Biomarkers as Predictive Factors of Anti-VEGF Response
Abstract
1. Introduction
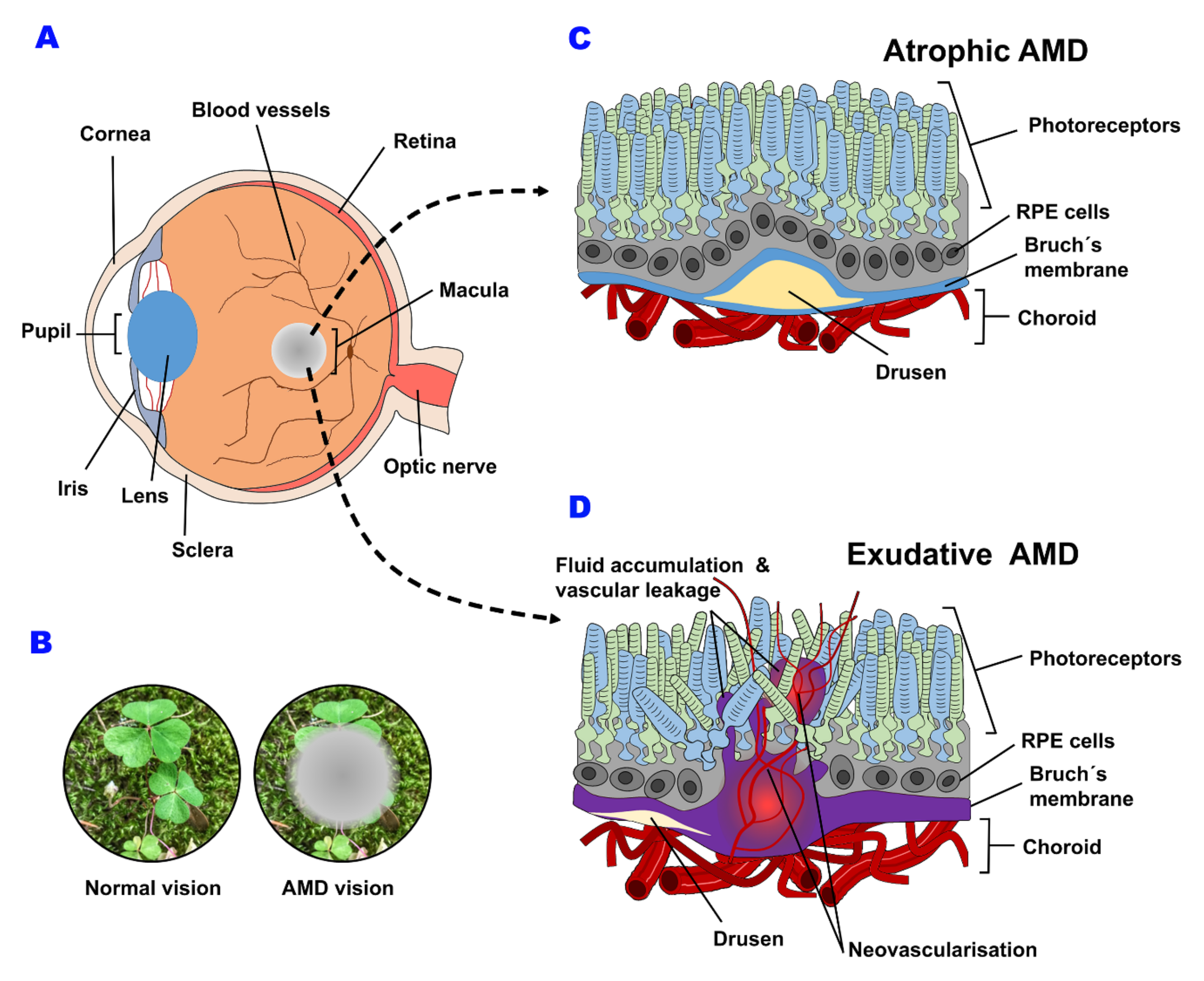
2. Macular Degeneration
3. Incidence, Prevalence and Risk Factors
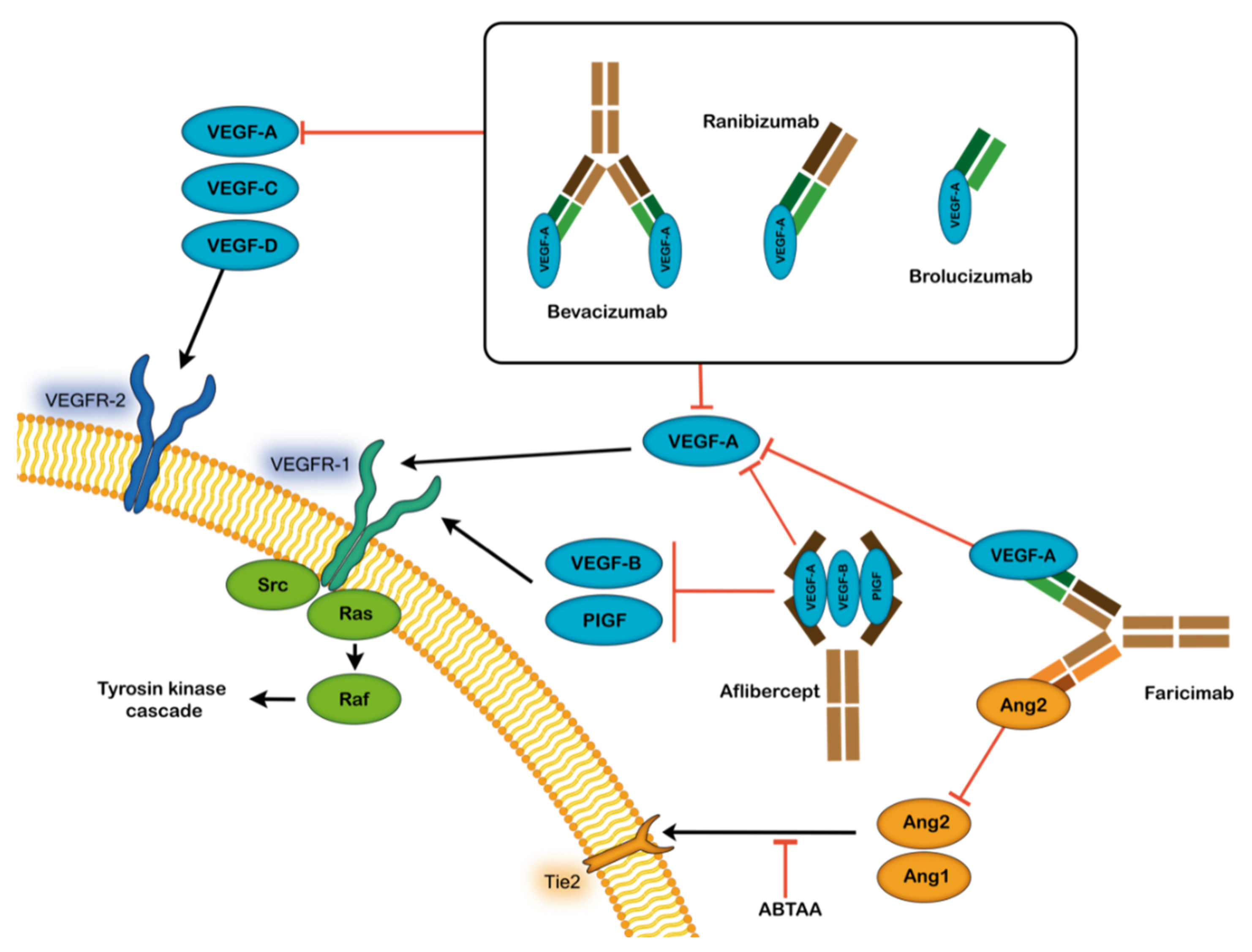
4. Treatment Options for AMD
5. Predictive Factors of Anti-VEGF Resistance
5.1. Predictive Ranibizumab Biomarkers Based on Clinical Characteristics of Patients
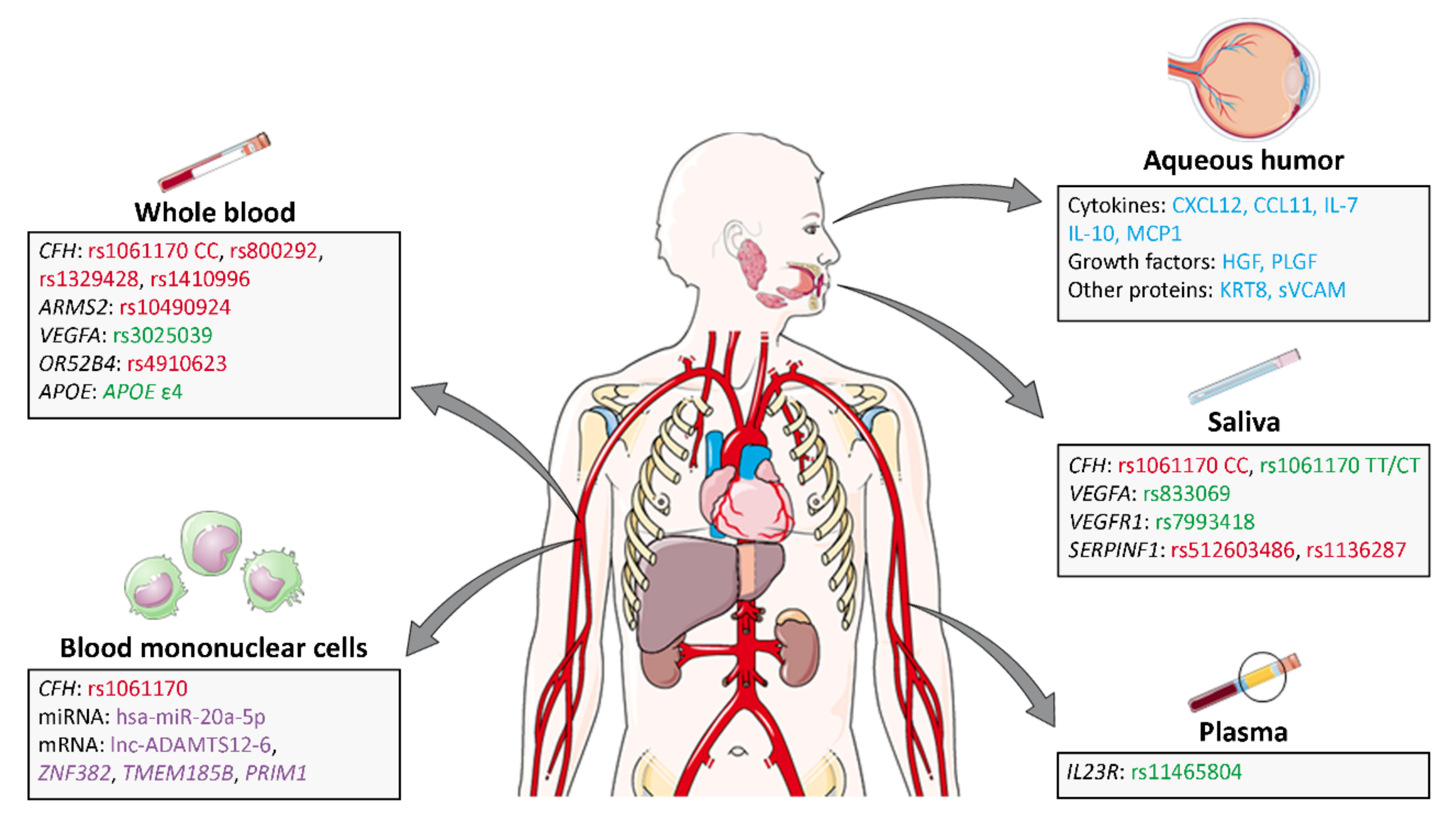
5.2. Genomics, Proteomic and Metabolomic Predictive Ranibizumab Biomarkers
5.2.1. Predictive Ranibizumab Biomarkers in Saliva
- Complement System
- ARMS2
- VEGFA and related genes
- SERPINF1
- Other genes
5.2.2. Predictive Ranibizumab Biomarkers in Whole Blood
- Complement Factor H
- ARMS2
- VEGFA and related genes
- HTRA1
- OR52B4
- ApoE
- Other genes
5.2.3. Predictive Ranibizumab Biomarkers in Plasma
5.2.4. Blood Mononuclear Cells as a Source of Ranibizumab Predictive Biomarkers
- Complement System
- ARMS2
- Other genes
- MicroRNAs
- mRNAs
5.2.5. Predictive Ranibizumab Biomarkers in Aqueous Humor
| Summary Predictive Biomarkers | |||||
|---|---|---|---|---|---|
| Sample | Biomarker Type | Symbol | Biomarker | Correlation Ranibizumab Response | Reference |
| Saliva | SNPs | CFH | rs1061170 CC | Negative | [99,100,102,107] |
| rs1061170 TT/CT | Positive | ||||
| Saliva | SNPs | VEGFA | rs833069 | Positive | [101] |
| Saliva | SNPs | VEGFR1 | rs7993418 | Positive | [103,104] |
| Saliva | SNPs | SERPINF1 | rs512603486 | Negative | [103] |
| rs1136287 | Negative | ||||
| Whole blood | SNPs | CFH | rs1061170 CC | Negative | [19,118,119,120] |
| rs1061170 TT/CT | Positive | ||||
| rs800292 | Negative | ||||
| rs1329428 | Negative | ||||
| rs1410996 | Negative | ||||
| Whole blood | SNPs | ARMS2 | rs10490924 | Negative | [19,124] |
| Whole blood | SNPs | VEGFA | rs3025039 TT | Positive | [122] |
| Whole blood | SNPs | OR52B4 | rs4910623 | Negative | [128] |
| Whole blood | SNPs | ApoE | Ɛ4 | Positive | [126,130] |
| Plasma | SNPs | IL23R | rs11465804 | Positive | [134] |
| Blood mononuclear cells | SNPs | CFH | rs1061170 CC | Negative | [142] |
| rs1061170 TT/CT | Positive | ||||
| Blood mononuclear cells | miRNA | has-miR-20a-5p | ↑ has-miR-20a-5p | Positive | [7] |
| ↓ has-miR-20a-5p | Negative | ||||
| Blood mononuclear cells | mRNA | Lnc-ADAMTS12-6 | ↓ Lnc-ADAMTS12-6 | Positive | [7] |
| ↑ Lnc-ADAMTS12-6 | Negative | ||||
| Aqueous humor | Protein | CXCL12 | CXCL12 | Negative | [158] |
| Aqueous humor | Protein | CCL11 | CCL11 | Positive | [151] |
| Aqueous humor | Protein | IL-7 | IL-7 | Positive | [156,157] |
| Aqueous humor | Protein | IL-10 | IL-10 | Negative | [158] |
| Aqueous humor | Protein | MCP1 | MCP1 | Negative | [158] |
| Aqueous humor | Protein | HGF | HGF | Negative | [157] |
| Aqueous humor | Protein | PLGF | PLGF | Negative | [156,157] |
| Aqueous humor | Protein | KRT8 | KRT8 | Negative | [159] |
| Aqueous humor | Protein | sVCAM | sVCAM | Negative | [157] |
6. Limitations and Possible Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kocur, I.; Resnikoff, S. Visual impairment and blindness in Europe and their prevention. Br. J. Ophthalmol. 2002, 86, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Finger, R.P.; Fleckenstein, M.; Holz, F.G.; Scholl, H.P.N. Quality of life in age-related macular degeneration: A review of available vision-specific psychometric tools. Qual. Life Res. 2008, 17, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Fauser, S.; Lambrou, G.N. Genetic predictive biomarkers of anti-VEGF treatment response in patients with neovascular age-related macular degeneration. Surv. Ophthalmol. 2015, 60, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, R.; Yasuda, M.; Song, S.J.; Chen, S.-J.; Jonas, J.B.; Wang, J.J.; Mitchell, P.; Wong, T.Y. The Prevalence of Age-Related Macular Degeneration in Asians: A Systematic Review and Meta-Analysis. Ophthalmology 2010, 117, 921–927. [Google Scholar] [CrossRef]
- Klein, R.; Klein, B.E.; Cruickshanks, K.J. The prevalence of age-related maculopathy by geographic region and ethnicity. Prog. Retin. Eye Res. 1999, 18, 371–389. [Google Scholar] [CrossRef]
- Yang, K.; Wang, F.-H.; Liang, Y.-B.; Wong, T.-Y.; Wang, J.-J.; Zhan, S.-Y.; Wang, N.-L. Associations between cardiovascular risk factors and early age-related macular degeneration in a rural chinese adult population. Retina 2014, 34, 1539–1553. [Google Scholar] [CrossRef]
- Oca, A.I.; Pérez-Sala, A.; Ochoa, R.; Velilla, S.; Peláez, R.; Larráyoz, I.M. Predictive Biomarkers of Age-Related Macular Degeneration Response to Anti-VEGF Treatment. J. Pers. Med. 2021, 11, 1329. [Google Scholar] [CrossRef]
- Chen, G.; Tzekov, R.; Li, W.; Jiang, F.; Mao, S.; Tong, Y. Pharmacogenetics of Complement Factor H Y402H Polymorphism and Treatment of Neovascular AMD with Anti-VEGF Agents: A Meta-Analysis. Sci. Rep. 2015, 5, 14517. [Google Scholar] [CrossRef]
- Finger, R.P.; Wickremasinghe, S.S.; Baird, P.; Guymer, R. Predictors of anti-VEGF treatment response in neovascular age-related macular degeneration. Surv. Ophthalmol. 2014, 59, 1–18. [Google Scholar] [CrossRef]
- Hagstrom, S.A.; Ying, G.-S.; Maguire, M.G.; Martin, D.F.; Gibson, J.; Lotery, A.; Chakravarthy, U. VEGFR2 Gene Polymorphisms and Response to Anti–Vascular Endothelial Growth Factor Therapy in Age-Related Macular Degeneration. Ophthalmology 2015, 122, 1563–1568. [Google Scholar] [CrossRef]
- Gorin, M.B.; Dasilva, M.J. Predictive genetics for AMD: Hype and hopes for genetics-based strategies for treatment and prevention. Exp. Eye Res. 2020, 191, 107894. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, U.; Augood, C.; Bentham, G.; de Jong, P.; Rahu, M.; Seland, J.; Soubrane, G.; Tomazzoli, L.; Topouzis, F.; Vingerling, J.; et al. Cigarette Smoking and Age-Related Macular Degeneration in the EUREYE Study. Ophthalmology 2007, 114, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Francis, P.J. The influence of genetics on response to treatment with ranibizumab (Lucentis) for age-related macular degeneration: The Lucentis Genotype Study (an American Ophthalmological Society thesis). Trans. Am. Ophthalmol. Soc. 2011, 109, 115–156. [Google Scholar] [PubMed]
