Raman Microspectroscopy Identifies Biochemical Activation Fingerprints in THP-1- and PBMC-Derived Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peripheral Blood Mononuclear Cell-Derived Monocyte Isolation and Culture
2.2. Macrophage Maturation and Polarization
2.3. THP-1 Culture
2.4. Flow Cytometry
2.5. Raman Microspectroscopy
2.6. Data Processing
2.7. Statistical Analysis
3. Results
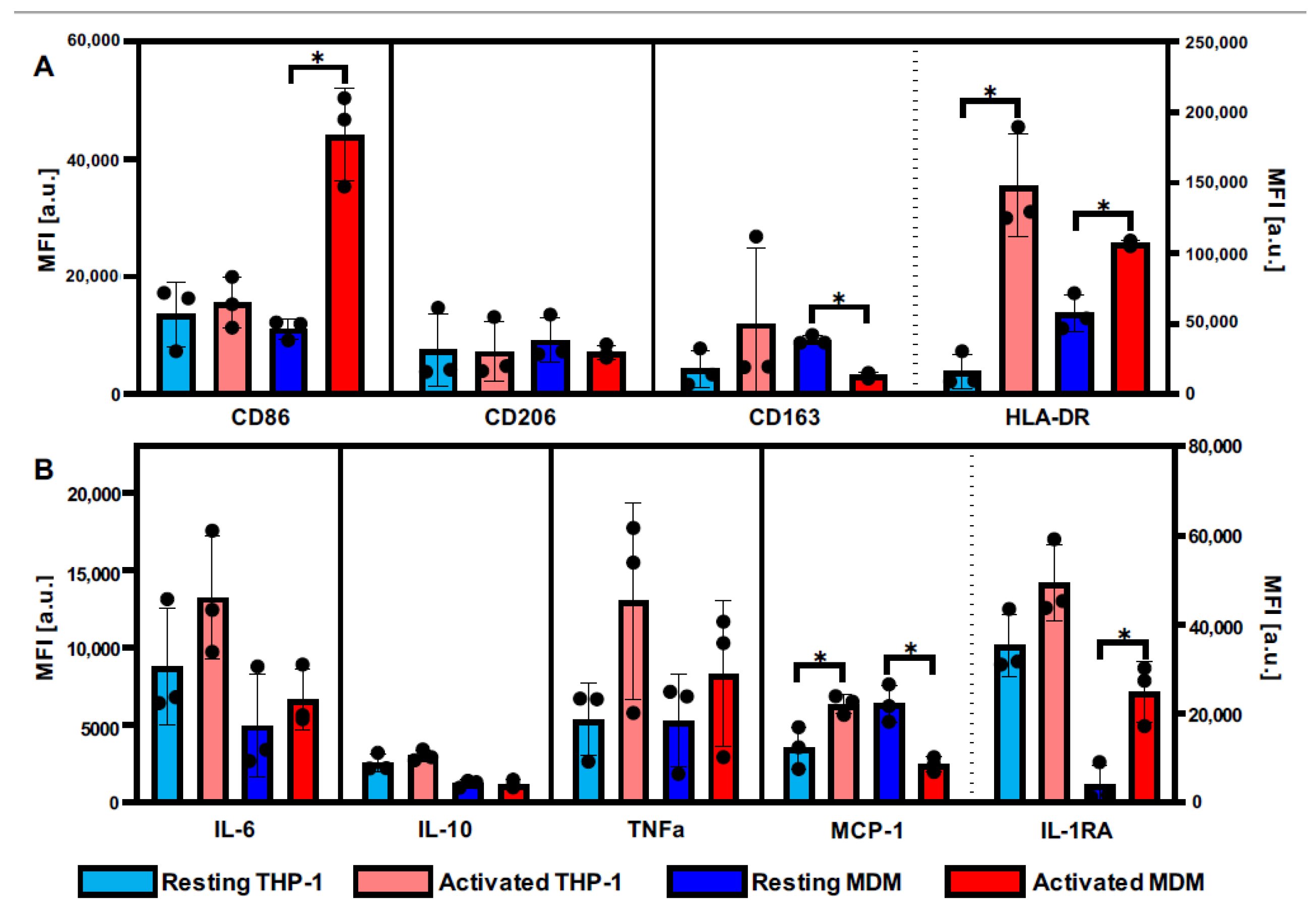
3.1. Definition of Activation Patterns of THP-1 Macrophages and MDMs by Flow Cytometry
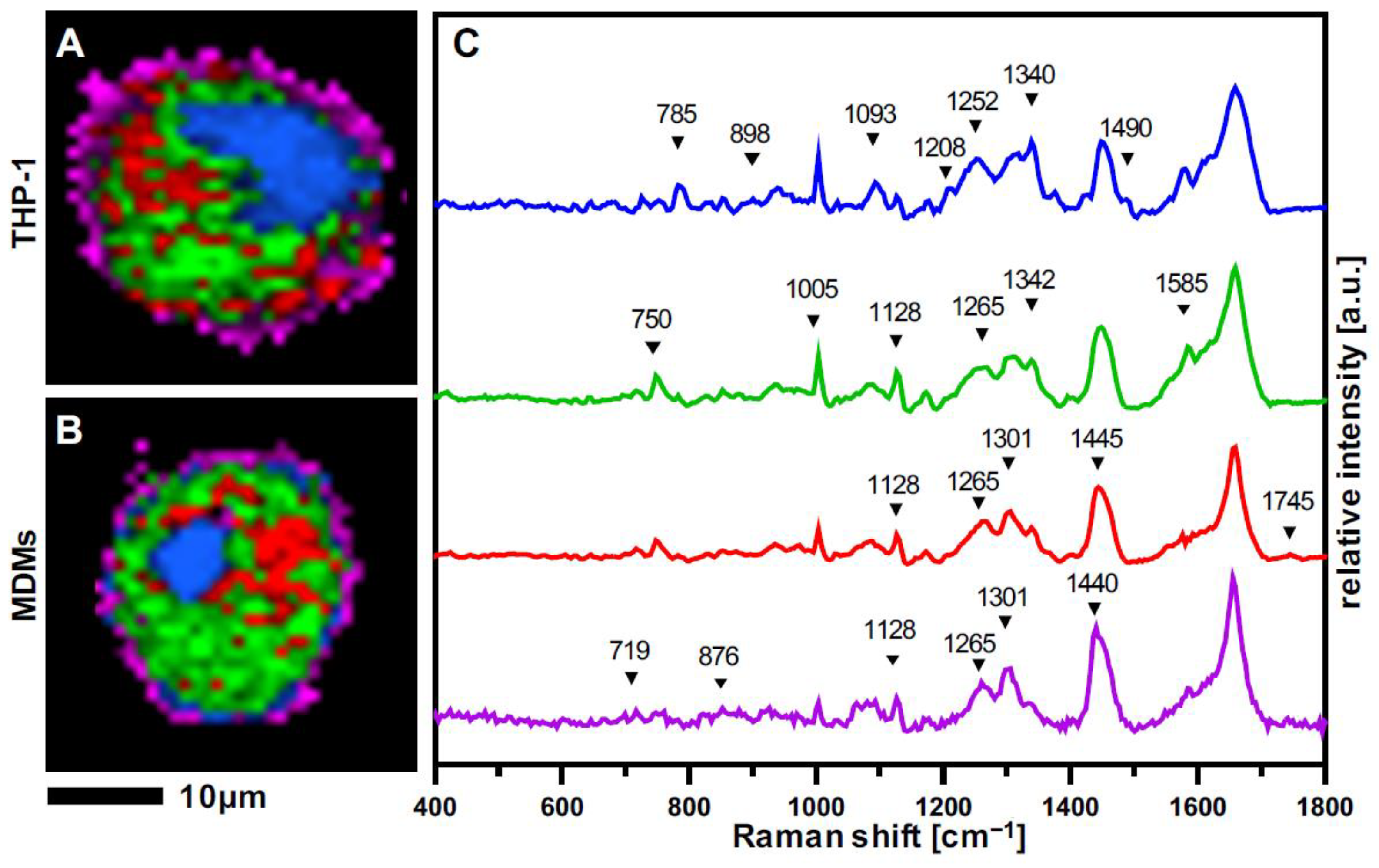
3.2. Raman Imaging Provides Spatial Resolution of Subcellular Structures in Macrophages
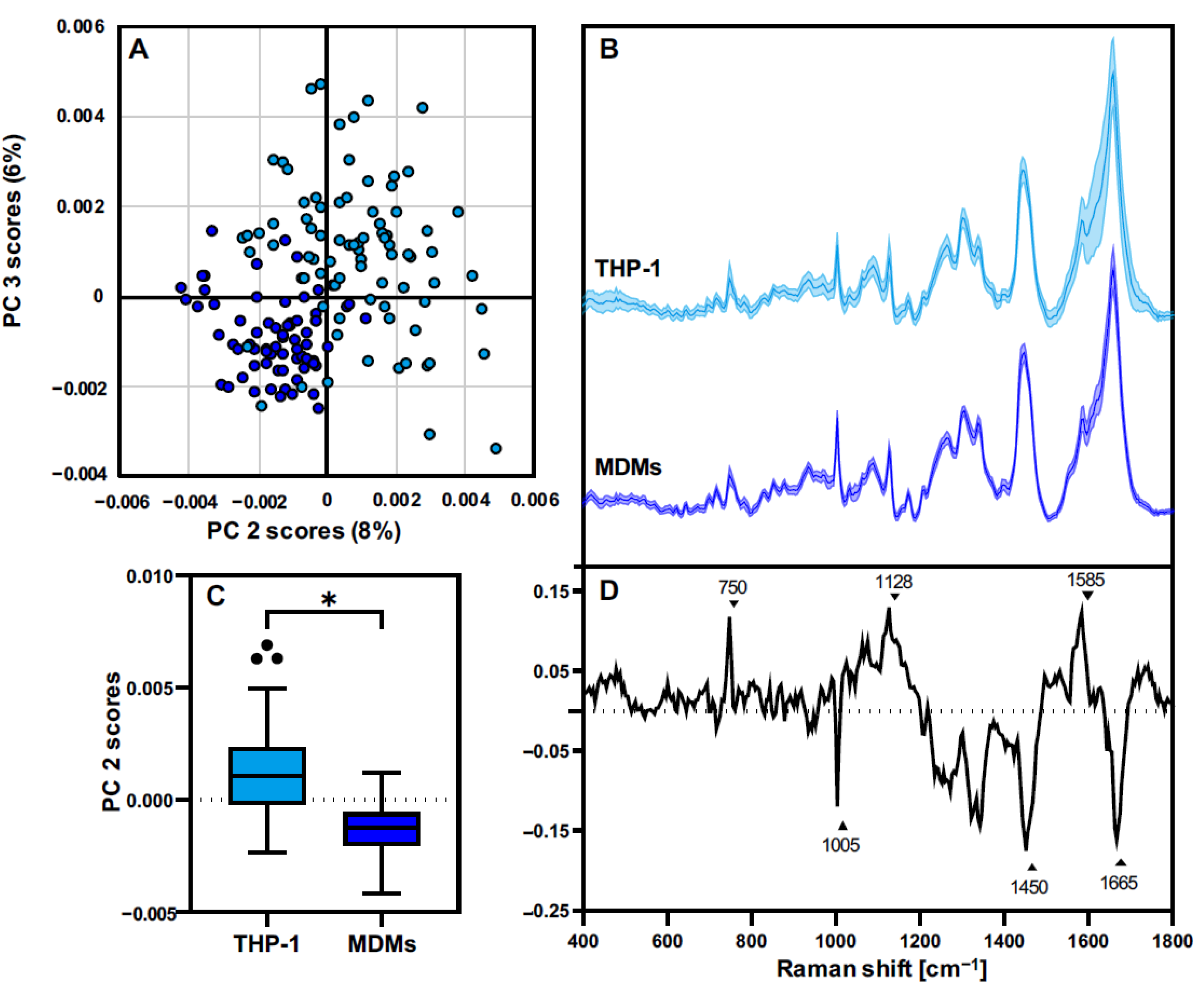
3.3. THP-1 Macrophages and MDMs Differ in Their Molecular Composition
3.4. THP-1 and MDMs Have a Different Response to Proinflammatory Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godwin, J.W.; Pinto, A.R.; Rosenthal, N.A. Macrophages are required for adult salamander limb regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 9415–9420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerrick, K.Y.; Gerrick, E.R.; Gupta, A.; Wheelan, S.J.; Yegnasubramanian, S.; Jaffee, E.M. Transcriptional profiling identifies novel regulators of macrophage polarization. PLoS ONE 2018, 13, e0208602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-C.; Zou, X.-B.; Chai, Y.-F.; Yao, Y.-M. Macrophage Polarization in Inflammatory Diseases. Int. J. Biol. Sci. 2014, 10, 520–529. [Google Scholar] [CrossRef]
- Okabe, Y.; Medzhitov, R. Tissue-Specific Signals Control Reversible Program of Localization and Functional Polarization of Macrophages. Cell 2014, 157, 832–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.; Wright, K.L.; Ashton, L. Raman spectroscopy: An evolving technique for live cell studies. Analyst 2016, 141, 3590–3600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beattie, J.R.; Bell, S.E.J.; Moss, B.W. A critical evaluation of Raman spectroscopy for the analysis of lipids: Fatty acid methyl esters. Lipids 2004, 39, 407–419. [Google Scholar] [CrossRef]
