Deciphering the Role of Histone Modifications in Uterine Leiomyoma: Acetylation of H3K27 Regulates the Expression of Genes Involved in Proliferation, Cell Signaling, Cell Transport, Angiogenesis and Extracellular Matrix Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data/Samples Acquisition
2.2. CHIP-Seq Analysis
2.3. RNA-Seq Analysis
2.4. Correlation of H3K27ac and Gene Expression
2.5. Functional Enrichment Analysis
2.6. Sample Collection
2.7. Validation of Gene Expression: qRT-PCR
2.8. SAHA Treatment and Gene Expression Analysis in Human Uterine Leiomyoma Primary Cells
2.9. Statistical Analysis
3. Results
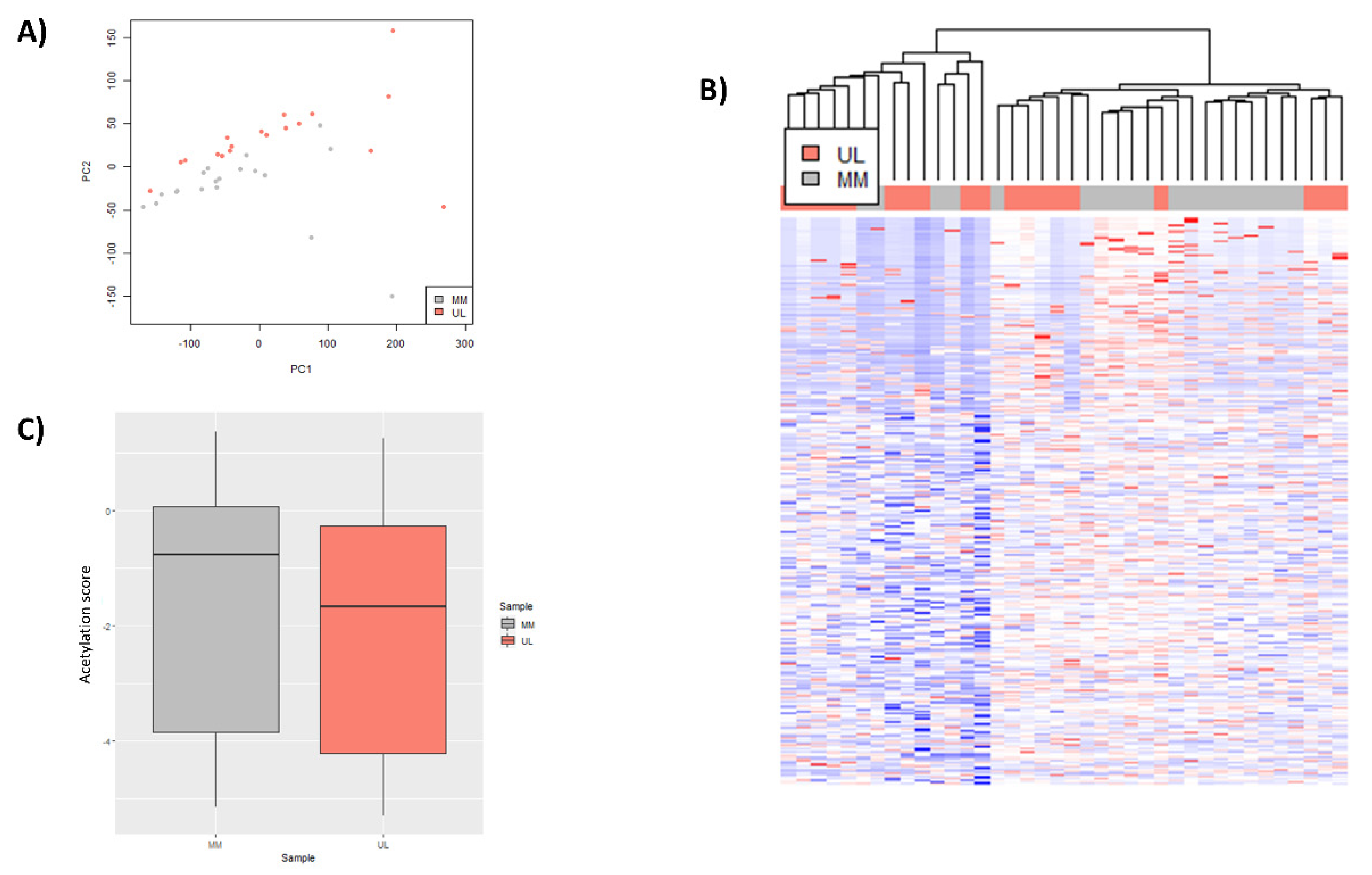
3.1. Global H3K27ac CHIP-Seq Peak Profile in Uterine Leiomyoma Tissue Compared to Adjacent Myometrium
3.2. Selection of Relevant Differentially Expressed Genes
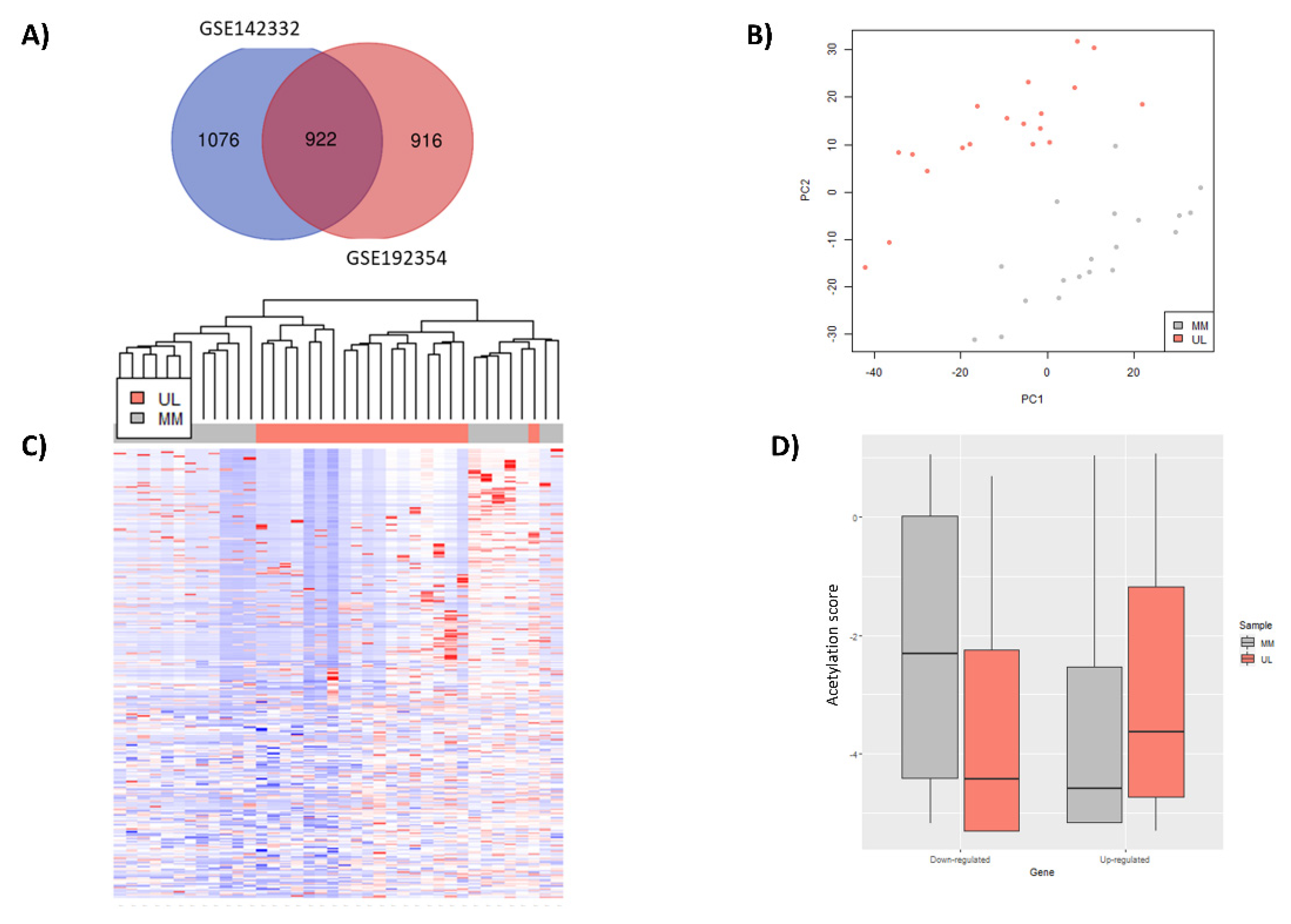
3.3. Identification Differentially Expressed Genes with an Aberrant H3K27ac Mark in Uterine Leiomyoma Tissue Compared to Adjacent Myometrium
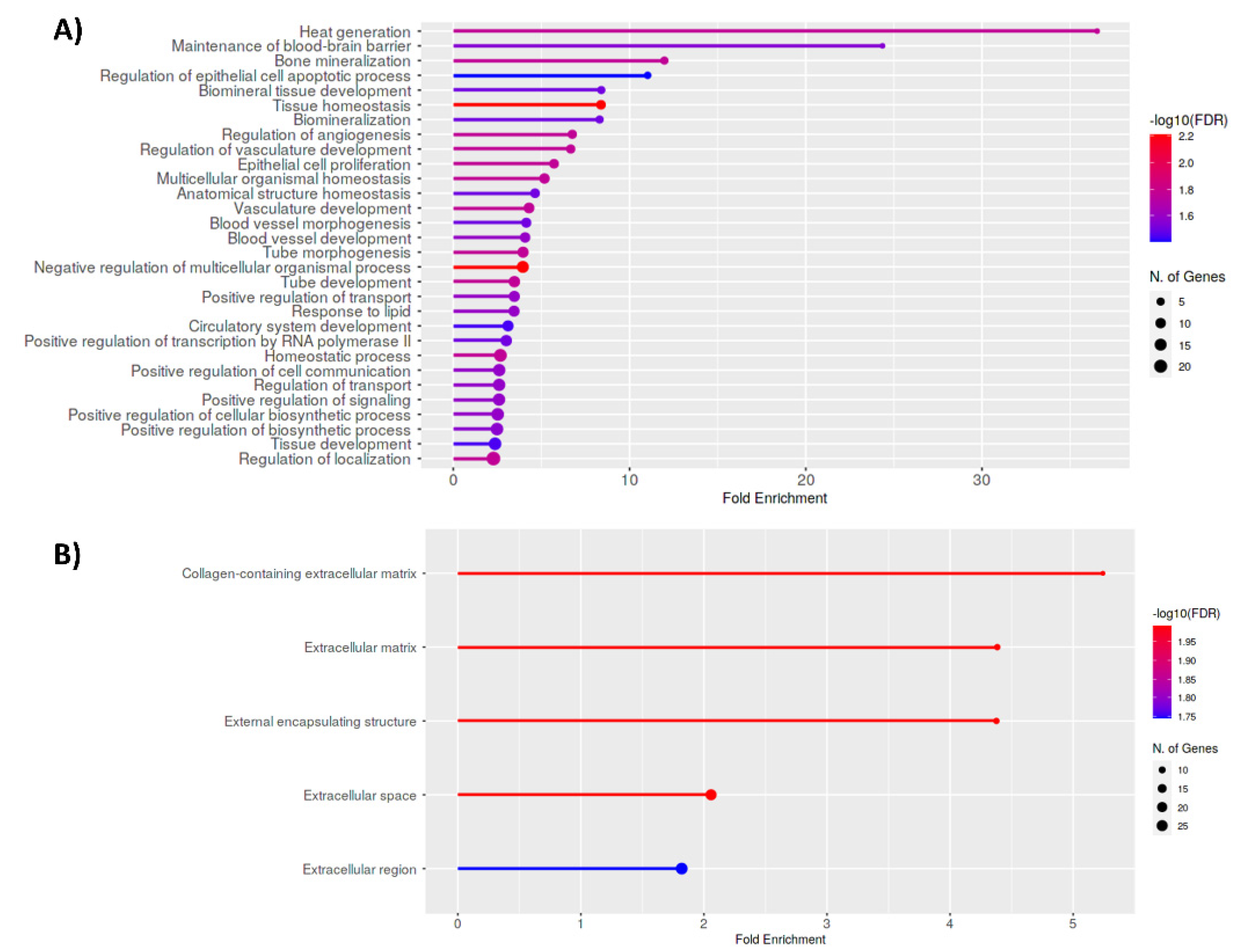
3.4. Functional Implications of Differentially Expressed Genes Associated with Aberrant H3K27 Acetylation in Uterine Leiomyoma Tissue Compared to Adjacent Myometrium
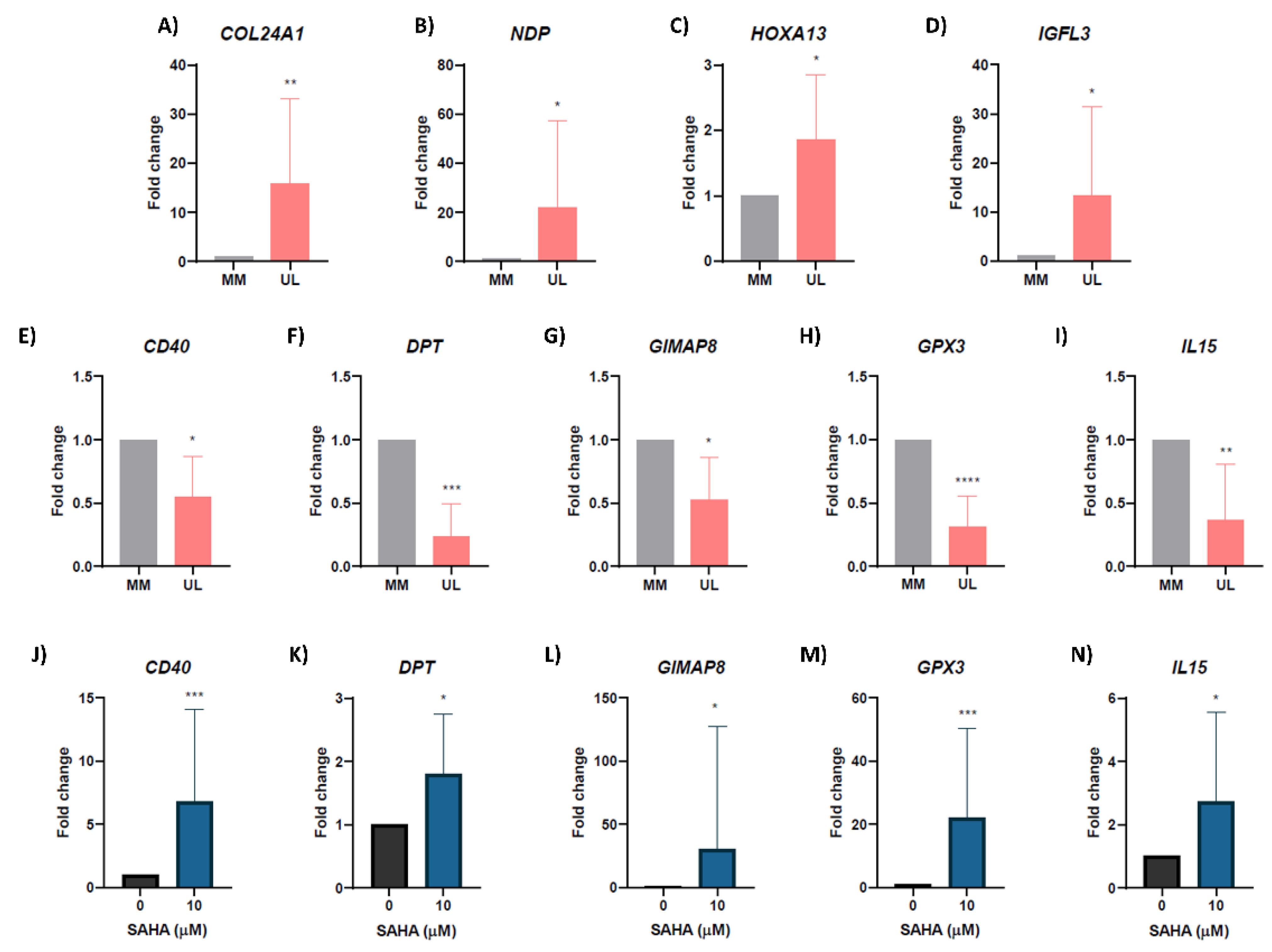
3.5. Validation of Hypoacetylated/Downregulated and Hyperacetylated/Upregulated Genes
3.6. Inhibiting Histone Deacetylases Reverses Expression of Hypoacetylated/Downregulated Genes in Human Uterine Leiomyoma Primary Cells In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stewart, E.A.; Laughlin-Tommaso, S.K.; Catherino, W.H.; Lalitkumar, S.; Gupta, D.; Vollenhoven, B. Uterine Fibroids. Nat. Rev. Dis. Prim. 2016, 2, 16043. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.H. Etiology, Symptomatology, and Diagnosis of Uterine Myomas. Fertil. Steril. 2007, 87, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Jose, J.; Manyonda, I. Clinical Presentation of Fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2008, 22, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Bulun, S.E. Uterine Fibroids. N. Engl. J. Med. 2013, 369, 1344–1355. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.D.; Malik, M.; Catherino, W.H. Human Myometrium and Leiomyomas Express Gonadotropin-Releasing Hormone 2 and Gonadotropin-Releasing Hormone 2 Receptor. Fertil. Steril. 2007, 88, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Manyonda, I.; Sinthamoney, E.; Belli, A.M. Controversies and Challenges in the Modern Management of Uterine Fibroids. BJOG An Int. J. Obstet. Gynaecol. 2004, 111, 95–102. [Google Scholar] [CrossRef]
