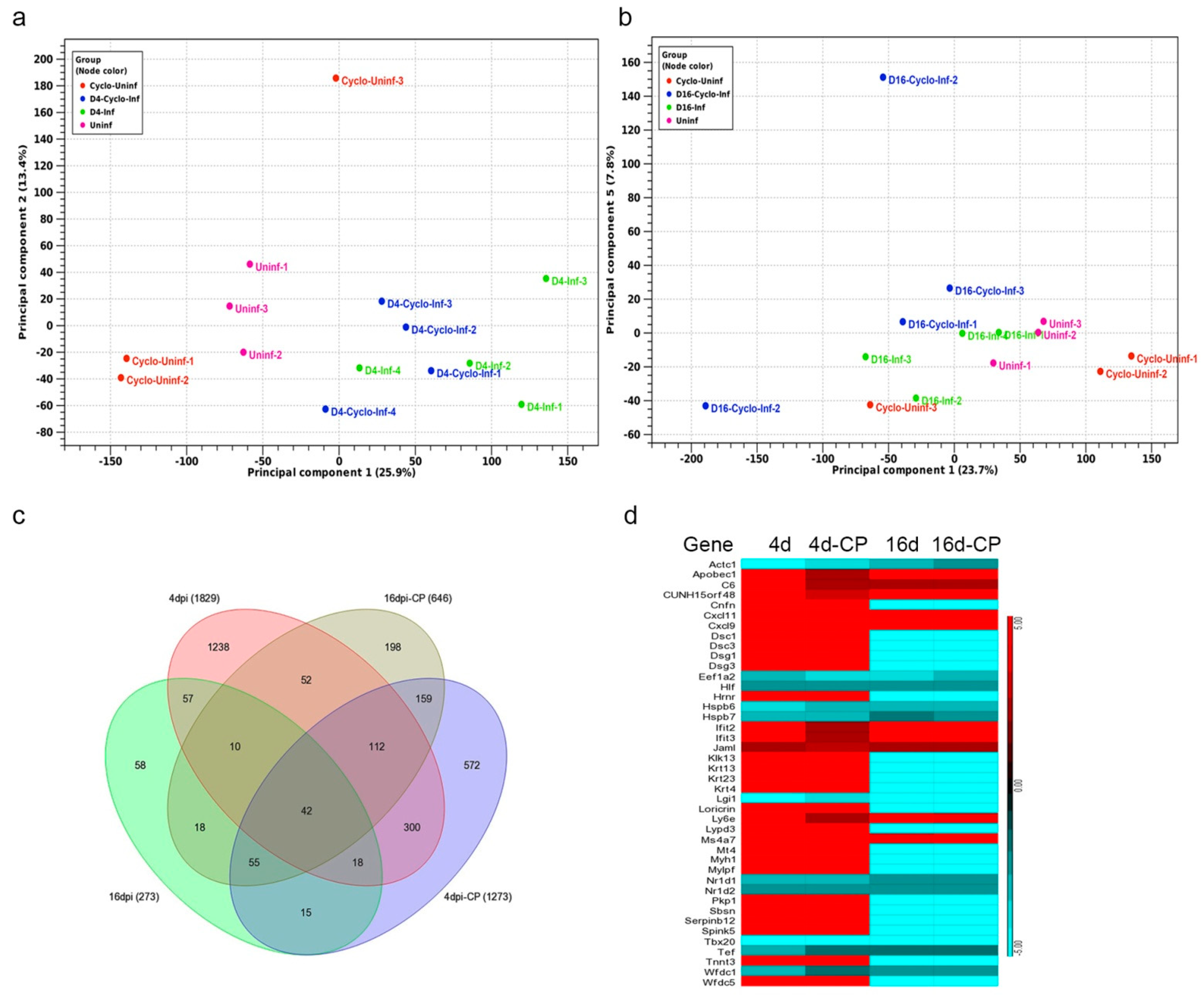
3.10. SARS-CoV-2 Infection Elicits Distinct Transcriptome Profiles in the Lungs
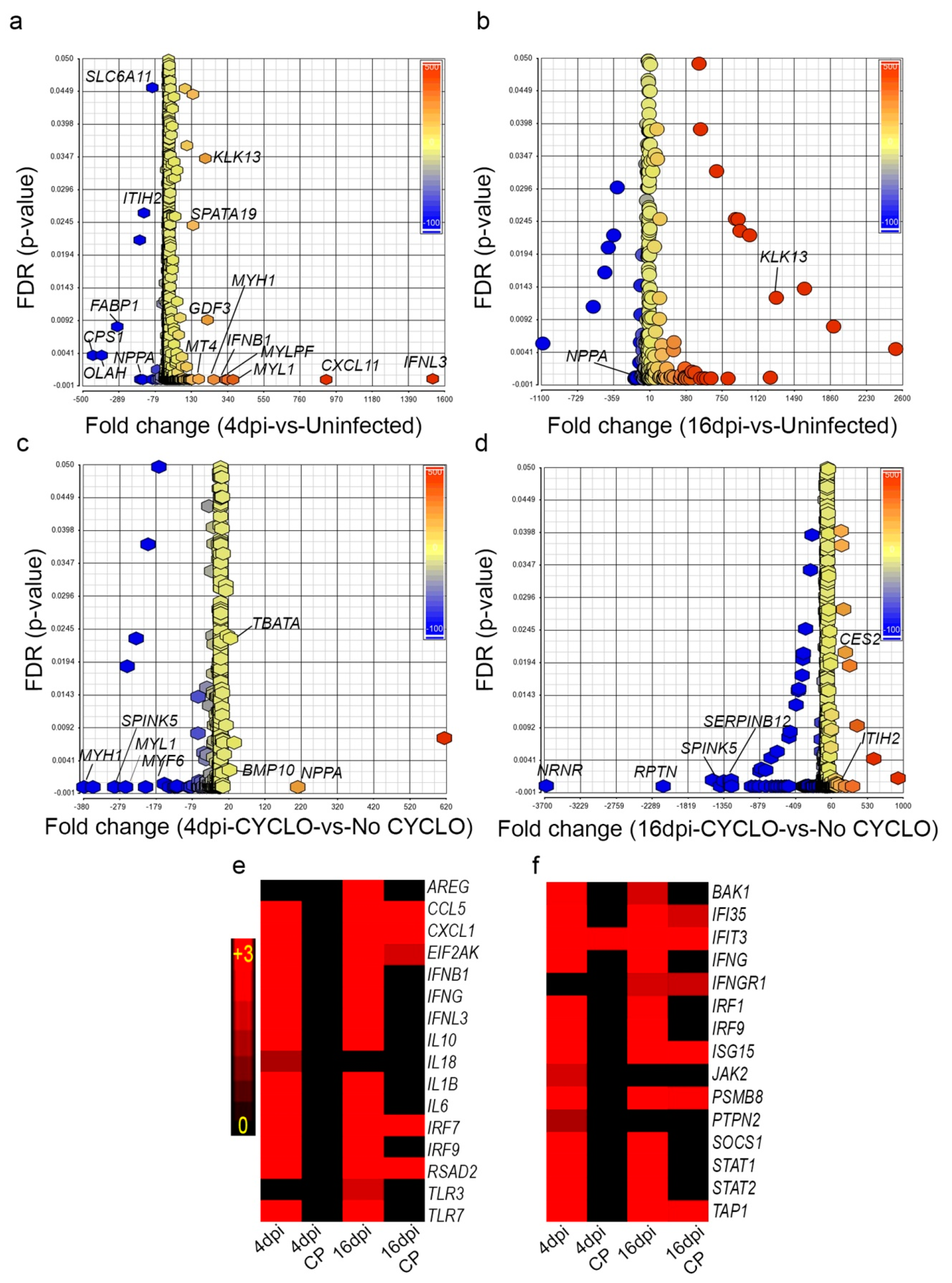