- Krafft, C. Raman spectroscopy and microscopy of cells and tissues. In Encyclopedia of Biophysics; Springer: Berlin/Heidelberg, Germany, 2013; p. 2178. [Google Scholar] [CrossRef]
- Zbinden, A.; Marzi, J.; Schlünder, K.; Probst, C.; Urbanczyk, M.; Black, S.; Brauchle, E.M.; Layland, S.L.; Kraushaar, U.; Duffy, G.; et al. Non-invasive marker-independent high content analysis of a microphysiological human pancreas-on-a-chip model. Matrix Biol. 2020, 85–86, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Fore, S.; Chan, J.; Taylor, D.; Huser, T. Raman spectroscopy of individual monocytes reveals that single-beam optical trapping of mononuclear cells occurs by their nucleus. J. Opt. 2011, 13, 044021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzi, J.; Brauchle, E.M.; Schenke-Layland, K.; Rolle, M.W. Non-invasive functional molecular phenotyping of human smooth muscle cells utilized in cardiovascular tissue engineering. Acta Biomater. 2019, 89, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Sabbatini, S.; Pro, C.; Belloni, A.; Orilisi, G.; Rubini, C.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Exploiting fourier transform infrared and Raman microspectroscopies on cancer stem cells from oral squamous cells carcinoma: New evidence of acquired cisplatin chemoresistance. Analyst 2020, 145, 8038–8049. [Google Scholar] [CrossRef] [PubMed]
- Brauchle, E.; Thude, S.; Brucker, S.Y.; Schenke-Layland, K. Cell death stages in single apoptotic and necrotic cells monitored by Raman microspectroscopy. Sci. Rep. 2014, 4, 4698. [Google Scholar] [CrossRef] [Green Version]
- Daigneault, M.; Preston, J.A.; Marriott, H.M.; Whyte, M.K.B.; Dockrell, D.H. The Identification of Markers of Macrophage Differentiation in PMA-Stimulated THP-1 Cells and Monocyte-Derived Macrophages. PLoS ONE 2010, 5, e8668. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef]