- Friedman, A.J. Treatment of Leiomyomata Uteri with Short-Term Leuprolide Followed by Leuprolide plus Estrogen-Progestin Hormone Replacement Therapy for 2 Years: A Pilot Study. Fertil. Steril. 1989, 51, 526–528. [Google Scholar] [CrossRef]
- El Sabeh, M.; Borahay, M. The Future of Uterine Fibroid Management: A More Preventive and Personalized Paradigm. Reprod. Sci. 2021. [Google Scholar] [CrossRef]
- Sant’Anna, G.d.S.; Brum, I.S.; Branchini, G.; Pizzolato, L.S.; Capp, E.; Corleta, H. von E. Ovarian Steroid Hormones Modulate the Expression of Progesterone Receptors and Histone Acetylation Patterns in Uterine Leiomyoma Cells. Gynecol. Endocrinol. 2017, 33, 629–633. [Google Scholar] [CrossRef]
- Maruo, T.; Ohara, N.; Wang, J.; Matsuo, H. Sex Steroidal Regulation of Uterine Leiomyoma Growth and Apoptosis. Hum. Reprod. Update 2004, 10, 207–220. [Google Scholar] [CrossRef] [Green Version]
- Ono, M.; Qiang, W.; Serna, V.A.; Yin, P.; Coon, J.S.; Navarro, A.; Monsivais, D.; Kakinuma, T.; Dyson, M.; Druschitz, S.; et al. Role of Stem Cells in Human Uterine Leiomyoma Growth. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, N.; Mehine, M.; Tolvanen, J.; Kaasinen, E.; Li, Y.; Lehtonen, H.J.; Gentile, M.; Yan, J.; Enge, M.; Taipale, M.; et al. MED12, the Mediator Complex Subunit 12 Gene, Is Mutated at High Frequency in Uterine Leiomyomas. Science 2011, 334, 252–255. [Google Scholar] [CrossRef]
- Mehine, M.; Kaasinen, E.; Heinonen, H.R.; Mäkinen, N.; Kämpjärvi, K.; Sarvilinna, N.; Aavikko, M.; Vähärautio, A.; Pasanen, A.; Bützow, R.; et al. Integrated Data Analysis Reveals Uterine Leiomyoma Subtypes with Distinct Driver Pathways and Biomarkers. Proc. Natl. Acad. Sci. USA 2016, 113, 1315–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehtonen, R.; Kiuru, M.; Vanharanta, S.; Sjöberg, J.; Aaltonen, L.M.; Aittomäki, K.; Arola, J.; Butzow, R.; Eng, C.; Husgafvel-Pursiainen, K.; et al. Biallelic Inactivation of Fumarate Hydratase (FH) Occurs in Nonsyndromic Uterine Leiomyomas but Is Rare in Other Tumors. Am. J. Pathol. 2004, 164, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Stewart, E.; Cookson, C.; Gandolfo, R.; Schulze-Rath, R. Epidemiology of Uterine Fibroids: A Systematic Review. BJOG An Int. J. Obstet. Gynaecol. 