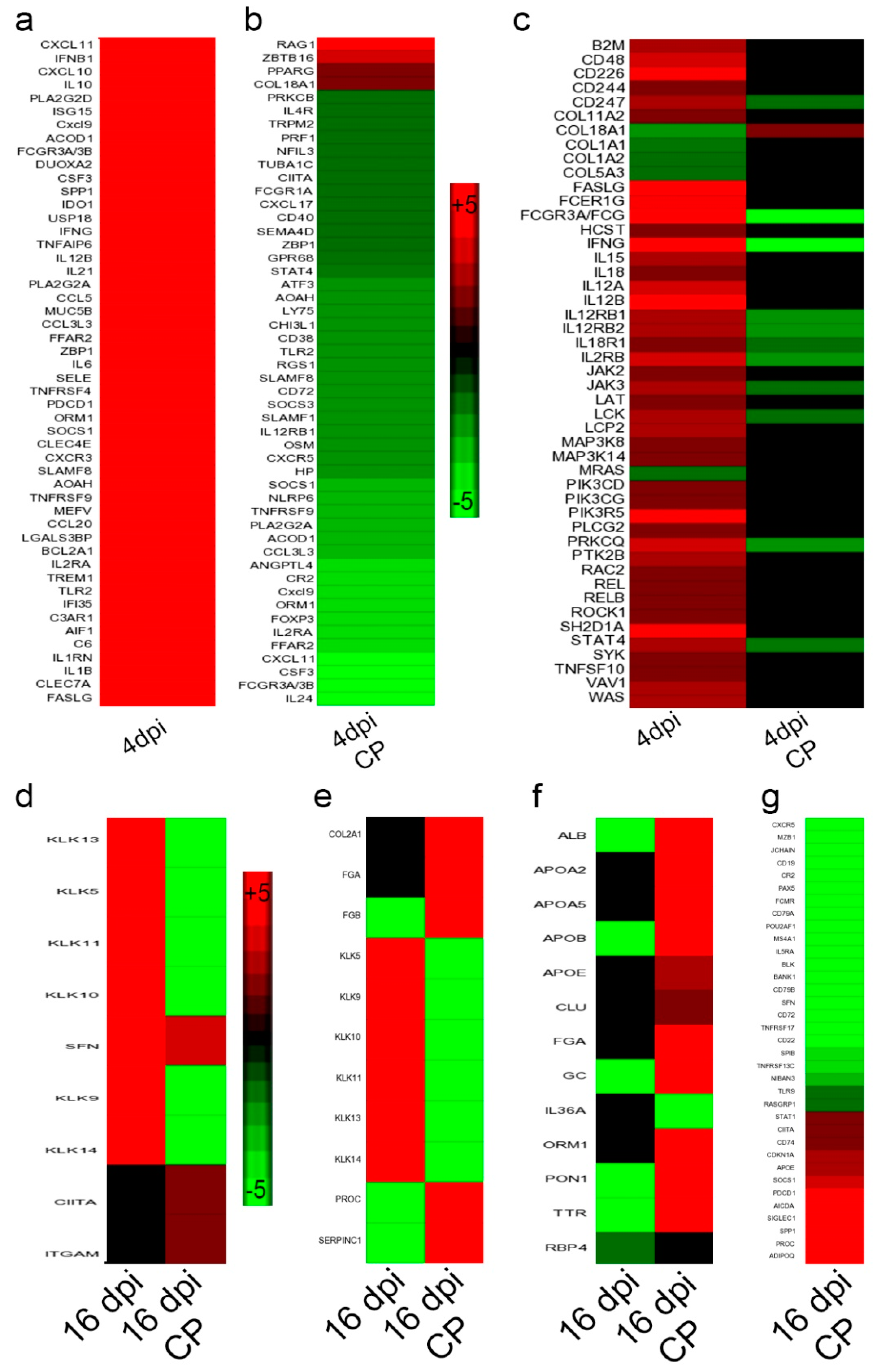
3.11. SARS-CoV-2 Infection Elicits Distinct Acute and Chronic Transcriptome Profiles in the Lungs of the Immunocompetent Host
3.12. Dampened Proinflammatory Response Pathways in the Lungs of SARS-CoV-2-Infected Immunocompromised Hamsters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 12 April 2022).
