Targeting CDK4/6 for Anticancer Therapy
Abstract
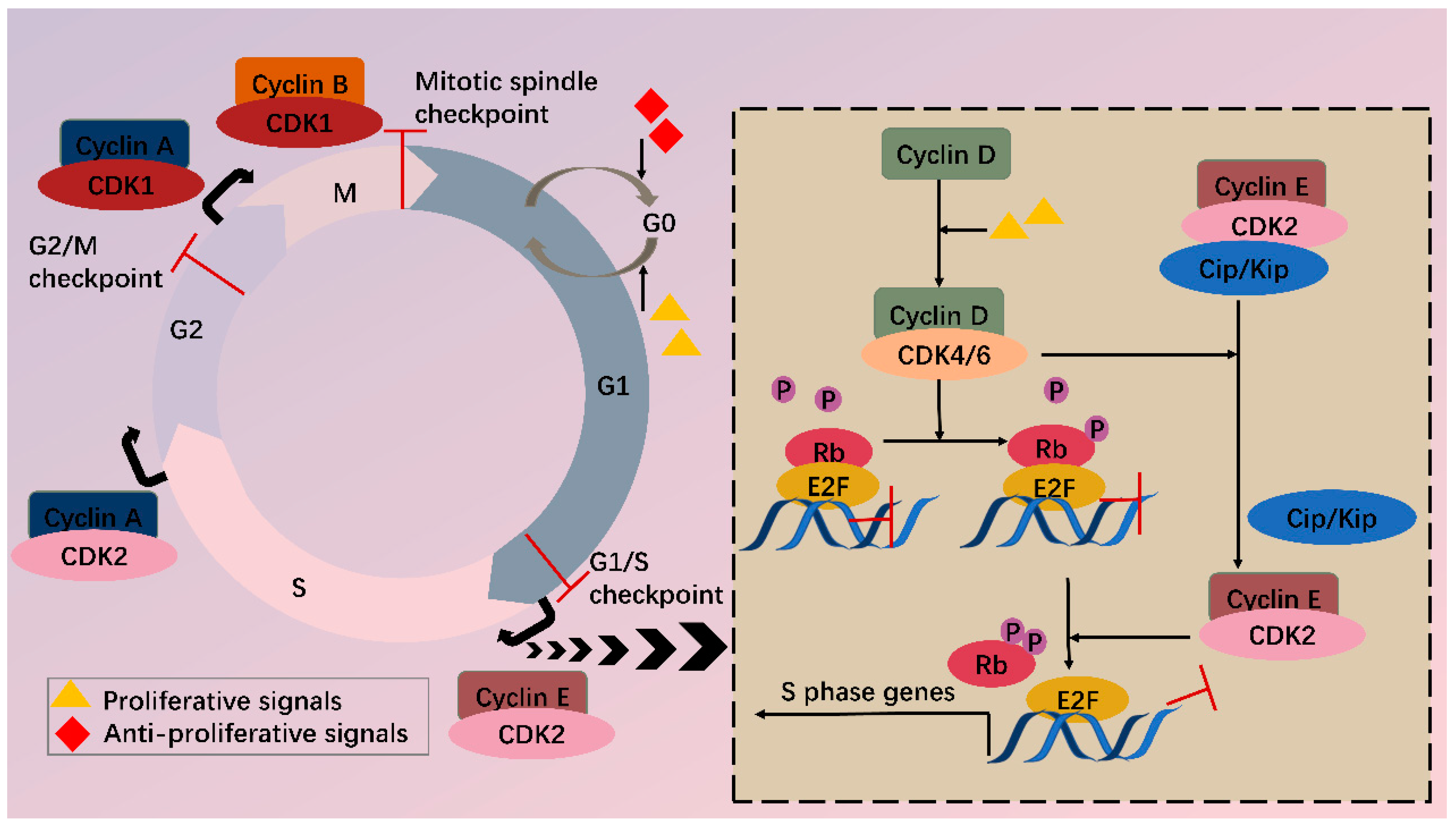
1. Introduction
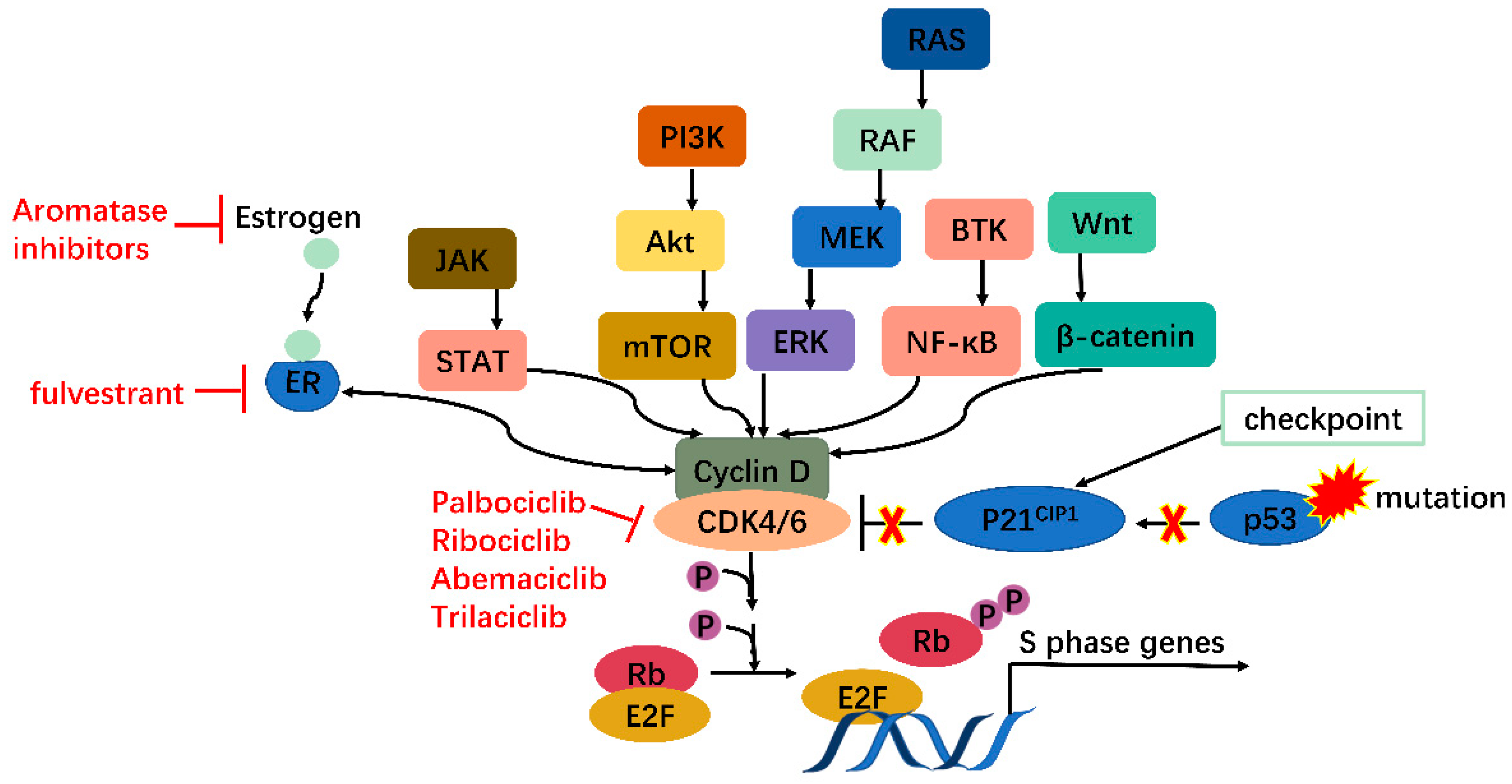
2. CDK4/6 Are Attractive Targets for Anticancer Treatment
3. CRISPR and RNAi
4. Small Molecular Inhibitors
4.1. One Drug Works Alone
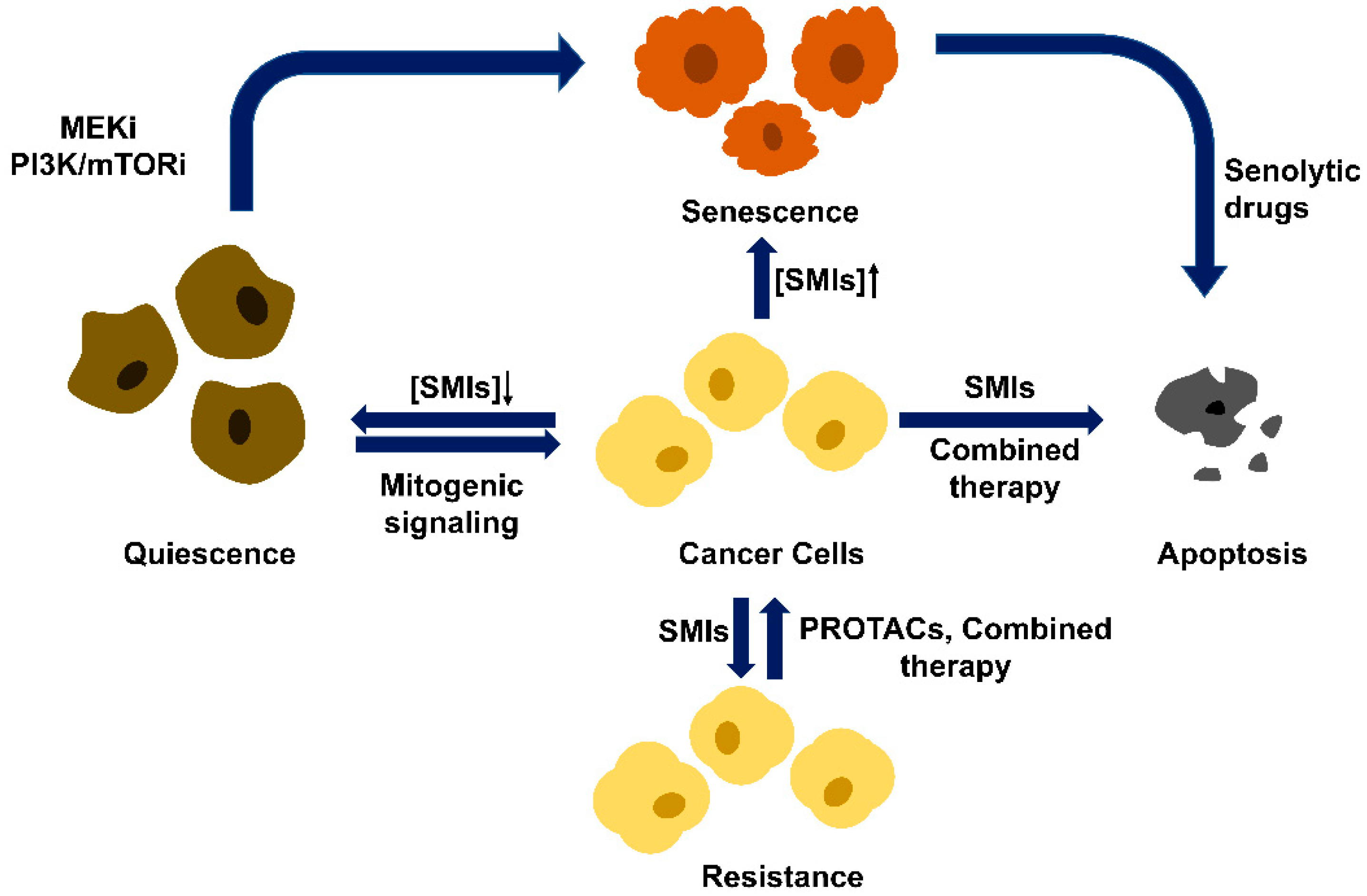
4.2. Therapeutic Mechanisms beyond G1/S Arrest
4.3. Drug Resistance Problem
4.4. Two Drugs Are Better Than One
4.5. Kill Two Birds with One Stone
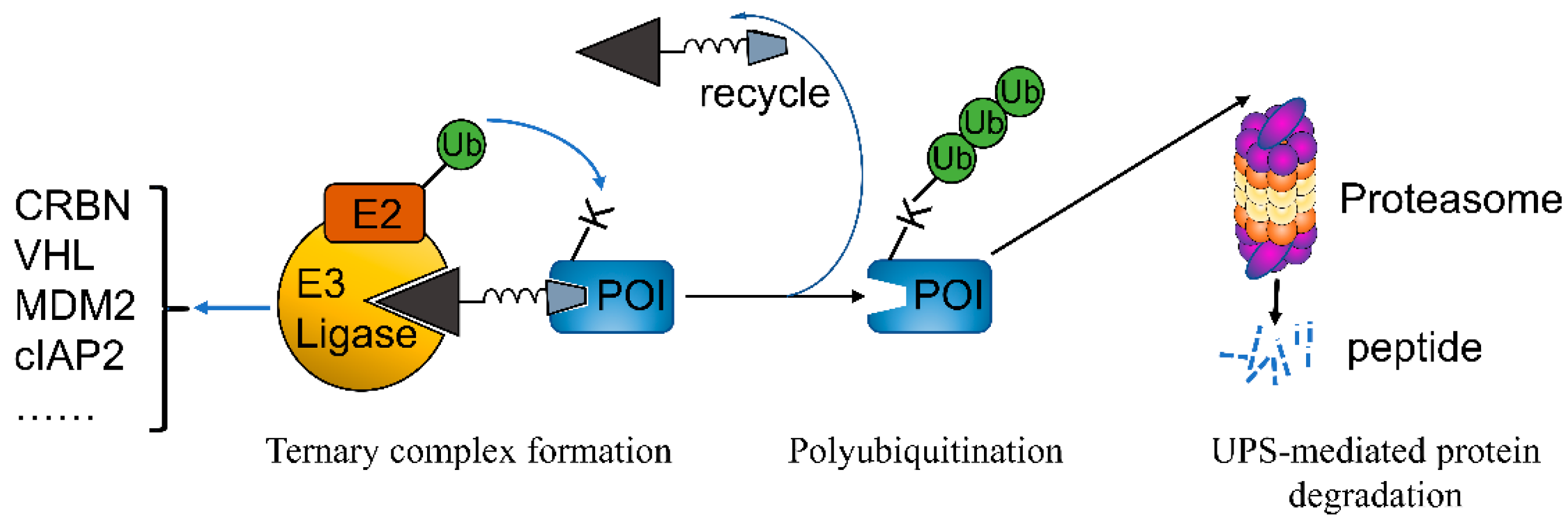
5. PROTACs
5.1. PROTACs Targeting Both CDK4/6
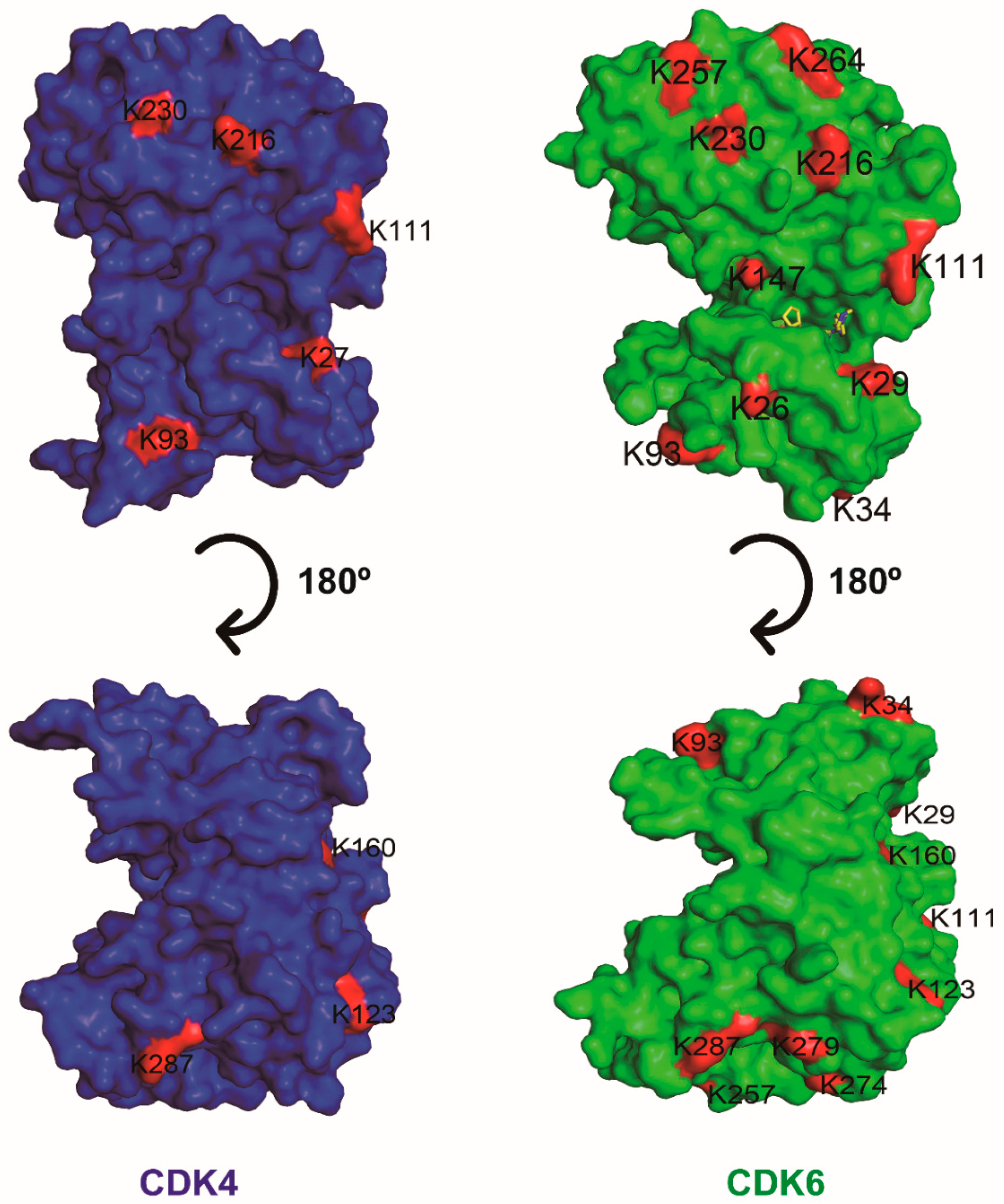
5.2. PROTACs Targeting CDK4 or CDK6 Alone
5.3. PROTACs Targeting Multiple CDKs
5.4. PROTACs Targeting CDK4/6 as Well as Other Targets
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, M.; Zhang, L.; Hei, R.; Li, X.; Cai, H.; Wu, X.; Zheng, Q.; Cai, C. CDK inhibitors in cancer therapy, an overview of recent development. Am. J. Cancer Res. 2021, 11, 1913–1935. [Google Scholar] [PubMed]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. To cycle or not to cycle: A critical decision in cancer. Nat. Rev. Cancer 2001, 1, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef]
- Shapiro, G.I. Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. 2006, 24, 1770–1783. [Google Scholar] [CrossRef]
- Li, M.; Xiao, A.; Floyd, D.; Olmez, I.; Lee, J.; Godlewski, J.; Bronisz, A.; Bhat, K.P.L.; Sulman, E.P.; Nakano, I.; et al. CDK4/6 inhibition is more active against the glioblastoma proneural subtype. Oncotarget 2017, 8, 55319–55331. [Google Scholar] [CrossRef]
- Sheppard, K.E.; McArthur, G.A. The Cell-Cycle Regulator CDK4: An Emerging Therapeutic Target in Melanoma. Clin. Cancer Res. 2013, 19, 5320–5328. [Google Scholar] [CrossRef]
- Malumbres, M. Oncogene-Induced Mitotic Stress: p53 and pRb Get Mad Too. Cancer Cell 2011, 19, 691–692. [Google Scholar] [CrossRef]
- Dall’Acqua, A.; Sonego, M.; Pellizzari, I.; Pellarin, I.; Canzonieri, V.; D’Andrea, S.; Benevol, S.; Sorio, R.; Giorda, G.; Califano, D.; et al. CDK6 protects epithelial ovarian cancer from platinum-induced death via FOXO3 regulation. Embo. Mol. Med. 2017, 9, 1415–1433. [Google Scholar] [CrossRef]
- Alvarez-Fernandez, M.; Malumbres, M. Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition. Cancer Cell 2020, 37, 514–529. [Google Scholar] [CrossRef]
- Dai, M.; Boudreault, J.; Wang, N.; Poulet, S.; Daliah, G.; Yan, G.; Moamer, A.; Burgos, S.A.; Sabri, S.; Ali, S.; et al. Differential Regulation of Cancer Progression by CDK4/6 Plays a Central Role in DNA Replication and Repair Pathways. Cancer Res. 2021, 81, 1332–1346. [Google Scholar] [CrossRef]
- Nebenfuehr, S.; Kollmann, K.; Sexl, V. The role of CDK6 in cancer. Int. J. Cancer 2020, 147, 2988–2995. [Google Scholar] [CrossRef]
- O’Sullivan, C.C. Overcoming Endocrine Resistance in Hormone-Receptor Positive Advanced Breast Cancer-The Emerging Role of CDK4/6 Inhibitors. Int. J. Cancer Clin. Res. 2015, 2, 29. [Google Scholar] [CrossRef]
- Malumbres, M.; Sotillo, R.; Santamaria, D.; Galan, J.; Cerezo, A.; Ortega, S.; Dubus, P.; Barbacid, M. Mammalian cells cycle without the D-type cyclin-elependent kinases Cdk4 and Cdk6. Cell 2004, 118, 493–504. [Google Scholar] [CrossRef]
- Xing, Z.; Zhang, Y.; Liang, K.; Yan, L.; Xiang, Y.; Li, C.; Hu, Q.; Jin, F.; Putluri, V.; Putluri, N.; et al. Expression of Long Noncoding RNA YIYA Promotes Glycolysis in Breast Cancer. Cancer Res. 2018, 78, 4524–4532. [Google Scholar] [CrossRef]