2017, 124, 1501–1512. [Google Scholar] [CrossRef]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Navarro, A.; Yin, P.; Monsivais, D.; Lin, S.M.; Du, P.; Wei, J.; Bulun, S.E. Genome-wide DNA methylation indicates silencing of tumor suppressor genes in uterine leiomyoma. PLoS ONE 2012, 7, e33284. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, R.; Sato, S.; Yamagata, Y.; Asada, H.; Tamura, I.; Lee, L.; Okada, M.; Tamura, H.; Takaki, E.; Nakai, A.; et al. Genome-Wide DNA Methylation Analysis Reveals a Potential Mechanism for the Pathogenesis and Development of Uterine Leiomyomas. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Carbajo-García, M.C.; Corachán, A.; Juárez-Barber, E.; Monleón, J.; Payá, V.; Trelis, A.; Quiñonero, A.; Pellicer, A.; Ferrero, H. Integrative Analysis of the DNA Methylome and Transcriptome in Uterine Leiomyoma Shows Altered Regulation of Genes Involved in Metabolism, Proliferation, Extracellular Matrix and Vesicles. J. Pathol. 2022. [Google Scholar] [CrossRef]
- Liu, S.; Yin, P.; Xu, J.; Dotts, A.J.; Kujawa, S.A.; Coon, V.J.S.; Zhao, H.; Shilatifard, A.; Dai, Y.; E Bulun, S. Targeting DNA Methylation Depletes Uterine Leiomyoma Stem Cell–enriched Population by Stimulating Their Differentiation. Endocrinology 2020, 161. [Google Scholar] [CrossRef]
- George, J.W.; Fan, H.; Johnson, B.; Carpenter, T.J.; Foy, K.K.; Chatterjee, A.; Patterson, A.L.; Koeman, J.; Adams, M.; Madaj, Z.B.; et al. Integrated Epigenome, Exome, and Transcriptome Analyses Reveal Molecular Subtypes and Homeotic Transformation in Uterine Fibroids. Cell Rep. 2019, 29, 4069–4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berta, D.G.; Kuisma, H.; Välimäki, N.; Räisänen, M.; Jäntti, M.; Pasanen, A.; Karhu, A.; Kaukomaa, J.; Taira, A.; Cajuso, T.; et al. Deficient H2A.Z Deposition Is Associated with Genesis of Uterine Leiomyoma. Nature 2021, 596, 398–403. [Google Scholar] [CrossRef]
- Yang, Q.; Mas, A.; Diamond, M.P.; Al-Hendy, A. The Mechanism and Function of Epigenetics in Uterine Leiomyoma Development. Reprod. Sci. 2016, 23, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Verdone, L.; Caserta, M.; Di Mauro, E. Role of Histone Acetylation in the Control of Gene Expression. Biochem. Cell Biol. 2005, 83, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallinari, P.; Di Marco, S.; Jones, P.; Pallaoro, M.; Steinkühler, C. HDACs, Histone Deacetylation and Gene Transcription: From Molecular Biology to Cancer Therapeutics. Cell Res. 2007, 17, 195–211. [Google Scholar] [CrossRef]