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Chang, M.G.; Yuan, X.; Tao, Y.; Peng, X.; Wang, F.S.; Xie, L.; Sharma, L.; Dela Cruz, C.S.; Qin, E. Time Kinetics of Viral Clearance and Resolution of Symptoms in Novel Coronavirus Infection. Am. J. Respir. Crit. Care Med. 2020, 201, 1150–1152. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhao, J.; Han, M.; Meng, F.; Zhou, J. SARS-CoV-2 infection in immunocompromised patients: Humoral versus cell-mediated immunity. J. Immunother. Cancer 2020, 8, e000862. [Google Scholar] [CrossRef]
- Lang, M.; Som, A.; Carey, D.; Reid, N.; Mendoza, D.P.; Flores, E.J.; Li, M.D.; Shepard, J.O.; Little, B.P. Pulmonary Vascular Manifestations of COVID-19 Pneumonia. Radiol. Cardiothorac. Imaging 2020, 2, e200277. [Google Scholar] [CrossRef]
- Roberts, C.M.; Levi, M.; McKee, M.; Schilling, R.; Lim, W.S.; Grocott, M.P.W. COVID-19: A complex multisystem disorder. Br. J. Anaesth. 2020, 125, 238–242. [Google Scholar] [CrossRef]
- Lacy, J.M.; Brooks, E.G.; Akers, J.; Armstrong, D.; Decker, L.; Gonzalez, A.; Humphrey, W.; Mayer, R.; Miller, M.; Perez, C.; et al. COVID-19: Postmortem Diagnostic and Biosafety Considerations. Am. J. Forensic. Med. Pathol. 2020, 41, 143–151. [Google Scholar] [CrossRef]
- Carsana, L.; Sonzogni, A.; Nasr, A.; Rossi, R.S.; Pellegrinelli, A.; Zerbi, P.; Rech, R.; Colombo, R.; Antinori, S.; Corbellino, M.; et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infect. Dis. 2020, 20, 1135–1140. [Google Scholar] [CrossRef]
- Elsoukkary, S.S.; Mostyka, M.; Dillard, A.; Berman, D.R.; Ma, L.X.; Chadburn, A.; Yantiss, R.K.; Jessurun, J.; Seshan, S.V.; Borczuk, A.C.; et al. Autopsy Findings in 32 Patients with COVID-19: A Single-Institution Experience. Pathobiology 2021, 88, 56–68. [Google Scholar] [CrossRef]
- Falasca, L.; Nardacci, R.; Colombo, D.; Lalle, E.; Di Caro, A.; Nicastri, E.; Antinori, A.; Petrosillo, N.; Marchioni, L.; Biava, G.; et al. Postmortem Findings in Italian Patients with COVID-19: A Descriptive Full Autopsy Study of Cases with and without Comorbidities. J. Infect. Dis. 2020, 222, 1807–1815. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef]
- Case, J.B.; Bailey, A.L.; Kim, A.S.; Chen, R.E.; Diamond, M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology 2020, 548, 39–48. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, E.J.; Manguiat, K.; Wood, H.; Drebot, M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57, ecpmc105. [Google Scholar] [CrossRef] [PubMed]
- Schaecher, S.R.; Stabenow, J.; Oberle, C.; Schriewer, J.; Buller, R.M.; Sagartz, J.E.; Pekosz, A. An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology 2008, 380, 312–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subbian, S.; Tsenova, L.; Holloway, J.; Peixoto, B.; O’Brien, P.; Dartois, V.; Khetani, V.; Zeldis, J.B.; Kaplan, G. Adjunctive Phosphodiesterase-4 Inhibitor Therapy Improves Antibiotic Response to Pulmonary Tuberculosis in a Rabbit Model. EBioMedicine 2016, 4, 104–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Subbian, S. Immune Correlates of Non-Necrotic and Necrotic Granulomas in Pulmonary Tuberculosis: A Pilot Study. J. Respir. 2021, 1, 248–259. [Google Scholar] [CrossRef]
- Perera, R.; Ko, R.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.M.; Brackman, C.J.; To, E.M.W.; Yen, H.L.; Leung, K.; Cheng, S.M.S.; et al. Evaluation of a SARS-CoV-2 Surrogate Virus Neutralization Test for Detection of Antibody in Human, Canine, Cat, and Hamster Sera. J. Clin. Microbiol. 2021, 59, e02504-20. [Google Scholar] [CrossRef]