- Lange, C.; Huttner, W.B.; Calegari, F. Cdk4/CyclinD1 Overexpression in Neural Stem Cells Shortens G1, Delays Neurogenesis, and Promotes the Generation and Expansion of Basal Progenitors. Cell Stem. Cell 2009, 5, 320–331. [Google Scholar] [CrossRef]
- Liu, H.; Li, Z.; Huo, S.; Wei, Q.; Ge, L. Induction of G0/G1 phase arrest and apoptosis by CRISPR/Cas9-mediated knockout of CDK2 in A375 melanocytes. Mol. Clin. Oncol. 2020, 12, 9–14. [Google Scholar] [CrossRef]
- Yoon, S.; Kawasaki, I.; Shim, Y.H. CDC-25.1 controls the rate of germline mitotic cell cycle by counteracting WEE-1.3 and by positively regulating CDK-1 in Caenorhabditis elegans. Cell Cycle 2012, 11, 1354–1363. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, L.; Leng, Y.; Dai, Y.; Orlowski, R.Z.; Grant, S. Positive transcription elongation factor b (P-TEFb) is a therapeutic target in human multiple myeloma. Oncotarget 2017, 8, 59476–59491. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Zhang, T.; Kwiatkowski, N.; Abraham, B.J.; Lee, T.I.; Xie, S.; Yuzugullu, H.; Thanh, V.; Li, H.; Lin, Z.; et al. CDK7-Dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell 2015, 163, 174–186. [Google Scholar] [CrossRef]
- Fukasawa, K.; Kadota, T.; Horie, T.; Tokumura, K.; Terada, R.; Kitaguchi, Y.; Park, G.; Ochiai, S.; Iwahashi, S.; Okayama, Y.; et al. CDK8 maintains stemness and tumorigenicity of glioma stem cells by regulating the c-MYC pathway. Oncogene 2021, 40, 2803–2815. [Google Scholar] [CrossRef]
- Zhong, S.; Zhang, Y.; Yin, X.; Di, W. CDK7 inhibitor suppresses tumor progression through blocking the cell cycle at the G2/M phase and inhibiting transcriptional activity in cervical cancer. Oncotargets Ther. 2019, 12, 2137–2147. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, F.; Shi, X.; Liu, X.; Yang, H.; Zhang, Z. Identification and Characterization of the Cyclin-Dependent Kinases Gene Family in Silkworm, Bombyx mori. DNA Cell Biol. 2016, 35, 13–23. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004, 18, 2699–2711. [Google Scholar] [CrossRef]
- Tetsu, O.; McCormick, F. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell 2003, 3, 233–245. [Google Scholar] [CrossRef]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef]
- Mitra, J.; Enders, G.H.; Azizkhan-Clifford, J.; Lengel, K.L. Dual regulation of the anaphase promoting complex in human cells by cyclin A-Cdk2 and cyclin A-Cdk1 complexes. Cell Cycle 2006, 5, 661–666. [Google Scholar] [CrossRef][Green Version]
- Jiang, B.; Wang, E.S.; Donovan, K.A.; Liang, Y.; Fischer, E.S.; Zhang, T.; Gray, N.S. Development of Dual and Selective Degraders of Cyclin-Dependent Kinases 4 and 6. Angew. Chem.-Int. Ed. 2019, 58, 6321–6326. [Google Scholar] [CrossRef]
- Asghar, U.; Witkiewicz, A.K.; Turner, N.C.; Knudsen, E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015, 14, 130–146. [Google Scholar] [CrossRef]
- Eggersmann, T.K.; Degenhardt, T.; Gluz, O.; Wuerstlein, R.; Harbeck, N. CDK4/6 Inhibitors Expand the Therapeutic Options in Breast Cancer: Palbociclib, Ribociclib and Abemaciclib. Biodrugs 2019, 33, 125–135. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol. Res. 2022, 175, 106037. [Google Scholar] [CrossRef] [PubMed]
- Fry, D.W.; Harvey, P.J.; Keller, P.R.; Elliott, W.L.; Meade, M.A.; Trachet, E.; Albassam, M.; Zheng, X.X.; Leopold, W.R.; Pryer, N.K.; et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004, 3, 1427–1437. [Google Scholar] [PubMed]