- Bitler, B.G.; Wu, S.; Park, P.H.; Hai, Y.; Aird, K.M.; Wang, Y.; Zhai, Y.; Kossenkov, A.V.; Vara-Ailor, A.; Rauscher, F.J.; et al. ARID1A-Mutated Ovarian Cancers Depend on HDAC6 Activity. Nat. Cell Biol. 2017, 19, 962–973. [Google Scholar] [CrossRef] [Green Version]
- Hervouet, E.; Claude-Taupin, A.; Gauthier, T.; Perez, V.; Fraichard, A.; Adami, P.; Despouy, G.; Monnien, F.; Algros, M.P.; Jouvenot, M.; et al. The Autophagy GABARAPL1 Gene Is Epigenetically Regulated in Breast Cancer Models. BMC Cancer 2015, 15. [Google Scholar] [CrossRef] [Green Version]
- Carbajo-García, M.C.; García-Alcázar, Z.; Corachán, A.; Monleón, J.; Trelis, A.; Faus, A.; Pellicer, A.; Ferrero, H. Histone Deacetylase Inhibition by Suberoylanilide Hydroxamic Acid: A Therapeutic Approach to Treat Human Uterine Leiomyoma. Fertil. Steril. 2022, 117, 433–443. [Google Scholar] [CrossRef]
- Laganà, A.S.; Vergara, D.; Favilli, A.; La Rosa, V.L.; Tinelli, A.; Gerli, S.; Noventa, M.; Vitagliano, A.; Triolo, O.; Rapisarda, A.M.C.; et al. Epigenetic and Genetic Landscape of Uterine Leiomyomas: A Current View over a Common Gynecological Disease. Arch. Gynecol. Obstet. 2017, 296, 855–867. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Yang, X.; Lu, W.; Chen, Y.; Lin, Y.; Wang, J.; Lin, S.; Yun, J.P. H3K27 Acetylation Activated-COL6A1 Promotes Osteosarcoma Lung Metastasis by Repressing STAT1 and Activating Pulmonary Cancer-Associated Fibroblasts. Theranostics 2021, 11, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, Y.; Zhang, X.; Cai, H.; Zhang, C.; Qu, H.; Liu, L.; Zhang, M.; Fu, J.; Zhang, J.; et al. Oxidative Stress Activates NORAD Expression by H3K27ac and Promotes Oxaliplatin Resistance in Gastric Cancer by Enhancing Autophagy Flux via Targeting the MiR-433-3p. Cell Death Dis. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zheng, W. H3K27ac-Induced LncRNA PAXIP1-AS1 Promotes Cell Proliferation, Migration, EMT and Apoptosis in Ovarian Cancer by Targeting MiR-6744-5p/PCBP2 Axis. J. Ovarian Res. 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Leistico, J.R.; Saini, P.; Futtner, C.R.; Hejna, M.; Omura, Y.; Soni, P.N.; Sandlesh, P.; Milad, M.; Wei, J.J.; Bulun, S.; et al. Epigenomic tensor predicts disease subtypes and reveals constrained tumor evolution. Cell Rep. 2021, 34, 108927. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Carbajo-García, M.C.; Corachán, A.; Segura-Benitez, M.; Monleón, J.; Escrig, J.; Faus, A.; Pellicer, A.; Cervelló, I.; Ferrero, H. 5-Aza-2′-Deoxycitidine Inhibits Cell Proliferation, Extracellular Matrix Formation and Wnt/β-Catenin Pathway in Human Uterine Leiomyomas. Reprod. Biol. Endocrinol. 2021, 19. [Google Scholar] [CrossRef]
- Sato, S.; Maekawa, R.; Tamura, I.; Shirafuta, Y.; Shinagawa, M.; Asada, H.; Taketani, T.; Tamura, H.; Sugino, N. SATB2 and NGR1: Potential Upstream Regulatory Factors in Uterine Leiomyomas. J. Assist. Reprod. Genet. 2019, 36, 2385–2397. [Google Scholar] [CrossRef]
- Moyo, M.B.; Parker, J.B.; Chakravarti, D. Altered chromatin landscape and enhancer engagement underlie transcriptional dysregulation in MED12 mutant uterine leiomyomas. Nat. Commun. 2020, 11, 1019. [Google Scholar] [CrossRef]
- Planutis, K.; Planutiene, M.; Holcombe, R.F. A Novel Signaling Pathway Regulates Colon Cancer Angiogenesis through Norrin. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- El-Sehemy, A.; Selvadurai, H.; Ortin-Martinez, A.; Pokrajac, N.; Mamatjan, Y.; Tachibana, N.; Rowland, K.; Lee, L.; Park, N.; Aldape, K.; et al. Norrin Mediates Tumor-Promoting and -Suppressive Effects in Glioblastoma via Notch and Wnt. J. Clin. Investig. 2020, 130, 3069–3086. [Google Scholar] [CrossRef] [Green Version]
- Duan, R.; Han, L.; Wang, Q.; Wei, J.; Chen, L.; Zhang, J.; Kang, C.; Wang, L. HOXA13 Is a Potential GBM Diagnostic Marker and Promotes Glioma Invasion by Activating the Wnt and TGF-β Pathways. Oncotarget 2015, 6, 27778–27793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Gu, J.; Shen, K.; Zhou, H.; Hao, J.; Li, F.; Yu, H.; Chen, Y.; Li, J.; Li, Y.; et al. HOXA13 Promotes Colon Cancer Progression through β-Catenin-Dependent WNT Pathway. Exp. Cell Res. 2020, 395, 112238. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.T.; Jia, W.D.; Yao, Q.Y.; Sun, Q.K.; Ren, W.H.; Huang, M.; Ma, J.; Li, J.S.; Ma, J.L.; Yu, J.H.; et al. Overexpression of HOXA13 as a Potential Marker for Diagnosis and Poor Prognosis of Hepatocellular Carcinoma. Tohoku J. Exp. Med. 2014, 234, 209–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamaluddin, M.F.B.; Nahar, P.; Tanwar, P.S. Proteomic Characterization of the Extracellular Matrix of Human Uterine Fibroids. Endocrinology 2018, 159, 2656–2669. [Google Scholar] [CrossRef]