- Yang, M.; So, K.-F.; Lam, W.C.; Lo, A.C.Y. Novel Programmed Cell Death as Therapeutic Targets in Age-Related Macular Degeneration? Int. J. Mol. Sci. 2020, 21, 7279. [Google Scholar] [CrossRef] [PubMed]
- Ozkiris, A. Anti-VEGF agents for age-related macular degeneration. Expert Opin. Ther. Patents 2009, 20, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, C.M.; Miller, J. Anti-vascular endothelial growth factor therapy for ocular neovascular disease. Curr. Opin. Ophthalmol. 2007, 18, 502–508. [Google Scholar] [CrossRef]
- Francis, P.J. Update on the role of genetics in the onset of age-related macular degeneration. Clin. Ophthalmol. 2011, 5, 1127–1133. [Google Scholar] [CrossRef]
- Restrepo, N.A.; Spencer, K.L.; Goodloe, R.; Garrett, T.A.; Heiss, G.; Bůžková, P.; Jorgensen, N.; Jensen, R.A.; Matise, T.C.; Hindorff, L.A.; et al. Genetic Determinants of Age-Related Macular Degeneration in Diverse Populations from the PAGE Study. Investig. Opthalmol. Vis. Sci. 2014, 55, 6839–6850. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, M.; Chen, A.; Liu, Z.; Young, C.A.; Wang, S.; Zheng, D.; Jin, G. Genetic associations of anti-vascular endothelial growth factor therapy response in age-related macular degeneration: A systematic review and meta-analysis. Acta Ophthalmol. 2021, 100, 669–680. [Google Scholar] [CrossRef]
- Hadziahmetovic, M.; Malek, G. Age-Related Macular Degeneration Revisited: From Pathology and Cellular Stress to Potential Therapies. Front. Cell Dev. Biol. 2021, 8, 612812. [Google Scholar] [CrossRef]
- Rodríguez, I.R.; Larrayoz, I.M. Cholesterol oxidation in the retina: Implications of 7KCh formation in chronic inflammation and age-related macular degeneration. J. Lipid Res. 2010, 51, 2847–2862. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.; Peto, T.; Bird, A.; VanNewkirk, M.R. The epidemiology of age-related macular degeneration. Am. J. Ophthalmol. 2004, 137, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Thornton, J.; Edwards, P.; Mitchell, P.; Harrison, R.A.; Buchan, I.; Kelly, S.P. Smoking and age-related macular degeneration: A review of association. Eye 2005, 19, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.R.; Fletcher, A.E.; Wormald, R.P.L. 28 000 Cases of age related macular degeneration causing visual loss in people aged 75 years and above in the United Kingdom may be attributable to smoking. Br. J. Ophthalmol. 2005, 89, 550–553. [Google Scholar] [CrossRef]
- Clemons, T.E.; Milton, R.C.; Klein, R.; Seddon, J.M.; Ferris, F. Age-Related Eye Disease Study Research Group Risk Factors for the Incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS): AREDS report no. 19. Ophthalmology 2005, 112, 533–539.e1. [Google Scholar] [CrossRef]
- Guymer, R.; Chong, E.W. Modifiable risk factors for age-related macular degeneration. Med. J. Aust. 2006, 184, 455–458. [Google Scholar] [CrossRef]
- Duan, Y.; Mo, J.; Klein, R.; Scott, I.U.; Lin, H.-M.; Caulfield, J.; Patel, M.; Liao, D. Age-Related Macular Degeneration Is Associated with Incident Myocardial Infarction among Elderly Americans. Ophthalmology 2007, 114, 732–737. [Google Scholar] [CrossRef]
- Seddon, J.M.; George, S.; Rosner, B. Cigarette Smoking, Fish Consumption, Omega-3 Fatty Acid Intake, and Associations with Age-Related Macular Degeneration. Arch. Ophthalmol. 2006, 124, 995–1001. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Hubbell, M.; Jairam, P.; Ambati, B. Neovascular Macular Degeneration: A Review of Etiology, Risk Factors, and Recent Advances in Research and Therapy. Int. J. Mol. Sci. 2021, 22, 1170. [Google Scholar] [CrossRef]
- Brown, C.N.; Green, B.D.; Thompson, R.B.; Hollander, A.I.D.; Lengyel, I.; on behalf of the EYE-RISK consortium. Metabolomics and Age-Related Macular Degeneration. Metabolites 2018, 9, 4. [Google Scholar] [CrossRef]
- Pennington, K.L.; DeAngelis, M.M. Epidemiology of age-related macular degeneration (AMD): Associations with cardiovascular disease phenotypes and lipid factors. Eye Vis. 2016, 3, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Pariente, A.; Pérez-Sala, Á.; Ochoa, R.; Peláez, R.; Larráyoz, I.M. Genome-Wide Transcriptomic Analysis Identifies Pathways Regulated by Sterculic Acid in Retinal Pigmented Epithelium Cells. Cells 2020, 9, 1187. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.-Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Hageman, G.S.; Anderson, D.H.; Johnson, L.V.; Hancox, L.S.; Taiber, A.J.; Hardisty, L.I.; Hageman, J.L.; Stockman, H.A.; Borchardt, J.D.; Gehrs, K.M.; et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 7227–7232. [Google Scholar] [CrossRef]
- Kanda, A.; Chen, W.; Othman, M.; Branham, K.E.H.; Brooks, M.; Khanna, R.; He, S.; Lyons, R.; Abecasis, G.R.; Swaroop, A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2007, 104, 16227–16232. [Google Scholar] [CrossRef]
- Gibson, J.; Cree, A.; Collins, A.; Lotery, A.; Ennis, S. Determination of a gene and environment risk model for age-related macular degeneration. Br. J. Ophthalmol. 2010, 94, 1382–1387. [Google Scholar] [CrossRef]
- Stahl, A. The Diagnosis and Treatment of Age-Related Macular Degeneration. Dtsch. Ärzteblatt Int. 2020, 117, 513–520. [Google Scholar] [CrossRef]
- Gold, B.; The AMD Genetics Clinical Study Group; Merriam, J.E.; Zernant, J.; Hancox, L.S.; Taiber, A.J.; Gehrs, K.; Cramer, K.; Neel, J.; Bergeron, J.; et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006, 38, 458–462. [Google Scholar] [CrossRef]
- Spencer, K.L.; Hauser, M.A.; Olson, L.M.; Schmidt, S.; Scott, W.K.; Gallins, P.; Agarwal, A.; Postel, E.A.; Pericak-Vance, M.A.; Haines, J.L. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Hum. Mol. Genet. 2007, 16, 1986–1992. [Google Scholar] [CrossRef]
- Yates, J.R.; Sepp, T.; Matharu, B.K.; Khan, J.C.; Thurlby, D.A.; Shahid, H.; Clayton, D.G.; Hayward, C.; Morgan, J.; Wright, A.F.; et al. Complement C3 Variant and the Risk of Age-Related Macular Degeneration. N. Engl. J. Med. 2007, 357, 553–561. [Google Scholar] [CrossRef]
- Maller, J.; Fagerness, J.A.; Reynolds, R.C.; Neale, B.M.; Daly, M.J.; Seddon, J.M. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat. Genet. 2007, 39, 1200–1201. [Google Scholar] [CrossRef] [PubMed]