- Gelbert, L.M.; Cai, S.; Lin, X.; Sanchez-Martinez, C.; del Prado, M.; Jose Lallena, M.; Torres, R.; Ajamie, R.T.; Wishart, G.N.; Flack, R.S.; et al. Preclinical characterization of the CDK4/6 inhibitor LY2835219: In-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Investig. New Drugs 2014, 32, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Dickler, M.N.; Tolaney, S.M.; Rugo, H.S.; Cortes, J.; Dieras, V.; Patt, D.; Wildiers, H.; Hudis, C.A.; O’Shaughnessy, J.; Zamora, E.; et al. MONARCH 1, A Phase II Study of Abemaciclib, a CDK4 and CDK6 Inhibitor, as a Single Agent, in Patients with Refractory HR+/HER2(-) Metastatic Breast Cancer. Clin. Cancer Res. 2017, 23, 5218–5224. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, D.; Bardia, A.; Sellers, W.R. Ribociclib (LEE011): Mechanism of Action and Clinical Impact of This Selective Cyclin-Dependent Kinase 4/6 Inhibitor in Various Solid Tumors. Clin. Cancer Res. 2017, 23, 3251–3262. [Google Scholar] [CrossRef]
- Corona, S.P.; Generali, D. Abemaciclib: A CDK4/6 inhibitor for the treatment of HR+/HER2−advanced breast cancer. Drug Des. Dev. Ther. 2018, 12, 321–330. [Google Scholar] [CrossRef]
- Hamilton, E.; Infante, J.R. Targeting CDK4/6 in patients with cancer. Cancer Treat. Rev. 2016, 45, 129–138. [Google Scholar] [CrossRef]
- Adon, T.; Shanmugarajan, D.; Kumar, H.Y. CDK4/6 inhibitors: A brief overview and prospective research directions. Rsc. Adv. 2021, 11, 29227–29246. [Google Scholar] [CrossRef]
- Hafner, M.; Mills, C.E.; Subramanian, K.; Chen, C.; Chung, M.; Boswell, S.A.; Everley, R.A.; Liu, C.; Walmsley, C.S.; Juric, D.; et al. Multiomics Profiling Establishes the Polypharmacology of FDA-Approved CDK4/6 Inhibitors and the Potential for Differential Clinical Activity. Cell Chem. Biol. 2019, 26, 1067–1080. [Google Scholar] [CrossRef]
- Lin, S.; Zhao, X.; Li, T.; Zhang, H.; Tan, H.; Wang, X.; Jiang, L.; Liu, Y.; Sun, J.; Li, L.; et al. FCN-437: A novel, potent and selective oral inhibitor of CDK4/6 for the treatment of solid tumors. Cancer Res. 2019, 79, 4425. [Google Scholar]
- Xu, B.; Wang, J.; Liu, X.; Fang, J.; Yao, Z.; Wang, J. A phase I study of XZP-3287, a novel oral CDK4/6 Inhibitor, administered on a continuous dosing schedule, in patients with advanced solid tumours. Ann. Oncol. 2019, 30, 123. [Google Scholar] [CrossRef]
- Bisi, J.E.; Sorrentino, J.A.; Jordan, J.L.; Darr, D.D.; Roberts, P.J.; Tavares, F.X.; Strum, J.C. Preclinical development of G1T38: A novel, potent and selective inhibitor of cyclin dependent kinases 4/6 for use as an oral antineoplastic in patients with CDK4/6 sensitive tumors. Oncotarget 2017, 8, 42343–42358. [Google Scholar] [CrossRef] [PubMed]
- Freeman-Cook, K.D.; Hoffman, R.L.; Behenna, D.C.; Boras, B.; Carelli, J.; Diehl, W.; Ferre, R.A.; He, Y.-A.; Hui, A.; Huang, B.; et al. Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer. J. Med. Chem. 2021, 64, 9056–9077. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Wang, X.; Dong, H.; Min, W.; Hao, H.; Yang, P. Selective inhibition of CDK4/6: A safe and effective strategy for developing anticancer drugs. Acta Pharm. Sin. B 2021, 11, 30–54. [Google Scholar] [CrossRef]
- Sobhani, N.; D’Angelo, A.; Pittacolo, M.; Roviello, G.; Miccoli, A.; Corona, S.P.; Bernocchi, O.; Generali, D.; Otto, T. Updates on the CDK4/6 Inhibitory Strategy and Combinations in Breast Cancer. Cells 2019, 8, 321. [Google Scholar] [CrossRef]
- O’Leary, B.; Finn, R.S.; Turner, N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016, 13, 417–430. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Rader, J.; Russell, M.R.; Hart, L.S.; Nakazawa, M.S.; Belcastro, L.T.; Martinez, D.; Li, Y.; Carpenter, E.L.; Attiyeh, E.F.; Diskin, S.J.; et al. Dual CDK4/CDK6 Inhibition Induces Cell-Cycle Arrest and Senescence in Neuroblastoma. Clin. Cancer Res. 2013, 19, 6173–6182. [Google Scholar] [CrossRef]