- Tedesco, S.; De Majo, F.; Kim, J.; Trenti, A.; Trevisi, L.; Fadini, G.P.; Bolego, C.; Zandstra, P.W.; Cignarella, A.; Vitiello, L. Convenience versus biological significance: Are PMA-differentiated THP-1 cells a reliable substitute for blood-derived macrophages when studying in vitro polarization? Front. Pharmacol. 2018, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Feuerer, N.; Morschl, J.; Daum, R.; Weiss, M.; Hinderer, S.; Schenke-Layland, K.; Shipp, C. Macrophage retrieval from 3D biomaterials: A detailed comparison of common dissociation methods. J. Immunol. Regen. Med. 2020, 11, 100035. [Google Scholar] [CrossRef]
- Feuerer, N.; Marzi, J.; Brauchle, E.M.; Berrio, D.A.C.; Billing, F.; Weiss, M.; Jakobi, M.; Schneiderhan-Marra, N.; Shipp, C.; Schenke-Layland, K. Lipidome profiling with Raman microspectroscopy identifies macrophage response to surface topographies of implant materials. Proc. Natl. Acad. Sci. USA 2021, 118, e2113694118. [Google Scholar] [CrossRef]
- Spiers, R.M.; Marzi, J.; Brauchle, E.M.; Cross, S.E.; Vaughan, R.H.; Bateman, P.A.; Hughes, S.J.; Schenke-Layland, K.; Johnson, P.R. Donor age significantly influences the Raman spectroscopic biomolecular fingerprint of human pancreatic extracellular matrix proteins following collagenase-based digestion. Acta Biomater. 2019, 99, 269–283. [Google Scholar] [CrossRef]
- Peticolas, W.L. [17] Raman spectroscopy of DNA and proteins. Methods Enzymol. 1995, 246, 389–416. [Google Scholar] [CrossRef] [PubMed]
- Prescott, B.; Steinmetz, W.; Thomas, G.J. Characterization of DNA structures by laser Raman spectroscopy. Biopolymers 1984, 23, 235–256. [Google Scholar] [CrossRef] [PubMed]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Spiro, T.G.; Strekas, T.C. Resonance Raman Spectra of Hemoglobin and Cytochrome c: Inverse Polarization and Vibronic Scattering. Proc. Natl. Acad. Sci. USA 1972, 69, 2622–2626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA 2011, 109, 28–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, R.; Yu, N.-T. Laser-excited Raman spectroscopy of biomolecules: I. Native lysozyme and its constituent amino acids. J. Mol. Biol. 1970, 50, 509–524. [Google Scholar] [CrossRef]