- Fagerness, J.A.; Maller, J.; Neale, B.M.; Reynolds, R.C.; Daly, M.J.; Seddon, J.M. Variation near complement factor I is associated with risk of advanced AMD. Eur. J. Hum. Genet. 2008, 17, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Hautamäki, A.; Seitsonen, S.; Holopainen, J.M.; Moilanen, J.A.; Kivioja, J.; Onkamo, P.; Järvelä, I.; Immonen, I. The genetic variant rs4073 A→T of the Interleukin-8 promoter region is associated with the earlier onset of exudative age-related macular degeneration. Acta Ophthalmol. 2015, 93, 726–733. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Lauer, N.; Hartmann, A.; Stippa, S.; Keilhauer, C.N.; Oppermann, M.; Pandey, M.K.; Köhl, J.; Zipfel, P.F.; Weber, B.H.; et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum. Mol. Genet. 2010, 19, 4694–4704. [Google Scholar] [CrossRef] [PubMed]
- Babanejad, M.; Moein, H.; Akbari, M.R.; Badiei, A.; Yaseri, M.; Soheilian, M.; Najmabadi, H. Investigating the CFH Gene Polymorphisms as a Risk Factor for Age-related Macular Degeneration in an Iranian Population. Ophthalmic Genet. 2015, 37, 144–149. [Google Scholar] [CrossRef]
- Sawitzke, J.; Gold, B.; Olsh, A.; Schlotterbeck, S.; Lemon, K.; Visvanathan, K.; Allikmets, R.; Dean, M. Multilocus analysis of age-related macular degeneration. Eur. J. Hum. Genet. 2009, 17, 1190–1199. [Google Scholar] [CrossRef]
- McKay, G.J.; Silvestri, G.; Patterson, C.C.; Hogg, R.E.; Chakravarthy, U.; Hughes, A.E. Further Assessment of the Complement Component 2 and Factor B Region Associated with Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2009, 50, 533–539. [Google Scholar] [CrossRef]
- Yang, X.; Hu, J.; Zhang, J.; Guan, H. Polymorphisms in CFH, HTRA1 and CX3CR1 confer risk to exudative age-related macular degeneration in Han Chinese. Br. J. Ophthalmol. 2010, 94, 1211–1214. [Google Scholar] [CrossRef]
- Kondo, N.; Honda, S.; Kuno, S.-I.; Negi, A. Positive association of common variants in CD36 with neovascular age-related macular degeneration. Aging 2009, 1, 266–274. [Google Scholar] [CrossRef][Green Version]
- Baird, P.N.; Guida, E.; Chu, D.T.; Vu, H.T.V.; Guymer, R.H. The 2 and 4 Alleles of the Apolipoprotein Gene Are Associated with Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1311–1315. [Google Scholar] [CrossRef]
- Baird, P.N.; Richardson, A.J.; Robman, L.D.; Dimitrov, P.N.; Tikellis, G.; McCarty, C.A.; Guymer, R.H. Apolipoprotein (APOE) gene is associated with progression of age-related macular degeneration (AMD). Hum. Mutat. 2006, 27, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Baird, P.N.; Chu, D.; Guida, E.; Vu, H.T.; Guymer, R. Association of the M55L and Q192R paraoxonase gene polymorphisms with age-related macular degeneration. Am. J. Ophthalmol. 2004, 138, 665–666. [Google Scholar] [CrossRef] [PubMed]
- Tuo, J.; Ning, B.; Bojanowski, C.M.; Lin, Z.-N.; Ross, R.J.; Reed, G.F.; Shen, D.; Jiao, X.; Zhou, M.; Chew, E.Y.; et al. Synergic effect of polymorphisms in ERCC6 5′ flanking region and complement factor H on age-related macular degeneration predisposition. Proc. Natl. Acad. Sci. USA 2006, 103, 9256–9261. [Google Scholar] [CrossRef] [PubMed]
- Fang, A.M.; Lee, A.Y.; Kulkarni, M.; Osborn, M.P.; Brantley, M.A., Jr. Polymorphisms in the VEGFA and VEGFR-2 genes and neovascular age-related macular degeneration. Mol. Vis. 2009, 15, 2710–2719. [Google Scholar]
- Qu, Y.; Dai, H.; Zhou, F.; Zhang, X.; Xu, X.; Zhang, X.; Bi, H.; Pan, X.; Wang, H.; Jiang, H.; et al. Vascular Endothelial Growth Factor Gene Polymorphisms and Risk of Neovascular Age-Related Macular Degeneration in a Chinese Cohort. Ophthalmic Res. 2011, 45, 142–148. [Google Scholar] [CrossRef]
- Combadiere, C.; Feumi, C.; Raoul, W.; Keller, N.; Rodero, M.; Pézard, A.; Lavalette, S.; Houssier, M.; Jonet, L.; Picard, E.; et al. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J. Clin. Investig. 2007, 117, 2920–2928. [Google Scholar] [CrossRef]
- Zareparsi, S.; Buraczynska, M.; Branham, K.E.; Shah, S.; Eng, D.; Li, M.; Pawar, H.; Yashar, B.M.; Moroi, S.E.; Lichter, P.R.; et al. Toll-like receptor 4 variant D299G is associated with susceptibility to age-related macular degeneration. Hum. Mol. Genet. 2005, 14, 1449–1455. [Google Scholar] [CrossRef]
- Fisher, S.A.; Abecasis, G.R.; Yashar, B.M.; Zareparsi, S.; Swaroop, A.; Iyengar, S.K.; Klein, B.E.; Klein, R.; Lee, K.E.; Majewski, J.; et al. Meta-analysis of genome scans of age-related macular degeneration. Hum. Mol. Genet. 2005, 14, 2257–2264. [Google Scholar] [CrossRef]
- Velilla, S.; Garcia-Medina, J.J.; García-Layana, A.; Dolz-Marco, R.; Pons-Vázquez, S.; Pinazo-Duran, M.D.; Gómez-Ulla, F.; Arevalo, J.F.; Díaz-Llopis, M.; Gallego-Pinazo, R. Smoking and Age-Related Macular Degeneration: Review and Update. J. Ophthalmol. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Cho, Y.-K.; Park, D.-H.; Jeon, I.-C. Medication Trends for Age-Related Macular Degeneration. Int. J. Mol. Sci. 2021, 22, 11837. [Google Scholar] [CrossRef]
- Merle, B.M.J.; Colijn, J.M.; Cougnard-Grégoire, A.; De Koning-Backus, A.P.M.; Delyfer, M.-N.; Kiefte-de Jong, J.C.; Meester-Smoor, M.; Féart, C.; Verzijden, T.; Samieri, C.; et al. Mediterranean Diet and Incidence of Advanced Age-Related Macular Degeneration: The EYE-RISK Consortium. Ophthalmology 2019, 126, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Bressler, S.B. Introduction: Understanding the Role of Angiogenesis and Antiangiogenic Agents in Age-Related Macular Degeneration. Ophthalmology 2009, 116, S1–S7. [Google Scholar] [CrossRef] [PubMed]
- Kijlstra, A.; La Heij, E.C.; Hendrikse, F. REVIEW ARTICLE, Immunological Factors in the Pathogenesis and Treatment of Age-Related Macular Degeneration. Ocul. Immunol. Inflamm. 2005, 13, 3–11. [Google Scholar] [CrossRef]
- Hernández-Zimbrón, L.F.; Zamora-Alvarado, R.; La Paz, L.O.-D.; Velez-Montoya, R.; Zenteno, E.; Gulias-Cañizo, R.; Quiroz-Mercado, H.; Gonzalez-Salinas, R. Age-Related Macular Degeneration: New Paradigms for Treatment and Management of AMD. Oxidative Med. Cell. Longev. 2018, 2018, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.; Katta, S.; Narayanan, R.; Mathai, A.; Majji, A.B.; Chakrabarti, S.; Reddy, R.K. The Involvement of Complement Factor B and Complement Component C2 in an Indian Cohort with Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2010, 51, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.M.; Arepalli, S.; Ehlers, J.P. Current and Future Anti-VEGF Agents for Neovascular Age-Related Macular Degeneration. J. Exp. Pharmacol. 2021, 13, 905–912. [Google Scholar] [CrossRef]