- Bonelli, M.A.; Digiacomo, G.; Fumarola, C.; Alfieri, R.; Quainia, F.; Falco, A.; Madeddu, D.; La Monica, S.; Cretella, D.; Ravelli, A.; et al. Combined Inhibition of CDK4/6 and PI3K/AKT/mTOR Pathways Induces a Synergistic Anti-Tumor Effect in Malignant Pleural Mesothelioma Cells. Neoplasia 2017, 19, 637–648. [Google Scholar] [CrossRef]
- Bollard, J.; Miguela, V.; de Galarreta, M.R.; Venkatesh, A.; Bian, C.B.; Roberto, M.P.; Tovar, V.; Sia, D.; Molina-Sanchez, P.; Nguyen, C.B.; et al. Palbociclib (PD-0332991), a selective CDK4/6 inhibitor, restricts tumour growth in preclinical models of hepatocellular carcinoma. Gut 2017, 66, 1286–1296. [Google Scholar] [CrossRef]
- Yoshida, A.; Lee, E.K.; Diehl, J.A. Induction of Therapeutic Senescence in Vemurafenib-Resistant Melanoma by Extended Inhibition of CDK4/6. Cancer Res. 2016, 76, 2990–3002. [Google Scholar] [CrossRef]
- Vijayaraghavan, S.; Karakas, C.; Doostan, I.; Chen, X.; Bui, T.; Yi, M.; Raghavendra, A.S.; Zhao, Y.; Bashour, S.I.; Ibrahim, N.K.; et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat. Commun. 2017, 8, 15916. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.; Gil, J. Senescence as a therapeutically relevant response to CDK4/6 inhibitors. Oncogene 2020, 39, 5165–5176. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva, M.; Liu, D.D.; Dickson, M.A.; Klein, M.E.; O’Connor, R.; Wilder, F.O.; Socci, N.D.; Tap, W.D.; Schwartz, G.K.; Singer, S.; et al. MDM2 turnover and expression of ATRX determine the choice between quiescence and senescence in response to CDK4 inhibition. Oncotarget 2015, 6, 8226–8243. [Google Scholar] [CrossRef] [PubMed]
- Leontieva, O.V.; Natarajan, V.; Demidenko, Z.N.; Burdelya, L.G.; Gudkov, A.V.; Blagosklonny, M.V. Hypoxia suppresses conversion from proliferative arrest to cellular senescence. Proc. Natl. Acad. Sci. USA 2012, 109, 13314–13318. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.; Micevic, G.; Meeth, K.; Muthusamy, V.; Curley, D.P.; Santhanakrishnan, M.; Erdelyi, I.; Platt, J.T.; Huang, L.; Theodosakis, N.; et al. mTORC1 Activation Blocks Braf(V600E)-Induced Growth Arrest but Is Insufficient for Melanoma Formation. Cancer Cell 2015, 27, 41–56. [Google Scholar] [CrossRef]
- Miettinen, T.P.; Peltier, J.; Hartlova, A.; Gierlinski, M.; Jansen, V.M.; Trost, M.; Bjorklund, M. Thermal proteome profiling of breast cancer cells reveals proteasomal activation by CDK4/6 inhibitor palbociclib. Embo. J. 2018, 37, e98359. [Google Scholar] [CrossRef]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Kang, T.-W.; Yevsa, T.; Woller, N.; Hoenicke, L.; Wuestefeld, T.; Dauch, D.; Hohmeyer, A.; Gereke, M.; Rudalska, R.; Potapova, A.; et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 2011, 479, 547–551. [Google Scholar] [CrossRef]
- Iannello, A.; Thompson, T.W.; Ardolino, M.; Lowe, S.W.; Raulet, D.H. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J. Exp. Med. 2013, 210, 2057–2069. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wang, E.S.; Jenkins, R.W.; Li, S.; Dries, R.; Yates, K.; Chhabra, S.; Huang, W.; Liu, H.; Aref, A.R.; et al. CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov. 2018, 8, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Hara, E. Roles and mechanisms of cellular senescence in regulation of tissue homeostasis. Cancer Sci. 2013, 104, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Coppe, J.-P.; Kauser, K.; Campisi, J.; Beausejour, C.M. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J. Biol. Chem. 2006, 281, 29568–29574. [Google Scholar] [CrossRef]