- Subbian, S.; Bandyopadhyay, N.; Tsenova, L.; O’Brien, P.; Khetani, V.; Kushner, N.L.; Peixoto, B.; Soteropoulos, P.; Bader, J.S.; Karakousis, P.C.; et al. Early innate immunity determines outcome of Mycobacterium tuberculosis pulmonary infection in rabbits. Cell Commun. Signal. 2013, 11, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subbian, S.; O’Brien, P.; Kushner, N.L.; Yang, G.; Tsenova, L.; Peixoto, B.; Bandyopadhyay, N.; Bader, J.S.; Karakousis, P.C.; Fallows, D.; et al. Molecular immunologic correlates of spontaneous latency in a rabbit model of pulmonary tuberculosis. Cell Commun. Signal. 2013, 11, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karwaciak, I.; Salkowska, A.; Karas, K.; Dastych, J.; Ratajewski, M. Nucleocapsid and Spike Proteins of the Coronavirus SARS-CoV-2 Induce IL6 in Monocytes and Macrophages-Potential Implications for Cytokine Storm Syndrome. Vaccines 2021, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Suess, C.; Hausmann, R. Gross and histopathological pulmonary findings in a COVID-19 associated death during self-isolation. Int. J. Leg. Med. 2020, 134, 1285–1290. [Google Scholar] [CrossRef]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA 2020, 117, 16587–16595. [Google Scholar] [CrossRef]
- Sia, S.F.; Yan, L.M.; Chin, A.W.H.; Fung, K.; Choy, K.T.; Wong, A.Y.L.; Kaewpreedee, P.; Perera, R.; Poon, L.L.M.; Nicholls, J.M.; et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [Google Scholar] [CrossRef]
- Rosenke, K.; Meade-White, K.; Letko, M.; Clancy, C.; Hansen, F.; Liu, Y.; Okumura, A.; Tang-Huau, T.L.; Li, R.; Saturday, G.; et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2673–2684. [Google Scholar] [CrossRef]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clin. Infect. Dis. 2020, 71, 2428–2446. [Google Scholar] [CrossRef] [PubMed]
- Deinhardt-Emmer, S.; Wittschieber, D.; Sanft, J.; Kleemann, S.; Elschner, S.; Haupt, K.F.; Vau, V.; Haring, C.; Rodel, J.; Henke, A.; et al. Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage. eLife 2021, 10, e60361. [Google Scholar] [CrossRef]
- Dong, M.; Zhang, J.; Ma, X.; Tan, J.; Chen, L.; Liu, S.; Xin, Y.; Zhuang, L. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed. Pharmacother. 2020, 131, 110678. [Google Scholar] [CrossRef] [PubMed]
- Wolfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Muller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Tostanoski, L.H.; Wegmann, F.; Martinot, A.J.; Loos, C.; McMahan, K.; Mercado, N.B.; Yu, J.; Chan, C.N.; Bondoc, S.; Starke, C.E.; et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020, 26, 1694–1700. [Google Scholar] [CrossRef]
- O’Donnell, K.L.; Pinski, A.N.; Clancy, C.S.; Gourdine, T.; Shifflett, K.; Fletcher, P.; Messaoudi, I.; Marzi, A. Pathogenic and transcriptomic differences of emerging SARS-CoV-2 variants in the Syrian golden hamster model. EBioMedicine 2021, 73, 103675. [Google Scholar] [CrossRef] [PubMed]
- Brocato, R.L.; Principe, L.M.; Kim, R.K.; Zeng, X.; Williams, J.A.; Liu, Y.; Li, R.; Smith, J.M.; Golden, J.W.; Gangemi, D.; et al. Disruption of Adaptive Immunity Enhances Disease in SARS-CoV-2-Infected Syrian Hamsters. J. Virol. 2020, 94, e01683-20. [Google Scholar] [CrossRef]