- de Guimaraes, T.A.C.; Georgiou, M.; Bainbridge, J.W.B.; Michaelides, M. Gene therapy for neovascular age-related macular degeneration: Rationale, clinical trials and future directions. Br. J. Ophthalmol. 2021, 105, 151–157. [Google Scholar] [CrossRef]
- D’Amico, D.J. Pegaptanib Sodium for Neovascular Age-Related Macular Degeneration: Two-Year Safety Results of the Two Prospective, Multicenter, Controlled Clinical Trials. Ophthalmology 2006, 113, 992–1001.e6. [Google Scholar] [CrossRef]
- Doggrell, S.A. Pegaptanib: The first antiangiogenic agent approved for neovascular macular degeneration. Expert Opin. Pharmacother. 2005, 6, 1421–1423. [Google Scholar] [CrossRef]
- Ferrara, N.; HillanK, J.; Gerber, H.-P.; Novotny, W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004, 3, 391–400. [Google Scholar] [CrossRef]
- Avery, R.L.; Castellarin, A.A.; Steinle, N.C.; Dhoot, D.S.; Pieramici, D.J.; See, R.; Couvillion, S.; Nasir, M.A.; Rabena, M.D.; Maia, M.; et al. Systemic pharmacokinetics and pharmacodynamics of intravitreal aflibercept, bevacizumab, and ranibizumab. Retina 2017, 37, 1847–1858. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.F.; Maguire, M.G.; Fine, S.L.; Ying, G.-S.; Jaffe, G.J.; Grunwald, J.E.; Toth, C.; Redford, M.; Ferris, F.L., 3rd; Comparison of Age-related Macular Degeneration Treatments Trials (CATT) Research Group. Ranibizumab and Bevacizumab for Treatment of Neovascular Age-related Macular Degeneration: Two-Year Results. Ophthalmology 2012, 119, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.F.; Maguire, M.G.; Ying, G.-S.; Grunwald, E.J.; Fine, S.L.; Jaffe, G.J. Ranibizumab and Bevacizumab for Neovascular Age-Related Macular Degeneration. N. Engl. J. Med. 2011, 364, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Wykoff, C.C.; Eichenbaum, D.A.; Roth, D.B.; Hill, L.; Fung, A.E.; Haskova, Z. Ranibizumab Induces Regression of Diabetic Retinopathy in Most Patients at High Risk of Progression to Proliferative Diabetic Retinopathy. Ophthalmol. Retin. 2018, 2, 997–1009. [Google Scholar] [CrossRef]
- Ferrara, N.; Damico, L.; Shams, N.; Lowman, H.; Kim, R. Development of ranibizumab, an anti–vascular endothelial growth factor antigen binding fragment, as therapy for neovascular age-related macular degeneration. Retina 2006, 26, 859–870. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Martin, J.; Ruan, Q.; Rafique, A.; Rosconi, M.P.; Shi, E.; Pyles, E.A.; Yancopoulos, G.D.; Stahl, N.; Wiegand, S.J. Binding and neutralization of vascular endothelial growth factor (VEGF) and related ligands by VEGF Trap, ranibizumab and bevacizumab. Angiogenesis 2012, 15, 171–185. [Google Scholar] [CrossRef]
- Semeraro, F.; Morescalchi, M.; Duse, S.; Parmeggiani, F.; Gambicorti, G.; Costagliola, C. Aflibercept in wet AMD: Specific role and optimal use. Drug Des. Dev. Ther. 2013, 7, 711–722. [Google Scholar] [CrossRef]
- Hussain, R.; Savant, V. Fine-Needle Diathermy with Simultaneous Subconjunctival Bevacizumab. Semin. Ophthalmol. 2017, 32, 550–552. [Google Scholar] [CrossRef]
- Tadayoni, R.; Sararols, L.; Weissgerber, G.; Verma, R.; Clemens, A.; Holz, F.G. Brolucizumab: A Newly Developed Anti-VEGF Molecule for the Treatment of Neovascular Age-Related Macular Degeneration. Ophthalmology 2020, 244, 93–101. [Google Scholar] [CrossRef]
- Miller, J. Treatment of age-related macular degeneration: Beyond VEGF. Jpn. J. Ophthalmol. 2010, 54, 523–528. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, N.; Kuppermann, B.D.; Bandello, F.; Loewenstein, A. Faricimab: Expanding horizon beyond VEGF. Eye 2019, 34, 802–804. [Google Scholar] [CrossRef] [PubMed]
- Nicolò, M.; Desideri, L.F.; Vagge, A.; Traverso, C.E. Faricimab: An investigational agent targeting the Tie-2/angiopoietin pathway and VEGF-A for the treatment of retinal diseases. Expert Opin. Investig. Drugs 2021, 30, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, S.; Zhao, J. Resistance to anti-VEGF therapy in neovascular age-related macular degeneration: A comprehensive review. Drug Des. Dev. Ther. 2016, 10, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Broadhead, G.K.; Hong, T.; Chang, A.A. Treating the untreatable patient: Current options for the management of treatment-resistant neovascular age-related macular degeneration. Acta Ophthalmol. 2014, 92, 713–723. [Google Scholar] [CrossRef]
- Bucolo, C.; Gozzo, L.; Longo, L.; Mansueto, S.; Vitale, D.C.; Drago, F. Long-term efficacy and safety profile of multiple injections of intravitreal dexamethasone implant to manage diabetic macular edema: A systematic review of real-world studies. J. Pharmacol. Sci. 2018, 138, 219–232. [Google Scholar] [CrossRef]
- Giancipoli, E.; Pinna, A.; Boscia, F.; Zasa, G.; Sotgiu, G.; Dore, S.; Ricci, G.D. Intravitreal Dexamethasone in Patients with Wet Age-Related Macular Degeneration Resistant to Anti-VEGF: A Prospective Pilot Study. J. Ophthalmol. 2018, 2018, 1–8. [Google Scholar] [CrossRef]
- Barikian, A.; Salti, H.; Safar, A.; Mahfoud, Z.; Bashshur, Z.F. Intravitreal dexamethasone implant as adjuvant treatment for bevacizumab- and ranibizumab-resistant neovascular age-related macular degeneration. Retina 2017, 37, 1337–1344. [Google Scholar] [CrossRef]
- Montemagno, C.; Pagès, G. Resistance to Anti-angiogenic Therapies: A Mechanism Depending on the Time of Exposure to the Drugs. Front. Cell Dev. Biol. 2020, 8, 584. [Google Scholar] [CrossRef]
- Matsuda, Y.; Nonaka, Y.; Futakawa, S.; Imai, H.; Akita, K.; Nishihata, T.; Fujiwara, M.; Ali, Y.; Bhisitkul, R.B.; Nakamura, Y. Anti-Angiogenic and Anti-Scarring Dual Action of an Anti-Fibroblast Growth Factor 2 Aptamer in Animal Models of Retinal Disease. Mol. Ther. Nucleic Acids 2019, 17, 819–828. [Google Scholar] [CrossRef]
- Kersten, E.; Geerlings, M.J.; Pauper, M.; Corominas, J.; Bakker, B.; Altay, L.; Fauser, S.; De Jong, E.K.; Hoyng, C.B.; Hollander, A.I.D. Genetic screening for macular dystrophies in patients clinically diagnosed with dry age-related macular degeneration. Clin. Genet. 2018, 94, 569–574. [Google Scholar] [CrossRef]