- Krtolica, A.; Parrinello, S.; Lockett, S.; Desprez, P.Y.; Campisi, J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc. Natl. Acad. Sci. USA 2001, 98, 12072–12077. [Google Scholar] [CrossRef]
- Ruhland, M.K.; Loza, A.J.; Capietto, A.-H.; Luo, X.; Knolhoff, B.L.; Flanagan, K.C.; Belt, B.A.; Alspach, E.; Leahy, K.; Luo, J.; et al. Stromal senescence establishes an immunosuppressive microenvironment that drives tumorigenesis. Nat. Commun. 2016, 7, 11762. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Dabritz, J.H.M.; Zhao, Z.; Yu, Y.; Doerr, J.R.; Dimitrova, L.; Lenze, D.; Barbosa, I.A.M.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of p16(Ink4a)-positive senescent cells delays ageing-associated disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- McHugh, D.; Gil, J. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 2018, 217, 65–77. [Google Scholar] [CrossRef]
- Wang, H.; Nicolay, B.N.; Chick, J.M.; Gao, X.; Geng, Y.; Ren, H.; Gao, H.; Yang, G.; Williams, J.A.; Suski, J.M.; et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 2017, 546, 426–430. [Google Scholar] [CrossRef]
- Li, J.; Xu, M.; Yang, Z.; Li, A.; Dong, J. Simultaneous Inhibition of MEK and CDK4 Leads to Potent Apoptosis in Human Melanoma Cells. Cancer Investig. 2010, 28, 350–356. [Google Scholar] [CrossRef]
- Franco, J.; Balaji, U.; Freinkman, E.; Witkiewicz, A.K.; Knudsen, E.S. Metabolic Reprogramming of Pancreatic Cancer Mediated by CDK4/6 Inhibition Elicits Unique Vulnerabilities. Cell Rep. 2016, 14, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Vilgelm, A.E.; Saleh, N.; Shattuck-Brandt, R.; Riemenschneider, K.; Slesur, L.; Chen, S.-C.; Johnson, C.A.; Yang, J.; Blevins, A.; Yan, C.; et al. MDM2 antagonists overcome intrinsic resistance to CDK4/6 inhibition by inducing p21. Sci. Transl. Med. 2019, 11, eaav7171. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Bhatt, T.; Dickler, M.; Giri, D.; Scaltriti, M.; Baselga, J.; Rosen, N.; Chandarlapaty, S. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene 2017, 36, 2255–2264. [Google Scholar] [CrossRef]
- Cen, L.; Carlson, B.L.; Schroeder, M.A.; Ostrem, J.L.; Kitange, G.J.; Mladek, A.C.; Fink, S.R.; Decker, P.A.; Wu, W.; Kim, J.-S.; et al. p16-Cdk4-Rb axis controls sensitivity to a cyclin-dependent kinase inhibitor PD0332991 in glioblastoma xenograft cells. Neuro-Oncology 2012, 14, 870–881. [Google Scholar] [CrossRef]
- Sammons, S.L.; Topping, D.L.; Blackwell, K.L. HR+, HER2−Advanced Breast Cancer and CDK4/6 Inhibitors: Mode of Action, Clinical Activity, and Safety Profiles. Curr. Cancer Drug Targets 2017, 17, 637–649. [Google Scholar] [CrossRef]
- Palleschi, M.; Maltoni, R.; Ravaioli, S.; Vagheggini, A.; Mannozzi, F.; Fanini, F.; Pirini, F.; Tumedei, M.M.; Barzotti, E.; Cecconetto, L.; et al. Ki67 and PR in Patients Treated with CDK4/6 Inhibitors: A Real-World Experience. Diagnostics 2020, 10, 573. [Google Scholar] [CrossRef]
- Dean, J.L.; Thangavel, C.; McClendon, A.K.; Reed, C.A.; Knudsen, E.S. Therapeutic CDK4/6 inhibition in breast cancer: Key mechanisms of response and failure. Oncogene 2010, 29, 4018–4032. [Google Scholar] [CrossRef]
- Sun, B.; Mason, S.; Wilson, R.C.; Hazard, S.E.; Wang, Y.; Fang, R.; Wang, Q.; Yeh, E.S.; Yang, M.; Roberts, T.M.; et al. Inhibition of the transcriptional kinase CDK7 overcomes therapeutic resistance in HER2-positive breast cancers. Oncogene 2020, 39, 50–63. [Google Scholar] [CrossRef]
- Herrera-Abreu, M.T.; Palafox, M.; Asghar, U.; Rivas, M.A.; Cutts, R.J.; Garcia-Murillas, I.; Pearson, A.; Guzman, M.; Rodriguez, O.; Grueso, J.; et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016, 76, 2301–2313. [Google Scholar] [CrossRef]