- Chen, M.C.; Lord, R.C. Laser-excited Raman spectroscopy of biomolecules. VI. Polypeptides as conformational models. J. Am. Chem. Soc. 1974, 96, 4750–4752. [Google Scholar] [CrossRef]
- Notingher, I.; Verrier, S.; Haque, S.; Polak, J.M.; Hench, L.L. Spectroscopic study of human lung epithelial cells (A549) in culture: Living cells versus dead cells. Biopolymers 2003, 72, 230–240. [Google Scholar] [CrossRef]
- He, Z.; Han, Z.; Kizer, M.; Linhardt, R.J.; Wang, X.; Sinyukov, A.M.; Wang, J.; Deckert, V.; Sokolov, A.V.; Hu, J.; et al. Tip-Enhanced Raman Imaging of Single-Stranded DNA with Single Base Resolution. J. Am. Chem. Soc. 2018, 141, 753–757. [Google Scholar] [CrossRef]
- Kohro, T.; Tanaka, T.; Murakami, T.; Wada, Y.; Aburatani, H.; Hamakubo, T.; Kodama, T. A Comparison of Differences in the Gene Expression Profiles of Phorbol 12-myristate 13-acetate Differentiated THP-1 Cells and Human Monocyte-derived Macrophage. J. Atheroscler. Thromb. 2004, 11, 88–97. [Google Scholar] [CrossRef] [Green Version]
- Sharif, O.; Bolshakov, V.N.; Raines, S.; Newham, P.; Perkins, N.D. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol. 2007, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Shiratori, H.; Feinweber, C.; Luckhardt, S.; Linke, B.; Resch, E.; Geisslinger, G.; Weigert, A.; Parnham, M.J. THP-1 and human peripheral blood mononuclear cell-derived macrophages differ in their capacity to polarize in vitro. Mol. Immunol. 2017, 88, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Pavillon, N.; Hobro, A.J.; Akira, S.; Smith, N.I. Noninvasive detection of macrophage activation with single-cell resolution through machine learning. Proc. Natl. Acad. Sci. USA 2018, 115, E2676–E2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Töpfer, N.; Müller, M.; Dahms, M.; Ramoji, A.; Popp, J.; Slevogt, H.; Neugebauer, U. Raman spectroscopy reveals LPS-induced changes of biomolecular composition in monocytic THP-1 cells in a label-free manner. Integr. Biol. 2019, 11, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Ugurel, S.; Uhlig, D.; Pföhler, C.; Tilgen, W.; Schadendorf, D.; Reinhold, U. Down-regulation of HLA class II and costimulatory CD86/B7-2 on circulating monocytes from melanoma patients. Cancer Immunol. Immunother. 