- Suzuki, M.; Nagai, N.; Izumi-Nagai, K.; Shinoda, H.; Koto, T.; Uchida, A.; Mochimaru, H.; Yuki, K.; Sasaki, M.; Tsubota, K.; et al. Predictive factors for non-response to intravitreal ranibizumab treatment in age-related macular degeneration. Br. J. Ophthalmol. 2014, 98, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Ritter, M.; Elledge, J.; Simader, C.; Deak, G.G.; Beneschv, T.; Blodi, B.A.; Schmidt-Erfurth, U.M. Evaluation of optical coherence tomography findings in age-related macular degeneration: A reproducibility study of two independent reading centres. Br. J. Ophthalmol. 2010, 95, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Keane, P.; Liakopoulos, S.; Chang, K.T.; Heussen, F.M.; Ongchin, S.C.; Walsh, A.C.; Sadda, S.R. Comparison of the optical coherence tomographic features of choroidal neovascular membranes in pathological myopia versus age-related macular degeneration, using quantitative subanalysis. Br. J. Ophthalmol. 2008, 92, 1081–1085. [Google Scholar] [CrossRef]
- Strelkova, N.I. Physical methods in the treatment of patients with radiculitis. Zhurnal Nevropatol. Psikhiatrii Im. S.S. Korsakova 1974, 74, 1654–1659. [Google Scholar]
- Weng, C.Y.; Gregori, N.Z.; Moysidis, S.N.; Shi, W.; Smiddy, W.E.; Flynn, H.W. Visual and anatomical outcomes of macular epiretinal membrane peeling after previous rhegmatogenous retinal detachment repair. Retina 2015, 35, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Fursova, A.; Derbeneva, A.S.; Tarasov, M.S.; Nikulich, I.F.; Devyatkin, V.A.; Telegina, D.V.; Kozhevnikova, O.S. Leukocyte telomere length and response to antiangiogenic therapy in patients with neovascular age-related macular degeneration. Adv. Gerontol. = Uspekhi Gerontol. 2022, 34, 823–830. [Google Scholar] [CrossRef]
- Diack, C.; Schwab, D.; Cosson, V.; Buchheit, V.; Mazer, N.; Frey, N. A Baseline Score to Predict Response to Ranibizumab Treatment in Neovascular Age-Related Macular Degeneration. Transl. Vis. Sci. Technol. 2021, 10, 11. [Google Scholar] [CrossRef]
- Lorés-Motta, L.; Riaz, M.; Grunin, M.; Corominas, J.; Van Asten, F.; Pauper, M.; Leenders, M.; Richardson, A.J.; Muether, P.; Cree, A.; et al. Association of Genetic Variants With Response to Anti–Vascular Endothelial Growth Factor Therapy in Age-Related Macular Degeneration. JAMA Ophthalmol. 2018, 136, 875–884. [Google Scholar] [CrossRef]
- Brantley, M.A.; Edelstein, S.L.; King, J.M.; Apte, R.S.; Kymes, S.M.; Shiels, A. Clinical Phenotypes Associated with the Complement Factor H Y402H Variant in Age-related Macular Degeneration. Am. J. Ophthalmol. 2007, 144, 404–408.e1. [Google Scholar] [CrossRef]
- Brantley, M.A.; Edelstein, S.L.; King, J.M.; Plotzke, M.R.; Apte, R.S.; Kymes, S.M.; Shiels, A. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to photodynamic therapy. Eye 2008, 23, 626–631. [Google Scholar] [CrossRef]
- Chang, W.; Noh, D.H.; Sagong, M.; Kim, I.T. Pharmacogenetic association with early response to intravitreal ranibizumab for age-related macular degeneration in a Korean population. Mol. Vis. 2013, 19, 702–709. [Google Scholar] [PubMed]
- Chaudhary, V.; Brent, M.; Lam, W.-C.; Devenyi, R.; Teichman, J.; Mak, M.; Barbosa, J.; Kaur, H.; Carter, R.; Farrokhyar, F. Genetic Risk Evaluation in Wet Age-Related Macular Degeneration Treatment Response. Ophthalmology 2016, 236, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Cobos, E.; Recalde, S.; Anter, J.; Hernandez-Sanchez, M.; Barreales, C.; Olavarrieta, L.; Valverde, A.; Suarez-Figueroa, M.; Cruz, F.; Abraldes, M.; et al. Association between CFH, CFB, ARMS2, SERPINF1, VEGFR1 and VEGF polymorphisms and anatomical and functional response to ranibizumab treatment in neovascular age-related macular degeneration. Acta Ophthalmol. 2017, 96, e201–e212. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.; Kitchens, J.W.; Kassem, N.; Stone, T.W.; Isernhagen, R.; Hancock, B.A.; Radovich, M.; Waymire, J.; Li, L.; Schneider, B.P. A pharmacogenetics study to predict outcome in patients receiving anti-VEGF therapy in age related macular degeneration. Clin. Ophthalmol. 2013, 7, 1987–1993. [Google Scholar] [CrossRef]
- Lee, A.; Raya, A.K.; Kymes, S.M.; Shiels, A.; Brantley, M.A. Pharmacogenetics of complement factor H (Y402H) and treatment of exudative age-related macular degeneration with ranibizumab. Br. J. Ophthalmol. 2008, 93, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Matušková, V.; Balcar, V.J.; Khan, N.A.; Bonczek, O.; Ewerlingová, L.; Zeman, T.; Kolář, P.; Vysloužilová, D.; Vlková, E.; Šerý, O. CD36 gene is associated with intraocular pressure elevation after intravitreal application of anti-VEGF agents in patients with age-related macular degeneration: Implications for the safety of the therapy. Ophthalmic Genet. 2017, 39, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Piermarocchi, S.; Miotto, S.; Colavito, D.; Leon, A.; Segato, T. Combined effects of genetic and non-genetic risk factors affect response to ranibizumab in exudative age-related macular degeneration. Acta Ophthalmol. 2014, 93, e451–e457. [Google Scholar] [CrossRef] [PubMed]
- Fauser, S.; Schwabecker, V.; Muether, P.S. Suppression of Intraocular Vascular Endothelial Growth Factor During Aflibercept Treatment of Age-Related Macular Degeneration. Am. J. Ophthalmol. 2014, 158, 532–536. [Google Scholar] [CrossRef]
- Dawson, D.W.; Volpert, O.V.; Gillis, P.; Crawford, S.E.; Xu, H.-J.; Benedict, W.; Bouck, N.P. Pigment Epithelium-Derived Factor: A Potent Inhibitor of Angiogenesis. Science 1999, 285, 245–248. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, S.; Li, X.; Huang, X.; Zhang, Y.; Chang, T.; Cai, Z.; Zhang, M. Efficacy and Safety of Anti–Vascular Endothelial Growth Factor Monotherapies for Neovascular Age-Related Macular Degeneration: A Mixed Treatment Comparison. Front. Pharmacol. 2021, 12, 797108. [Google Scholar] [CrossRef]