- Yan, L.; Xu, F.; Dai, C. Overexpression of COL24A1 in Hepatocellular Carcinoma Predicts Poor Prognosis: A Study Based on Multiple Databases, Clinical Samples and Cell Lines. Onco. Targets. Ther. 2020, 13, 2819–2832. [Google Scholar] [CrossRef] [Green Version]
- Vishnubalaji, R.; Alajez, N.M. Epigenetic Regulation of Triple Negative Breast Cancer (TNBC) by TGF-β Signaling. Sci. Rep. 2021, 11, 15410. [Google Scholar] [CrossRef]
- Pan, P.Y.; Ma, G.; Weber, K.J.; Ozao-Choy, J.; Wang, G.; Yin, B.; Divino, C.M.; Chen, S.H. Immune Stimulatory Receptor CD40 Is Required for T-Cell Suppression and T Regulatory Cell Activation Mediated by Myeloid-Derived Suppressor Cells in Cancer. Cancer Res. 2010, 70, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Ragusa, S.; Prat-Luri, B.; González-Loyola, A.; Nassiri, S.; Squadrito, M.L.; Guichard, A.; Cavin, S.; Gjorevski, N.; Barras, D.; Marra, G.; et al. Antiangiogenic Immunotherapy Suppresses Desmoplastic and Chemoresistant Intestinal Tumors in Mice. J. Clin. Investig. 2020, 130, 1199–1216. [Google Scholar] [CrossRef] [Green Version]
- Mégarbané, A.; Piquemal, D.; Rebillat, A.S.; Stora, S.; Pierrat, F.; Bruno, R.; Noguier, F.; Mircher, C.; Ravel, A.; Vilaire-Meunier, M.; et al. Transcriptomic Study in Women with Trisomy 21 Identifies a Possible Role of the GTPases of the Immunity-Associated Proteins (GIMAP) in the Protection of Breast Cancer. Sci. Rep. 2020, 10, 9447. [Google Scholar] [CrossRef]
- Protic, O.; Toti, P.; Islam, M.S.; Occhini, R.; Giannubilo, S.R.; Catherino, W.H.; Cinti, S.; Petraglia, F.; Ciavattini, A.; Castellucci, M.; et al. Possible Involvement of Inflammatory/Reparative Processes in the Development of Uterine Fibroids. Cell Tissue Res. 2016, 364, 415–427. [Google Scholar] [CrossRef]
- Rohena-Rivera, K.; Sanchez-Vazquez, M.M.; Aponte-Colon, D.A.; Forestier-Roman, I.S.; Quintero-Aguilo, M.E.; Martanez-Ferrer, M. IL-15 Regulates Migration, Invasion, Angiogenesis and Genes Associated with Lipid Metabolism and Inflammation in Prostate Cancer. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [Green Version]
- Cai, M.; Sikong, Y.; Wang, Q.; Zhu, S.; Pang, F.; Cui, X. Gpx3 Prevents Migration and Invasion in Gastric Cancer by Targeting NFкB/Wnt5a/JNK Signaling. Int. J. Clin. Exp. Pathol. 2019, 12, 1194. [Google Scholar]