- Li, Z.; Razavi, P.; Li, Q.; Toy, W.; Liu, B.; Ping, C.; Hsieh, W.; Sanchez-Vega, F.; Brown, D.N.; Paula, A.F.D.C.; et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 2018, 34, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, N.; Fassl, A.; Mondani, G.; Generali, D.; Otto, T. Targeting Aberrant FGFR Signaling to Overcome CDK4/6 Inhibitor Resistance in Breast Cancer. Cells 2021, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, N.A.; McDermott, M.S.J.; Conklin, D.; Luo, T.; Ayala, R.; Salgar, S.; Chau, K.; DiTomaso, E.; Babbar, N.; Su, F.; et al. Targeting activated PI3K/mTOR signaling overcomes acquired resistance to CDK4/6-based therapies in preclinical models of hormone receptor-positive breast cancer. Breast Cancer Res. 2020, 22, 89. [Google Scholar] [CrossRef] [PubMed]
- McCartney, A.; Migliaccio, I.; Bonechi, M.; Biagioni, C.; Romagnoli, D.; De Luca, F.; Galardi, F.; Risi, E.; De Santo, I.; Benelli, M.; et al. Mechanisms of Resistance to CDK4/6 Inhibitors: Potential Implications and Biomarkers for Clinical Practice. Front. Oncol. 2019, 9, 666. [Google Scholar] [CrossRef]
- Olmez, I.; Zhang, Y.; Manigat, L.; Benamar, M.; Brenneman, B.; Nakano, I.; Godlewski, J.; Bronisz, A.; Lee, J.; Abbas, T.; et al. Combined c-Met/Trk Inhibition Overcomes Resistance to CDK4/6 Inhibitors in Glioblastoma. Cancer Res. 2018, 78, 4360–4369. [Google Scholar] [CrossRef]
- Wander, S.A.; Cohent, O.; Gong, X.; Johnson, G.N.; Buendia-Buendia, J.E.; Lloyd, M.R.; Kim, D.; Luo, F.; Mao, P.; Helvie, K.; et al. The Genomic Landscape of Intrinsic and Acquired Resistance to Cyclin-Dependent Kinase 4/6 Inhibitors in Patients with Hormone Receptor-Positive Metastatic Breast Cancer. Cancer Discov. 2020, 10, 1174–1193. [Google Scholar] [CrossRef]
- Patnaik, A.; Rosen, L.S.; Tolaney, S.M.; Tolcher, A.W.; Goldman, J.W.; Gandhi, L.; Papadopoulos, K.P.; Beeram, M.; Rasco, D.W.; Hilton, J.F.; et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov. 2016, 6, 740–753. [Google Scholar] [CrossRef]
- Gong, X.; Litchfield, L.M.; Webster, Y.; Chio, L.-C.; Wong, S.S.; Stewart, T.R.; Dowless, M.; Dempsey, J.; Zeng, Y.; Torres, R.; et al. Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib. Cancer Cell 2017, 32, 761–776. [Google Scholar] [CrossRef]
- Kettner, N.M.; Vijayaraghavan, S.; Durak, M.G.; Bui, T.; Kohansal, M.; Ha, M.J.; Liu, B.; Rao, X.; Wang, J.; Yi, M.; et al. Combined Inhibition of STAT3 and DNA Repair in Palbociclib-Resistant ER-Positive Breast Cancer. Clin. Cancer Res. 2019, 25, 3996–4013. [Google Scholar] [CrossRef]
- Turner, N.C.; Liu, Y.; Zhu, Z.; Loi, S.; Colleoni, M.; Loibl, S.; DeMichele, A.; Harbeck, N.; Andre, F.; Bayar, M.A.; et al. Cyclin E1 Expression and Palbociclib Efficacy in Previously Treated Hormone Receptor-Positive Metastatic Breast Cancer. J. Clin. Oncol. 2019, 37, 1169–1178. [Google Scholar] [CrossRef]
- Kollmann, K.; Heller, G.; Schneckenleithner, C.; Warsch, W.; Scheicher, R.; Ott, R.G.; Schaefer, M.; Fajmann, S.; Schlederer, M.; Schiefer, A.-I.; et al. A Kinase-Independent Function of CDK6 Links the Cell Cycle to Tumor Angiogenesis. Cancer Cell 2013, 24, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Tigan, A.S.; Bellutti, F.; Kollmann, K.; Tebb, G.; Sexl, V. CDK6-a review of the past and a glimpse into the future: From cell-cycle control to transcriptional regulation. Oncogene 2016, 35, 3083–3091. [Google Scholar] [CrossRef] [PubMed]