- Wu, W.-C.; Shih, C.-P.; Lien, R.; Wang, N.-K.; Chen, Y.-P.; Chao, A.-N.; Chen, K.-J.; Chen, T.-L.; Hwang, Y.-S.; Lai, C.-C. Serum vascular endothelial growth factor after bevacizumab or ranibizumab treatment for retinopathy of prematurity. Retina 2017, 37, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Hagstrom, S.A.; Ying, G.-S.; Pauer, G.J.; Sturgill-Short, G.M.; Huang, J.; Callanan, D.G.; Kim, I.; Klein, M.L.; Maguire, M.G.; Martin, D.F. Pharmacogenetics for Genes Associated with Age-related Macular Degeneration in the Comparison of AMD Treatments Trials (CATT). Ophthalmology 2013, 120, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Kloeckener-Gruissem, B.; Barthelmes, D.; Labs, S.; Schindler, C.; Kurz-Levin, M.; Michels, S.; Fleischhauer, J.; Berger, W.; Sutter, F.; Menghini, M. Genetic Association with Response to Intravitreal Ranibizumab in Patients with Neovascular AMD. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4694–4702. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, K.; Tomita, K.; Tsujikawa, A.; Nakata, I.; Akagi-Kurashige, Y.; Miyake, M.; Ooto, S.; Tamura, H.; Yoshimura, N. Factors Associated with the Response of Age-Related Macular Degeneration to Intravitreal Ranibizumab Treatment. Am. J. Ophthalmol. 2012, 154, 125–136. [Google Scholar] [CrossRef]
- Smailhodzic, D.; Muether, P.S.; Chen, J.; Kwestro, A.; Zhang, A.Y.; Omar, A.; Van de Ven, J.P.; Keunen, J.E.; Kirchhof, B.; Hoyng, C.B.; et al. Cumulative Effect of Risk Alleles in CFH, ARMS2, and VEGFA on the Response to Ranibizumab Treatment in Age-related Macular Degeneration. Ophthalmology 2012, 119, 2304–2311. [Google Scholar] [CrossRef]
- Orlin, A.; Hadley, D.; Chang, W.; Ho, A.C.; Brown, G.; Kaiser, R.S.; Regillo, C.D.; Godshalk, A.N.; Lier, A.; Kaderli, B.; et al. Association between high-risk disease loci and response to anti–vascular endothelial growth factor treatment for wet age-related macular degeneration. Retina 2012, 32, 4–9. [Google Scholar] [CrossRef]
- Mohamad, N.A.; Ramachandran, V.; Ismail, P.; Isa, H.M.; Chan, Y.M.; Ngah, N.F.; Bakri, N.M.; Ching, S.M.; Hoo, F.K.; Sulaiman, W.A.W.; et al. Analysis of the association between CFH Y402H polymorphism and response to intravitreal ranibizumab in patients with neovascular age-related macular degeneration (nAMD). Bosn. J. Basic Med. Sci. 2018, 18, 260–267. [Google Scholar] [CrossRef]
- Rodríguez, F.J.; Rios, H.; Aguilar, M.C.; Rosenstiehl, S.M.; Gelvez, N.; Lopez, G.; Tamayo, M.L. Genetic association with intravitreal ranibizumab response for neovascular age-related macular degeneration in Hispanic population. Taiwan J. Ophthalmol. 2019, 9, 243–248. [Google Scholar] [CrossRef]
- Dikmetas, O.; Kadayıfcılar, S.; Eldem, B. The effect of CFH polymorphisms on the response to the treatment of age-related macular degeneration (AMD) with intravitreal ranibizumab. Mol. Vis. 2013, 19, 2571–2578. [Google Scholar]
- Medina, F.M.C.; Da Motta, A.A.L.; Takahashi, W.Y.; Carricondo, P.C.; Motta, M.M.D.S.; Melo, M.B.; Vasconcellos, J.P.C. Association of the CFH Y402H Polymorphism with the 1-Year Response of Exudative AMD to Intravitreal Anti-VEGF Treatment in the Brazilian Population. Ophthalmic Res. 2017, 61, 168–173. [Google Scholar] [CrossRef]
- Teper, S.J.; Nowinska, A.; Pilat, J.; Palucha, A.; Wylegala, E. Involvement of genetic factors in the response to a variable-dosing ranibizumab treatment regimen for age-related macular degeneration. Mol. Vis. 2010, 16, 2598–2604. [Google Scholar]
- Park, U.C.; Shin, J.Y.; Kim, S.J.; Shin, E.S.; Lee, J.E.; McCarthy, L.C.; Newcombe, P.J.; Xu, C.-F.; Chung, H.; Yu, H.G. Genetic factors associated with response to intravitreal ranibizumab in korean patients with neovascular age-related macular degeneration. Retina 2014, 34, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Gonzalez, F.; Cabrillo-Estevez, L.; Gutiérrez, V.R.; Sanchez-Jara, A.; De Juan-Marcos, L.; Gonzalez-Sarmiento, R. Influence of CFH, HTRA1 and ARMS2 polymorphisms in the response to intravitreal ranibizumab treatment for wet age-related macular degeneration in a Spanish population. Int. J. Ophthalmol. 2016, 9, 1304–1309. [Google Scholar] [CrossRef] [PubMed]
- Abedi, F.; Wickremasinghe, S.; Richardson, A.J.; Islam, F.M.A.; Guymer, R.; Baird, P. Genetic Influences on the Outcome of Anti-Vascular Endothelial Growth Factor Treatment in Neovascular Age-related Macular Degeneration. Ophthalmology 2013, 120, 1641–1648. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.A.; Ramachandran, V.; Isa, H.M.; Chan, Y.M.; Ngah, N.F.; Ching, S.M.; Hoo, F.K.; Sulaiman, W.A.W.; Mat, L.N.I.; Mohamed, M.H. Association of HTRA1 and ARMS2 gene polymorphisms with response to intravitreal ranibizumab among neovascular age-related macular degenerative subjects. Hum. Genom. 2019, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- Wickremasinghe, S.S.; Xie, J.; Lim, J.; Chauhan, D.S.; Robman, L.; Richardson, A.J.; Hageman, G.; Baird, P.N.; Guymer, R. Variants in theAPOEGene Are Associated with Improved Outcome after Anti-VEGF Treatment for Neovascular AMD. Investig. Ohpthalmol. Vis. Sci. 2011, 52, 4072–4079. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, S.; Figus, M.; Orlandi, P.; Fioravanti, A.; Di Desidero, T.; Agosta, E.; Sartini, M.S.; Posarelli, C.; Nardi, M.; Danesi, R.; et al. VEGF-A polymorphisms predict short-term functional response to intravitreal ranibizumab in exudative age-related macular degeneration. Pharmacogenomics 2013, 14, 623–630. [Google Scholar] [CrossRef]
- Riaz, M.; Lorés-Motta, L.; Richardsonv, A.J.; Lu, Y.; Montgomery, G.; Omar, A.; Koenekoop, R.K.; Chen, J.; Muether, P.; Altay, L.; et al. GWAS study using DNA pooling strategy identifies association of variant rs4910623 in OR52B4 gene with anti-VEGF treatment response in age-related macular degeneration. Sci. Rep. 2016, 6, 37924. [Google Scholar] [CrossRef]
- Bojanowski, C.M.; Shen, D.; Chew, E.Y.; Ning, B.; Csaky, K.G.; Green, W.R.; Chan, C.-C.; Tuo, J. Anapolipoprotein E variant may protect against age-related macular degeneration through cytokine regulation. Environ. Mol. Mutagen. 2006, 47, 594–602. [Google Scholar] [CrossRef]
- Bakbak, B.; Ozturk, B.T.; Zamani, A.G.; Gonul, S.; Iyit, N.; Gedik, S.; Yıldırım, M.S. Association of Apolipoprotein E Polymorphism with Intravitreal Ranibizumab Treatment Outcomes in Age-Related Macular Degeneration. Curr. Eye Res. 2015, 41, 862–866. [Google Scholar] [CrossRef]