2004, 53, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Daum, R.; Brauchle, E.M.; Berrio, D.A.C.; Jurkowski, T.P.; Schenke-Layland, K. Non-invasive detection of DNA methylation states in carcinoma and pluripotent stem cells using Raman microspectroscopy and imaging. Sci. Rep. 2019, 9, 7014. [Google Scholar] [CrossRef]
- Tur, J.; Vico, T.; Lloberas, J.; Zorzano, A.; Celada, A. Chapter One—Macrophages and Mitochondria: A Critical Interplay Between Metabolism, Signaling, and the Functional Activity. In Advances in Immunology; Alt, F.W., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 1–36. [Google Scholar]
- Huang, Z.; McWilliams, A.; Lui, H.; McLean, D.I.; Lam, S.; Zeng, H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer 2003, 107, 1047–1052. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Shepherd, N.; Crow, P.; Barr, H. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J. Raman Spectrosc. 2002, 33, 564–573. [Google Scholar] [CrossRef]
- Liu, Z.; Davis, C.; Cai, W.; He, L.; Chen, X.; Dai, H. Circulation and long-term fate of functionalized, biocompatible single-walled carbon nanotubes in mice probed by Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2008, 105, 1410–1415. [Google Scholar] [CrossRef] [Green Version]
- Cheng, W.-T.; Liu, M.-T.; Liu, H.-N.; Lin, S.-Y. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc. Res. Tech. 2005, 68, 75–79. [Google Scholar] [CrossRef]
- Pétrilli, V.; Dostert, C.; Muruve, A.D.; Tschopp, J. The inflammasome: A danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 2007, 19, 615–622. [Google Scholar] [CrossRef]
- Ruiz-Chica, A.J.; Medina, M.A.; Sánchez-Jiménez, F.; Ramírez, F.J. Characterization by Raman spectroscopy of conformational changes on guanine–cytosine and adenine–thymine oligonucleotides induced by aminooxy analogues of spermidine. J. Raman Spectrosc. 2004, 35, 93–100. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Y.; Wang, F.; Wang, Z.; Lu, Y.; Xu, Y.; Wang, K.; Shen, H.; Yang, P.; Li, S.; et al. Quantitative profiling of glycerophospholipids during mouse and human macrophage differentiation using targeted mass spectrometry. Sci. Rep. 2017, 7, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montenegro-Burke, J.R.; Sutton, J.A.; Rogers, L.M.; Milne, G.L.; McLean, J.A.; Aronoff, D.M. Lipid profiling of polarized human monocyte-derived macrophages. Prostaglandins Other Lipid Mediat. 2016, 127, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shirota, K.; Yagi, K.; Inaba, T.; Li, P.-C.; Murata, M.; Sugita, Y.; Kobayashi, T. Detection of Sphingomyelin Clusters by Raman Spectroscopy. Biophys. J. 2016, 111, 999–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, N.; Nguyen, T.N.Q.; Cullen, D.; Meade, A.D.; Wynne, C. Discrimination of immune cell activation using Raman micro-spectroscopy in an in-vitro & ex-vivo model. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 248, 119118. [Google Scholar] [CrossRef]
- Bosshart, H.; Heinzelmann, M. Lipopolysaccharide-mediated cell activation without rapid mobilization of cytosolic free calcium. Mol. Immunol. 2004, 41, 1023–1028. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef]


| Wavenumber [cm−1] | Vibration Mode | Assignment |
|---|---|---|
| 695 | DNA bases (G & T) | DNA [22,23] |
| 719 | Symmetric stretch vibration of choline group | Phospholipids [24] |
| 750 | Pyrrole ring breathing | Cytochrome C [25,26] |
| 776–780 | Symmetric breathing of tryptophan | Proteins [27,28] |
| 785–790 | Ring breathing modes (DNA/RNA bases) | DNA [22,23] |
| 868 | C-O-O skeletal vibration | Lipids [24] |
| 876 | Asymmetric vibration choline N(CH3)3 | Phospholipids [24] |
| 898 | Adenine | DNA [22,23] |
| 940 | C-C skeletal vibration (backbone) | Proteins [27,28] |
| 1005 | Symmetric ring breathing of phenylalanine | Proteins [27,28] |
| 1093 | Symmetric PO2− stretching vibration of the DNA backbone | DNA [22,23] |
| 1128 | C-N stretching (proteins); C-C vibration in fatty acids | Proteins; Lipids [24,27,28] |