- Kollmann, K.; Sexl, V. CDK6 and p16(INK4A) in lymphoid malignancies. Oncotarget 2013, 4, 1858–1859. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Zhao, X.; Zhao, L.; Yan, H.; Li, J.; Yang, H.; Huang, G.; Liu, J. Non-canonical roles of PFKFB3 in regulation of cell cycle through binding to CDK4. Oncogene 2018, 37, 1685–1698. [Google Scholar] [CrossRef] [PubMed]
- Scheicher, R.; Hoelbl-Kovacic, A.; Bellutti, F.; Tigan, A.-S.; Prchal-Murphy, M.; Heller, G.; Schneckenleithner, C.; Salazar-Roa, M.; Zoechbauer-Mueller, S.; Zuber, J.; et al. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood 2015, 125, 90–101. [Google Scholar] [CrossRef]
- Fassl, A.; Brain, C.; Abu-Remaileh, M.; Stukan, I.; Butter, D.; Stepien, P.; Feit, A.S.; Bergholz, J.; Michowski, W.; Otto, T.; et al. Increased lysosomal biomass is responsible for the resistance of triple-negative breast cancers to CDK4/6 inhibition. Sci. Adv. 2020, 6, eabb2210. [Google Scholar] [CrossRef]
- Foidart, P.; Yip, C.; Radermacher, J.; Blacher, S.; Lienard, M.; Montero-Ruiz, L.; Maquoi, E.; Montaudon, E.; Chateau-Joubert, S.; Collignon, J.; et al. Expression of MT4-MMP, EGFR, and RB in Triple-Negative Breast Cancer Strongly Sensitizes Tumors to Erlotinib and Palbociclib Combination Therapy. Clin. Cancer Res. 2019, 25, 1838–1850. [Google Scholar] [CrossRef]
- Vora, S.R.; Juric, D.; Kim, N.; Mino-Kenudson, M.; Huynh, T.; Costa, C.; Lockerman, E.L.; Pollack, S.F.; Liu, M.; Li, X.; et al. CDK 4/6 Inhibitors Sensitize PIK3CA Mutant Breast Cancer to PI3K Inhibitors. Cancer Cell 2014, 26, 136–149. [Google Scholar] [CrossRef]
- Formisano, L.; Lu, Y.; Servetto, A.; Hanker, A.B.; Jansen, V.M.; Bauer, J.A.; Sudhan, D.R.; Guerrero-Zotano, A.L.; Croessmann, S.; Guo, Y.; et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER plus breast cancer. Nat. Commun. 2019, 10, 1373. [Google Scholar] [CrossRef]
- Heilmann, A.M.; Perera, R.M.; Ecker, V.; Nicolay, B.N.; Bardeesy, N.; Benes, C.H.; Dyson, N.J. CDK4/6 and IGF1 Receptor Inhibitors Synergize to Suppress the Growth of p16(INK4A)-Deficient Pancreatic Cancers. Cancer Res. 2014, 74, 3947–3958. [Google Scholar] [CrossRef]
- Teh, J.L.F.; Cheng, P.F.; Purwin, T.J.; Nikbakht, N.; Patel, P.; Chervoneva, I.; Ertel, A.; Fortina, P.M.; Kleiber, I.; HooKim, K.; et al. In Vivo E2F Reporting Reveals Efficacious Schedules of MEK1/2-CDK4/6 Targeting and mTOR-S6 Resistance Mechanisms. Cancer Discov. 2018, 8, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; Wang, Q.; Watt, A.C.; Tolaney, S.M.; Dillon, D.A.; Li, W.; Ramm, S.; Palmer, A.C.; Yuzugullu, H.; Varadan, V.; et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell 2016, 29, 255–269. [Google Scholar] [CrossRef] [PubMed]
- AbuHammad, S.; Cullinane, C.; Martin, C.; Bacolas, Z.; Ward, T.; Chen, H.; Slater, A.; Ardley, K.; Kirby, L.; Chan, K.T.; et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl. Acad. Sci. USA 2019, 116, 17990–18000. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.; Xue, Y.; Cencic, R.; Zhu, X.; Monast, A.; Fu, Z.; Pilon, V.; Sangwan, V.; Guiot, M.-C.; Foulkes, W.D.; et al. eIF4A Inhibitors Suppress Cell-Cycle Feedback Response and Acquired Resistance to CDK4/6 Inhibition in Cancer. Mol. Cancer Ther. 2019, 18, 2158–2170. [Google Scholar] [CrossRef]
- Patel, P.; Tsiperson, V.; Gottesman, S.R.S.; Somma, J.; Blain, S.W. Dual Inhibition of CDK4 and CDK2 via Targeting p27 Tyrosine Phosphorylation Induces a Potent and Durable Response in Breast Cancer Cells. Mol. Cancer Res. 2018, 16, 361–377. [Google Scholar] [CrossRef]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Blackwell, K.L.; Andre, F.; Winer, E.P.; et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1738–1748. [Google Scholar] [CrossRef]