- Zhu, L.P.; Cupps, T.R.; Whalen, G.; Fauci, A.S. Selective effects of cyclophosphamide therapy on activation, proliferation, and differentiation of human B cells. J. Clin. Investig. 1987, 79, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Schramm, M.A.; Venhoff, N.; Wagner, D.; Thiel, J.; Huzly, D.; Craig-Mueller, N.; Panning, M.; Hengel, H.; Kern, W.V.; Voll, R.E. COVID-19 in a Severely Immunosuppressed Patient with Life-Threatening Eosinophilic Granulomatosis with Polyangiitis. Front. Immunol. 2020, 11, 2086. [Google Scholar] [CrossRef]
- Ramasamy, S.; Subbian, S. Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00299-20. [Google Scholar] [CrossRef]
- Di Filippo, L.; De Lorenzo, R.; D’Amico, M.; Sofia, V.; Roveri, L.; Mele, R.; Saibene, A.; Rovere-Querini, P.; Conte, C. COVID-19 is associated with clinically significant weight loss and risk of malnutrition, independent of hospitalisation: A post-hoc analysis of a prospective cohort study. Clin. Nutr. 2021, 40, 2420–2426. [Google Scholar] [CrossRef] [PubMed]
- Nouailles, G.; Wyler, E.; Pennitz, P.; Postmus, D.; Vladimirova, D.; Kazmierski, J.; Pott, F.; Dietert, K.; Muelleder, M.; Farztdinov, V.; et al. Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19. Nat. Commun. 2021, 12, 4869. [Google Scholar] [CrossRef]
- Cantwell, A.M.; Singh, H.; Platt, M.; Yu, Y.; Lin, Y.H.; Ikeno, Y.; Hubbard, G.; Xiang, Y.; Gonzalez-Juarbe, N.; Dube, P.H. Kinetic Multi-omic Analysis of Responses to SARS-CoV-2 Infection in a Model of Severe COVID-19. J. Virol. 2021, 95, e0101021. [Google Scholar] [CrossRef] [PubMed]
- Traverso, I.; Fenoglio, D.; Negrini, S.; Parodi, A.; Battaglia, F.; Kalli, F.; Conteduca, G.; Tardito, S.; Traverso, P.; Indiveri, F.; et al. Cyclophosphamide inhibits the generation and function of CD8(+) regulatory T cells. Hum. Immunol. 2012, 73, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Lutsiak, M.E.; Semnani, R.T.; De Pascalis, R.; Kashmiri, S.V.; Schlom, J.; Sabzevari, H. Inhibition of CD4(+)25+ T regulatory cell function implicated in enhanced immune response by low-dose cyclophosphamide. Blood 2005, 105, 2862–2868. [Google Scholar] [CrossRef] [PubMed]
- Motawea, A.M.; Omar, S.; Yasin, R. Imaging of COVID-19 simulators. Egypt. J. Radiol. Nucl. Med. 2021, 52, 4. [Google Scholar] [CrossRef]
- Ye, Z.; Zhang, Y.; Wang, Y.; Huang, Z.; Song, B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): A pictorial review. Eur. Radiol. 2020, 30, 4381–4389. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Das, S.; Mondal, R.; Abdullah, S.; Sultana, S.; Singh, S.; Sehgal, A.; Behl, T. A review on the effect of COVID-19 in type 2 asthma and its management. Int. Immunopharmacol. 2021, 91, 107309. [Google Scholar] [CrossRef]
- Rinaldi, L.F.; Marazzi, G.; Marone, E.M. Endovascular Treatment of a Ruptured Pararenal Abdominal Aortic Aneurysm in a Patient with Coronavirus Disease-2019: Suggestions and Case Report. Ann. Vasc. Surg. 2020, 66, 18–23. [Google Scholar] [CrossRef]
- Shih, M.; Swearingen, B.; Rhee, R. Ruptured Abdominal Aortic Aneurysm Treated with Endovascular Repair in a Patient with Active COVID-19 Infection during the Pandemic. Ann. Vasc. Surg. 2020, 66, 14–17. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Xu, Y.J.; Guan, W.J.; Lin, L.F. Regional, age and respiratory-secretion-specific prevalence of respiratory viruses associated with asthma exacerbation: A literature review. Arch. Virol. 2018, 163, 845–853. [Google Scholar] [CrossRef]