| 1173 | C-C vibrations fatty acids | Lipids [24] |
| 1208 | Adenine, Thymine (ring breathing modes) | DNA [22,23] |
| 1252 | Guanine, cytosine (NH2) | DNA [22,23] |
| 1265 | Amide III; =CH2 vibration in lipids | Proteins; Lipids [24,27,28] |
| 1301 | C-H vibration | (Phospho-) Lipids [24] |
| 1340 | Adenine, guanine & CH deformation in proteins | DNA [22,23], Proteins [29] |
| 1435–1445 | CH3/CH2 scissoring | Lipids [24] |
| 1450 | CH2 deformation | Proteins [27,28] |
| 1585 | C=C olefinic stretch | Proteins [27,28] |
| 1630 | DNA bases (C, G, T) | DNA [30] |
| 1640 | C=C vibrations (fatty acids) | Lipids [24] |
| 1655–1670 | Amide I, C=C vibrations | Lipids; Proteins [24,27,28] |
| 1745 | C=O vibrations triacylglycerids | Lipids [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feuerer, N.; Carvajal Berrio, D.A.; Billing, F.; Segan, S.; Weiss, M.; Rothbauer, U.; Marzi, J.; Schenke-Layland, K. Raman Microspectroscopy Identifies Biochemical Activation Fingerprints in THP-1- and PBMC-Derived Macrophages. Biomedicines 2022, 10, 989. https://doi.org/10.3390/biomedicines10050989
Feuerer N, Carvajal Berrio DA, Billing F, Segan S, Weiss M, Rothbauer U, Marzi J, Schenke-Layland K. Raman Microspectroscopy Identifies Biochemical Activation Fingerprints in THP-1- and PBMC-Derived Macrophages. Biomedicines. 2022; 10(5):989. https://doi.org/10.3390/biomedicines10050989
Chicago/Turabian StyleFeuerer, Nora, Daniel A. Carvajal Berrio, Florian Billing, Sören Segan, Martin Weiss, Ulrich Rothbauer, Julia Marzi, and Katja Schenke-Layland. 2022. "Raman Microspectroscopy Identifies Biochemical Activation Fingerprints in THP-1- and PBMC-Derived Macrophages" Biomedicines 10, no. 5: 989. https://doi.org/10.3390/biomedicines10050989
APA StyleFeuerer, N., Carvajal Berrio, D. A., Billing, F., Segan, S., Weiss, M., Rothbauer, U., Marzi, J., & Schenke-Layland, K. (2022). Raman Microspectroscopy Identifies Biochemical Activation Fingerprints in THP-1- and PBMC-Derived Macrophages. Biomedicines, 10(5), 989. https://doi.org/10.3390/biomedicines10050989