- Kersten, E.; Paun, C.C.; Schellevis, R.L.; Hoyng, C.B.; Delcourt, C.; Lengyel, I.; Peto, T.; Ueffing, M.; Klaver, C.; Dammeier, S.; et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv. Ophthalmol. 2018, 63, 9–39. [Google Scholar] [CrossRef]
- Chawla, R.; Kumar, A.; Tripathy, K. Intraocular use of bevacizumab in India: An issue resolved? Natl. Med. J. India 2017, 30, 345–347. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, B.K.; Ozdek, S.; Ergun, M.A.; Ergun, S.; Tuncay, F.Y.; Elbeg, S. CFH Y402H and VEGF Polymorphisms and Anti-VEGF Treatment Response in Exudative Age-Related Macular Degeneration. Ophthalmic Res. 2016, 56, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, V.M.; Chan, C.-C. The role of anti-inflammatory agents in age-related macular degeneration (AMD) treatment. Eye 2010, 25, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Chan, C.-C.; Tuo, J. Genetic mechanisms and age-related macular degeneration: Common variants, rare variants, copy number variations, epigenetics, and mitochondrial genetics. Hum. Genom. 2012, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Bittersohl, H.; Herbinger, J.; Wen, M.; Renders, L.; Steimer, W.; Luppa, P.B. Simultaneous Determination of Protein-Unbound Cyclosporine A and Mycophenolic Acid in Kidney Transplant Patients Using Liquid Chromatography–Tandem Mass Spectrometry. Ther. Drug Monit. 2017, 39, 211–219. [Google Scholar] [CrossRef] [PubMed]
- McKibbin, M.; Ali, M.; Bansal, S.; Baxter, P.D.; West, K.; Williams, G.; Cassidy, F.; Inglehearn, C.F. CFH, VEGF and HTRA1promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br. J. Ophthalmol. 2012, 96, 208–212. [Google Scholar] [CrossRef]
- Kuroda, Y.; Tsujikawa, A.; Ooto, S.; Yamashiro, K.; Oishi, A.; Nakanishi, H.; Kumagai, K.; Hata, M.; Arichika, S.; Ellabban, A.; et al. Association of Focal Choroidal Excavation with Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6046–6054. [Google Scholar] [CrossRef]
- Mouallem-Beziere, A.; Blanco-Garavito, R.; Richard, F.; Miere, A.; Jung, C.; Rozet, J.-M.; Souied, E.H. Genetics of large pigment epithelial detachments in neovascular age-related macular degeneratioN. Retina 2020, 40, 663–671. [Google Scholar] [CrossRef]
- Leveziel, N.; Puche, N.; Zerbib, J.; Benlian, P.; Coscas, G.; Soubrane, G.; Souied, E. Génétique de la dégénérescence maculaire liée à l’âge. Médecine Sci. 2010, 26, 505–515. [Google Scholar] [CrossRef][Green Version]
- Yamashiro, K.; Mori, K.; Honda, S.; Kano, M.; Yanagi, Y.; Obana, A.; Sakurada, Y.; Sato, T.; Nagai, Y.; Hikichi, T.; et al. A prospective multicenter study on genome wide associations to ranibizumab treatment outcome for age-related macular degeneration. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gourgouli, K.; Gourgouli, I.; Tsaousis, G.; Spai, S.; Niskopoulou, M.; Efthimiopoulos, S.; Lamnissou, K. Investigation of genetic base in the treatment of age-related macular degeneration. Int. Ophthalmol. 2020, 40, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Trotta, M.C.; Gesualdo, C.; Platania, C.B.M.; De Robertis, D.; Giordano, M.; Simonelli, F.; D’Amico, M.; Drago, F.; Bucolo, C.; Rossi, S. Circulating miRNAs in diabetic retinopathy patients: Prognostic markers or pharmacological targets? Biochem. Pharmacol. 2021, 186, 114473. [Google Scholar] [CrossRef]
- Romano, G.L.; Platania, C.B.M.; Forte, S.; Salomone, S.; Drago, F.; Bucolo, C. MicroRNA target prediction in glaucoma. Prog. Brain Res. 2015, 220, 217–240. [Google Scholar] [CrossRef] [PubMed]
- Hofmans, M.; Lammens, T.; Depreter, B.; Wu, Y.; Erlacher, M.; Caye, A.; Cavé, H.; Flotho, C.; de Haas, V.; Niemeyer, C.M.; et al. Long non-coding RNAs as novel therapeutic targets in juvenile myelomonocytic leukemia. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, N.; Wahid, F.; Azam, M.; Shah, K.; Hollander, A.I.D.; Qamar, R.; Ayub, H. Molecular Mechanisms of Complement System Proteins and Matrix Metalloproteinases in the Pathogenesis of Age-Related Macular Degeneration. Curr. Mol. Med. 2019, 19, 705–718. [Google Scholar] [CrossRef] [PubMed]
- García-Onrubia, L.; Valentín-Bravo, F.; Coco-Martin, R.; González-Sarmiento, R.; Pastor, J.; Usategui-Martín, R.; Pastor-Idoate, S. Matrix Metalloproteinases in Age-Related Macular Degeneration (AMD). Int. J. Mol. Sci. 2020, 21, 5934. [Google Scholar] [CrossRef]
- Farrar, G.J.; Carrigan, M.; Dockery, A.; Millington-Ward, S.; Palfi, A.; Chadderton, N.; Humphries, M.; Kiang, A.S.; Kenna, P.F.; Humphries, P. Toward an elucidation of the molecular genetics of inherited retinal degenerations. Hum. Mol. Genet. 2017, 26, R2–R11. [Google Scholar] [CrossRef]
- Wang, Q.; Thau, A.; Levin, A.V.; Lee, D. Ocular hypotony: A comprehensive review. Surv. Ophthalmol. 2019, 64, 619–638. [Google Scholar] [CrossRef]
- Sakamoto, S.; Takahashi, H.; Tan, X.; Inoue, Y.; Nomura, Y.; Arai, Y.; Fujino, Y.; Kawashima, H.; Yanagi, Y. Changes in multiple cytokine concentrations in the aqueous humour of neovascular age-related macular degeneration after 2 months of ranibizumab therapy. Br. J. Ophthalmol. 2017, 102, 448–454. [Google Scholar] [CrossRef]
- Sun, T.; Wei, Q.; Gao, P.; Zhang, Y.; Peng, Q. Cytokine and Chemokine Profile Changes in Patients with Neovascular Age-Related Macular Degeneration After Intravitreal Ranibizumab Injection for Choroidal Neovascularization. Drug Des. Dev. Ther. 2021, 15, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.R.F.; Castellanos, B.E.P.; Gatto, M.A.F.; Silva, A. Fator de risco: Enfoque na disciplina enfermagem em centro cirrgico. Rev. Bras. Enferm. 1986, 39, 26–32. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joyal, J.-S.; Gantner, M.L.; Smith, L.E. Retinal energy demands control vascular supply of the retina in development and disease: The role of neuronal lipid and glucose metabolism. Prog. Retin. Eye Res. 2018, 64, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Tao, Y.; Li, X.-X. Inflammatory cytokines in aqueous humor of patients with choroidal neovascularization. Mol. Vis. 2012, 18, 574–580. [Google Scholar]
- Arai, Y.; Takahashi, H.; Inoda, S.; Tan, X.; Sakamoto, S.; Inoue, Y.; Fujino, Y.; Kawashima, H.; Yanagi, Y. Aqueous humour proteins and treatment outcomes of anti-VEGF therapy in neovascular age-related macular degeneration. PLoS ONE 2020, 15, e0229342. [Google Scholar] [CrossRef]