- Morais-Almeida, M.; Bousquet, J. COVID-19 and asthma: To have or not to have T2 inflammation makes a difference? Pulmonology 2020, 26, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.J.; Nikolaienko, S.I.; Dibrova, V.A.; Dibrova, Y.V.; Vasylyk, V.M.; Novikov, M.Y.; Shults, N.V.; Gychka, S.G. SARS-CoV-2 spike protein-mediated cell signaling in lung vascular cells. Vasc. Pharmacol. 2021, 137, 106823. [Google Scholar] [CrossRef]
- Mehta, P.K.; Griendling, K.K. Angiotensin II cell signaling: Physiological and pathological effects in the cardiovascular system. Am. J. Physiol. Cell Physiol. 2007, 292, C82–C97. [Google Scholar] [CrossRef] [PubMed]
- Freire Santana, M.; Borba, M.G.S.; Baia-da-Silva, D.C.; Val, F.; Alexandre, M.A.A.; Brito-Sousa, J.D.; Melo, G.C.; Queiroga, M.V.O.; Leao Farias, M.E.; Camilo, C.C.; et al. Case Report: Adrenal Pathology Findings in Severe COVID-19: An Autopsy Study. Am. J. Trop. Med. Hyg. 2020, 103, 1604–1607. [Google Scholar] [CrossRef]
- Mondello, C.; Roccuzzo, S.; Malfa, O.; Sapienza, D.; Gualniera, P.; Ventura Spagnolo, E.; Di Nunno, N.; Salerno, M.; Pomara, C.; Asmundo, A. Pathological Findings in COVID-19 as a Tool to Define SARS-CoV-2 Pathogenesis. A Systematic Review. Front. Pharmacol. 2021, 12, 614586. [Google Scholar] [CrossRef] [PubMed]
- Kidambi, S.; Kotchen, J.M.; Grim, C.E.; Raff, H.; Mao, J.; Singh, R.J.; Kotchen, T.A. Association of adrenal steroids with hypertension and the metabolic syndrome in blacks. Hypertension 2007, 49, 704–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moneva, M.H.; Gomez-Sanchez, C.E. Pathophysiology of adrenal hypertension. Semin. Nephrol. 2002, 22, 44–53. [Google Scholar] [CrossRef]
- Ji, D.; Qin, E.; Xu, J.; Zhang, D.; Cheng, G.; Wang, Y.; Lau, G. Non-alcoholic fatty liver diseases in patients with COVID-19: A retrospective study. J. Hepatol. 2020, 73, 451–453. [Google Scholar] [CrossRef]
- Xu, L.; Liu, J.; Lu, M.; Yang, D.; Zheng, X. Liver injury during highly pathogenic human coronavirus infections. Liver Int. 2020, 40, 998–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; Yang, M.; Wan, C.; Yi, L.X.; Tang, F.; Zhu, H.Y.; Yi, F.; Yang, H.C.; Fogo, A.B.; Nie, X.; et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020, 98, 219–227. [Google Scholar] [CrossRef]
- Sharma, P.; Uppal, N.N.; Wanchoo, R.; Shah, H.H.; Yang, Y.; Parikh, R.; Khanin, Y.; Madireddy, V.; Larsen, C.P.; Jhaveri, K.D.; et al. COVID-19-Associated Kidney Injury: A Case Series of Kidney Biopsy Findings. J. Am. Soc. Nephrol. 2020, 31, 1948–1958. [Google Scholar] [CrossRef]
- Ahmadian, E.; HosseiniyanKhatibi, S.M.; RaziSoofiyani, S.; Abediazar, S.; Shoja, M.M.; Ardalan, M.; ZununiVahed, S. COVID-19 and kidney injury: Pathophysiology and molecular mechanisms. Rev. Med. Virol. 2021, 31, e2176. [Google Scholar] [CrossRef]



| Day 2 | Day 4 | Day 7 | Day 12 | Day 16 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organ | LD | HD | CP-LD | LD | HD | CP-LD | LD | HD | CP-LD | LD | HD | CP-LD | LD | HD | CP-LD |
| Nasal wash | × | × | × | × | × | × | × | × | - | - | × | - | - | × | |
| Nasal turbinates | × | × | × | × | × | × | × | × | × | - | - | × | - | - | × |
| Larynx and trachea | × | × | × | × | × | × | × | - | - | × | - | - | × | ||
| Lung | × | × | × | × | × | × | × | × | × | - | - | × | - | - | × |
| Heart | × | × | × | × | × | × | - | - | × | - | - | × | - | - | × |
| Adrenal | × | × | × | × | × | × | - | - | - | - | - | - | - | × | |
| Kidney | × | × | × | × | - | - | × | - | - | - | - | - | - | ||