- Chang, C.; Worley, B.L.; Phaëton, R.; Hempel, N. Extracellular Glutathione Peroxidase GPx3 and Its Role in Cancer. Cancers 2020, 12, 2197. [Google Scholar] [CrossRef] [PubMed]
- Arslan, A.A.; Gold, L.I.; Mittal, K.; Suen, T.-C.; Belitskaya-Levy, I.; Tang, M.-S.; Toniolo, P. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: New evidence and a systematic review. Hum. Reprod. 2005, 20, 852–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, M.; Yin, P.; Navarro, A.; Moravek, M.B.; Coon, V.J.S.; Druschitz, S.A.; Serna, V.A.; Qiang, W.; Brooks, D.C.; Malpani, S.S.; et al. Paracrine Activation of WNT/β-Catenin Pathway in Uterine Leiomyoma Stem Cells Promotes Tumor Growth. Proc. Natl. Acad. Sci. USA 2013, 110, 17053–17058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borahay, M.A.; Al Hendy, A.; Kilic, G.S.; Boehning, D. Signaling Pathways in Leiomyoma: Understanding Pathobiology and Implications for Therapy. Mol. Med. 2015, 21, 242–256. [Google Scholar] [CrossRef]
- Baranov, V.S.; Osinovskaya, N.S.; Yarmolinskaya, M.I. Pathogenomics of Uterine Fibroids Development. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Ciarmela, P.; Delli Carpini, G.; Greco, S.; Zannotti, A.; Montik, N.; Giannella, L.; Giuliani, L.; Grelloni, C.; Panfoli, F.; Paolucci, M.; et al. Uterine Fibroid Vascularization: From Morphological Evidence to Clinical Implications. Reprod. Biomed. Online 2022, 44, 281–294. [Google Scholar] [CrossRef]
- Leppert, P.C.; Baginski, T.; Prupas, C.; Catherino, W.H.; Pletcher, S.; Segars, J.H. Comparative Ultrastructure of Collagen Fibrils in Uterine Leiomyomas and Normal Myometrium. Fertil. Steril. 2004, 82, 1182–1187. [Google Scholar] [CrossRef]
- Islam, M.S.; Ciavattini, A.; Petraglia, F.; Castellucci, M.; Ciarmela, P. Extracellular Matrix in Uterine Leiomyoma Pathogenesis: A Potential Target for Future Therapeutics. Hum. Reprod. Update 2018, 24, 59–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyaw, M.T.H.; Yamaguchi, Y.; Choijookhuu, N.; Yano, K.; Takagi, H.; Takahashi, N.; Oo, P.S.; Sato, K.; Hishikawa, Y. The HDAC Inhibitor, SAHA, Combined with Cisplatin Synergistically Induces Apoptosis in Alpha-Fetoprotein-Producing Hepatoid Adenocarcinoma Cells. Acta Histochem. Cytochem. 