- Pongsachareonnont, P.; Mak, M.Y.K.; Hurst, C.P.; Lam, W.-C. Neovascular age-related macular degeneration: Intraocular inflammatory cytokines in the poor responder to ranibizumab treatment. Clin. Ophthalmol. 2018, 12, 1877–1885. [Google Scholar] [CrossRef]
- Mantel, I.; Borgo, A.; Guidotti, J.; Forestier, E.; Kirsch, O.; Derradji, Y.; Waridel, P.; Burdet, F.; Mehl, F.; Schweizer, C.; et al. Molecular Biomarkers of Neovascular Age-Related Macular Degeneration with Incomplete Response to Anti-Vascular Endothelial Growth Factor Treatment. Front. Pharmacol. 2020, 11, 594087. [Google Scholar] [CrossRef]
- Sakamoto, T.; Ito, S.; Yoshikawa, H.; Hata, Y.; Ishibashi, T.; Sueishi, K.; Inomata, H. Tissue factor increases in the aqueous humor of proliferative diabetic retinopathy. Graefe’s Arch. Clin. Exp. Ophthalmol. 2001, 239, 865–871. [Google Scholar] [CrossRef]
- Shin, J.Y.; Lee, J.; Park, J.; Kim, M.; Chung, H.; Byeon, S.H. Association of Keratin 8 Level in Aqueous Humor with Outcomes of Intravitreal Ranibizumab Treatment for Neovascular Age-Related Macular Degeneration. Transl. Vis. Sci. Technol. 2022, 11, 26. [Google Scholar] [CrossRef]
- Friedlander, M.; Theesfeld, C.L.; Sugita, M.; Fruttiger, M.; Thomas, M.A.; Chang, S.; Cheresh, D.A. Involvement of integrins alpha v beta 3 and alpha v beta 5 in ocular neovascular diseases. Proc. Natl. Acad. Sci. USA 1996, 93, 9764–9769. [Google Scholar] [CrossRef]
- Bhatwadekar, A.D.; Kansara, V.; Luo, Q.; Ciulla, T. Anti-integrin therapy for retinovascular diseases. Expert Opin. Investig. Drugs 2020, 29, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Ross, W.H.; Gottner, M.J. Peripheral Retinal Cryopexy for Subtotal Vitreous Hemorrhage. Am. J. Ophthalmol. 1988, 105, 377–382. [Google Scholar] [CrossRef]
- Friedlander, M.; Brooks, P.C.; Shaffer, R.W.; Kincaid, C.M.; Varner, J.A.; Cheresh, D.A. Definition of Two Angiogenic Pathways by Distinct αv Integrins. Science 1995, 270, 1500–1502. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.-Y.; Bang, J.Y.; Choi, A.J.; Yoon, J.; Lee, W.-C.; Choi, S.; Yoon, S.; Kim, H.C.; Baek, J.-H.; Park, H.S.; et al. Exosomal Proteins in the Aqueous Humor as Novel Biomarkers in Patients with Neovascular Age-related Macular Degeneration. J. Proteome Res. 2014, 13, 581–595. [Google Scholar] [CrossRef]
- Baek, A.; Yoon, S.; Kim, J.; Baek, Y.M.; Park, H.; Lim, D.; Chung, H.; Kim, D.-E. Autophagy and KRT8/keratin 8 protect degeneration of retinal pigment epithelium under oxidative stress. Autophagy 2017, 13, 248–263. [Google Scholar] [CrossRef]
- Babapoor-Farrokhran, S.; Jee, K.; Puchner, B.; Hassan, S.J.; Xin, X.; Rodrigues, M.; Kashiwabuchi, F.; Ma, T.; Hu, K.; Deshpande, M.; et al. Angiopoietin-like 4 is a potent angiogenic factor and a novel therapeutic target for patients with proliferative diabetic retinopathy. Proc. Natl. Acad. Sci. USA 2015, 112, E3030–E3039. [Google Scholar] [CrossRef]
- Kim, J.H.; Shin, J.P.; Kim, I.T.; Park, D.H. Angiopoietin-like 4 correlates with response to intravitreal ranibizumab injections in neovascular age-related macular degeneration. Retina 2018, 38, 523–530. [Google Scholar] [CrossRef]
- Han, G.; Wei, P.; He, M.; Teng, H. Glucose Metabolic Characterization of Human Aqueous Humor in Relation to Wet Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2020, 61, 49. [Google Scholar] [CrossRef]
- Kersten, E.; Dammeier, S.; Ajana, S.; Groenewoud, J.M.M.; Codrea, M.; Klose, F.; Lechanteur, Y.T.; Fauser, S.; Ueffing, M.; Delcourt, C.; et al. Metabolomics in serum of patients with non-advanced age-related macular degeneration reveals aberrations in the glutamine pathway. PLoS ONE 2019, 14, e0218457. [Google Scholar] [CrossRef]
- Lains, I.; Kelly, R.S.; Miller, J.B.; Silva, R.; Vavvas, D.; Kim, I.; Murta, J.; Lasky-Su, J.; Miller, J.W.; Husain, D. Human Plasma Metabolomics Study across All Stages of Age-Related Macular Degeneration Identifies Potential Lipid Biomarkers. Ophthalmology 2018, 125, 245–254. [Google Scholar] [CrossRef]
- Osborn, M.P.; Park, Y.; Parks, M.B.; Burgess, L.G.; Uppal, K.; Lee, K.; Jones, D.P.; Brantley, M.A., Jr. Metabolome-Wide Association Study of Neovascular Age-Related Macular Degeneration. PLoS ONE 2013, 8, e72737. [Google Scholar] [CrossRef] [PubMed]
| Genes Related to Susceptibility and Progression of AMD | ||
|---|---|---|
| Gene | Single Nucleotide Polymorphism | Reference |
| CFH | rs1410996; rs1061170 (Y402H); rs800292 (V62I); rs2274700 | [43,44,45] |
| C2 | rs1042663; rs3020644; rs2072632; rs9332739; rs547154 | [46,47] |
| ARMS2 | rs10490924; rs3750848; rs10490923 | [42,45,47,48] |
| HTRA1 | rs11200638; rs932275 | [48] |
| CD36 | rs3173798; rs3211883; rs10499862; rs3173800; rs17154232 | [49] |
| APOE | Ɛ2; Ɛ4 | [50,51] |
| PON1 | M55L; Q192R | [52] |
| ERCC6 | C−6530 > G | [53] |
| CFB | rs641153; rs4151657; rs4151672; rs4151667 | [46] |
| C3 | rs1047286; rs3745565; rs171094; rs2230199 (R102G); rs11569536 | [46] |
| VEGFA | rs699947; rs1413711; rs2010963 | [54] |
| CFI | rs10033900 | [42] |
| VEGFR | rs9319425; rs622227; rs2387632 | [54,55] |
| CXCR3CR1 | T280M; V249I | [56] |
| TLR4 | D299G | [57] |
| ELOVL4 | M299V | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobadilla, M.; Pariente, A.; Oca, A.I.; Peláez, R.; Pérez-Sala, Á.; Larráyoz, I.M. Biomarkers as Predictive Factors of Anti-VEGF Response. Biomedicines 2022, 10, 1003. https://doi.org/10.3390/biomedicines10051003
Bobadilla M, Pariente A, Oca AI, Peláez R, Pérez-Sala Á, Larráyoz IM. Biomarkers as Predictive Factors of Anti-VEGF Response. Biomedicines. 2022; 10(5):1003. https://doi.org/10.3390/biomedicines10051003
Chicago/Turabian StyleBobadilla, Miriam, Ana Pariente, Ana I. Oca, Rafael Peláez, Álvaro Pérez-Sala, and Ignacio M. Larráyoz. 2022. "Biomarkers as Predictive Factors of Anti-VEGF Response" Biomedicines 10, no. 5: 1003. https://doi.org/10.3390/biomedicines10051003
APA StyleBobadilla, M., Pariente, A., Oca, A. I., Peláez, R., Pérez-Sala, Á., & Larráyoz, I. M. (2022). Biomarkers as Predictive Factors of Anti-VEGF Response. Biomedicines, 10(5), 1003. https://doi.org/10.3390/biomedicines10051003