| Epididymal fat | × | × | × | × | × | × | - | - | × | - | - | - | - | - | × |
| Brain with olfactory bulb | × | × | × | × | × | × | × | × | |||||||
| Eye | × | × | × | × | × | × | × | ||||||||
| Colon | × | × | × | ||||||||||||
| Bone marrow | × | × | × | × | × | × | × | ||||||||
| Liver | × | × | × | × | |||||||||||
| Spleen | × | × | × | × | × | × | |||||||||
| Lesion Parameters (Scale) | Virus Dose/Treatment | Day 4 (Mean ± SD) | Day 7 (Mean ± SD) | Day 16 (Mean ± SD) |
|---|---|---|---|---|
| Mononuclear cells infiltration (0–5) | LD | 2.33 ± 0.577 | 3.667 ± 0.288 | 2 ± 0 |
| HD | 3.33 ± 0.577 | 4.33 ± 1.15 | 4 ± 0 | |
| CP-LD | 2 ± 0 | 2.33 ± 0.288 | 2 ± 0 | |
| Alveolar hyperplasia (0–5) | LD | 0.66 ± 0.577 | 1.833 ± 1.25 | 1.16 ± 0.76 |
| HD | 2.66 ± 0.57 | 5 ± 0 | 4 ± 0 | |
| CP-LD | 2.16 ± 0.288 | 2 ± 0 | 3 ± 0 | |
| Interstitial and alveolar edema (0–4) | LD | 0.66 ± 0.288 | 2.33 ± 0.577 | 0.166 ± 0.288 |
| HD | 1.33 ± 0.577 | 1.66 ± 0.577 | 0.5 ± 0.7 | |
| CP-LD | 0.66 ± 0.288 | 2.166 ± 0.288 | 0 | |
| Emphysema (0–4) | LD | 0.33 ± 0.577 | 1 ± 0 | 0 |
| HD | 1.33 ± 1.15 | 2.66 ± 0.577 | 0.5 ± 0.707 | |
| CP-LD | 1.66 ± 0.577 | 1 ± 0 | 1 ± 0 | |
| Bronchiolar epithelial hyperplasia and inflammation (0–4) | LD | 1.66 ± 0.577 | 2.66 ± 0.577 | 1.83 ± 0.28 |
| HD | 2.66 ± 1.15 | 4 ± 0 | 3.5 ± 0.7 | |
| CP-LD | 2.33 ± 0.577 | 2 ± 0 | 2.33 ± 1.52 | |
| Bronchiolar smooth muscle hyperplasia (0–4) | LD | 0.5 ± 0.866 | 0.66 ± 1.15 | 0.66 ± 1.15 |
| HD | 0.66 ± 1.15 | 1 ± 1 | 3 ± 0 | |
| CP-LD | 1 ± 1 | 0 | 0 | |
| Vascular lesions; vasculitis, congestion, perivasculitis (0–4) | LD | 2 ± 0 | 2.66 ± 0.577 | 1.33 ± 0.577 |
| HD | 2.66 ± 0.577 | 3.66 ± 0.577 | 2 ± 0 | |
| CP-LD | 1.33 ± 0.577 | 1.66 ± 1.15 | 1.5 ± 0.70 | |
| Hyperplasia/hypertrophy of vascular smooth muscle (0–4) | LD | 1.33 ± 1.15 | 1 ± 0.866 | 2 ± 1 |
| HD | 2.83 ± 0.76 | 2.16 ± 1.04 | 4 ± 0 | |
| CP-LD | 1 ± 1 | 1.66 ± 0.577 | 3 ± 0 | |
| Foamy macrophages (0–4) | LD | 0 ± 0 | 2 ± 0 | 1 ± 0 |
| HD | 0 ± 0 | 1.66 ± 0.577 | 0.5 ± 0.7 | |
| CP-LD | 0 ± 0 | 1 ± 0 | 0.5 ± 0.866 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramasamy, S.; Kolloli, A.; Kumar, R.; Husain, S.; Soteropoulos, P.; Chang, T.L.; Subbian, S. Comprehensive Analysis of Disease Pathology in Immunocompetent and Immunocompromised Hosts following Pulmonary SARS-CoV-2 Infection. Biomedicines 2022, 10, 1343. https://doi.org/10.3390/biomedicines10061343
Ramasamy S, Kolloli A, Kumar R, Husain S, Soteropoulos P, Chang TL, Subbian S. Comprehensive Analysis of Disease Pathology in Immunocompetent and Immunocompromised Hosts following Pulmonary SARS-CoV-2 Infection. Biomedicines. 2022; 10(6):1343. https://doi.org/10.3390/biomedicines10061343
Chicago/Turabian StyleRamasamy, Santhamani, Afsal Kolloli, Ranjeet Kumar, Seema Husain, Patricia Soteropoulos, Theresa L. Chang, and Selvakumar Subbian. 2022. "Comprehensive Analysis of Disease Pathology in Immunocompetent and Immunocompromised Hosts following Pulmonary SARS-CoV-2 Infection" Biomedicines 10, no. 6: 1343. https://doi.org/10.3390/biomedicines10061343
APA StyleRamasamy, S., Kolloli, A., Kumar, R., Husain, S., Soteropoulos, P., Chang, T. L., & Subbian, S. (2022). Comprehensive Analysis of Disease Pathology in Immunocompetent and Immunocompromised Hosts following Pulmonary SARS-CoV-2 Infection. Biomedicines, 10(6), 1343. https://doi.org/10.3390/biomedicines10061343