2019, 52, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou Najem, S.; Khawaja, G.; Hodroj, M.H.; Babikian, P.; Rizk, S. Adjuvant Epigenetic Therapy of Decitabine and Suberoylanilide Hydroxamic Acid Exerts Anti-Neoplastic Effects in Acute Myeloid Leukemia Cells. Cells 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Butler, L.M.; Agus, D.B.; Scher, H.I.; Higgins, B.; Rose, A.; Cordon-Cardo, C.; Thaler, H.T.; Rifkind, R.A.; Marks, P.A.; Richon, V.M. Suberoylanilide Hydroxamic Acid, an Inhibitor of Histone Deacetylase, Suppresses the Growth of Prostate Cancer Cells in Vitro and in Vivo. Cancer Res. 2000, 60, 5165–5170. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carbajo-García, M.C.; de Miguel-Gómez, L.; Juárez-Barber, E.; Trelis, A.; Monleón, J.; Pellicer, A.; Flanagan, J.M.; Ferrero, H. Deciphering the Role of Histone Modifications in Uterine Leiomyoma: Acetylation of H3K27 Regulates the Expression of Genes Involved in Proliferation, Cell Signaling, Cell Transport, Angiogenesis and Extracellular Matrix Formation. Biomedicines 2022, 10, 1279. https://doi.org/10.3390/biomedicines10061279
Carbajo-García MC, de Miguel-Gómez L, Juárez-Barber E, Trelis A, Monleón J, Pellicer A, Flanagan JM, Ferrero H. Deciphering the Role of Histone Modifications in Uterine Leiomyoma: Acetylation of H3K27 Regulates the Expression of Genes Involved in Proliferation, Cell Signaling, Cell Transport, Angiogenesis and Extracellular Matrix Formation. Biomedicines. 2022; 10(6):1279. https://doi.org/10.3390/biomedicines10061279
Chicago/Turabian StyleCarbajo-García, María Cristina, Lucia de Miguel-Gómez, Elena Juárez-Barber, Alexandra Trelis, Javier Monleón, Antonio Pellicer, James M. Flanagan, and Hortensia Ferrero. 2022. "Deciphering the Role of Histone Modifications in Uterine Leiomyoma: Acetylation of H3K27 Regulates the Expression of Genes Involved in Proliferation, Cell Signaling, Cell Transport, Angiogenesis and Extracellular Matrix Formation" Biomedicines 10, no. 6: 1279. https://doi.org/10.3390/biomedicines10061279
APA StyleCarbajo-García, M. C., de Miguel-Gómez, L., Juárez-Barber, E., Trelis, A., Monleón, J., Pellicer, A., Flanagan, J. M., & Ferrero, H. (2022). Deciphering the Role of Histone Modifications in Uterine Leiomyoma: Acetylation of H3K27 Regulates the Expression of Genes Involved in Proliferation, Cell Signaling, Cell Transport, Angiogenesis and Extracellular Matrix Formation. Biomedicines, 10(6), 1279. https://doi.org/10.3390/biomedicines10061279