- Finn, R.S.; Crown, J.P.; Lang, I.; Boer, K.; Bondarenko, I.M.; Kulyk, S.O.; Ettl, J.; Patel, R.; Pinter, T.; Schmidt, M.; et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study. Lancet Oncol. 2015, 16, 25–35. [Google Scholar] [CrossRef]
- Sledge, G.W., Jr.; Toi, M.; Neven, P.; Sohn, J.; Inoue, K.; Pivot, X.; Burdaeva, O.; Okera, M.; Masuda, N.; Kaufman, P.A.; et al. MONARCH 2: Abemaciclib in Combination With Fulvestrant in Women With HR+/HER2−-Advanced Breast Cancer Who Had Progressed While Receiving Endocrine Therapy. J. Clin. Oncol. 2017, 35, 2875–2884. [Google Scholar] [CrossRef]
- Zhang, J.; Bu, X.; Wang, H.; Zhu, Y.; Geng, Y.; Nihira, N.T.; Tan, Y.; Ci, Y.; Wu, F.; Dai, X.; et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018, 553, 91–95. [Google Scholar] [CrossRef]
- Schaer, D.A.; Beckmann, R.P.; Dempsey, J.A.; Huber, L.; Forest, A.; Amaladas, N.; Li, Y.; Wang, Y.C.; Rasmussen, E.R.; Chin, D.; et al. The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep. 2018, 22, 2978–2994. [Google Scholar] [CrossRef]
- Wang, C.-L.; Chiang, I.T.; Hsu, F.-T.; Chang, Y. Palbociclib enhances anti-PD-L1 therapeutic efficacy in oral squamous cell carcinoma was associated with CXCR4 inactivation. Cancer Res. 2021, 81, 1592. [Google Scholar]
- Tolaney, S.M.; Kabos, P.; Dickler, M.N.; Gianni, L.; Jansen, V.; Lu, Y.; Young, S.; Rugo, H.S. Updated efficacy, safety, & PD-L1 status of patients with HR+, HER2−metastatic breast cancer administered abemaciclib plus pembrolizumab. J. Clin. Oncol. 2018, 36, 1059. [Google Scholar] [CrossRef]
- Pujol, J.-L.; Vansteenkiste, J.; Paz-Ares Rodriguez, L.; Gregorc, V.; Mazieres, J.; Awad, M.; Janne, P.A.; Chisamore, M.; Hossain, A.M.; Chen, Y.; et al. Abemaciclib in Combination With Pembrolizumab for Stage IV KRAS-Mutant or Squamous NSCLC: A Phase 1b Study. JTO Clin. Res. Rep. 2021, 2, 100234. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.L.; McClendon, A.K.; Hickey, T.E.; Butler, L.M.; Tilley, W.D.; Witkiewicz, A.K.; Knudsen, E.S. Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle 2012, 11, 2756–2761. [Google Scholar] [CrossRef]
- McClendon, A.K.; Dean, J.L.; Rivadeneira, D.B.; Yu, J.E.; Reed, C.A.; Gao, E.; Farber, J.L.; Force, T.; Koch, W.J.; Knudsen, E.S. CDK4/6 inhibition antagonizes the cytotoxic response to anthracycline therapy. Cell Cycle 2012, 11, 2747–2755. [Google Scholar] [CrossRef]
- Pikman, Y.; Alexe, G.; Roti, G.; Conway, A.S.; Furman, A.; Lee, E.S.; Place, A.E.; Kim, S.; Saran, C.; Modiste, R.; et al. Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2017, 23, 1012–1024. [Google Scholar] [CrossRef]
- He, S.; Roberts, P.J.; Sorrentino, J.A.; Bisi, J.E.; Storrie-White, H.; Tiessen, R.G.; Makhuli, K.M.; Wargin, W.A.; Tadema, H.; van Hoogdalem, E.-J.; et al. Transient CDK4/6 inhibition protects hematopoietic stem cells from chemotherapy-induced exhaustion. Sci. Transl. Med. 2017, 9, eaal3986. [Google Scholar] [CrossRef]
- Purba, T.S.; Ng’andu, K.; Brunken, L.; Smart, E.; Mitchell, E.; Hassan, N.; O’Brien, A.; Mellor, C.; Jackson, J.; Shahmalak, A.; et al. CDK4/6 inhibition mitigates stem cell damage in a novel model for taxane-induced alopecia. EMBO Mol. Med. 2019, 11, e11031. [Google Scholar] [CrossRef]
- Weiss, J.M.; Csoszi, T.; Maglakelidze, M.; Hoyer, R.J.; Beck, J.T.; Domine Gomez, M.; Lowczak, A.; Aljumaily, R.; Lima, C.M.R.; Boccia, R.V.; et al. Myelopreservation with the CDK4/6 inhibitor trilaciclib in patients with small-cell lung cancer receiving first-line chemotherapy: A phase Ib/randomized phase II trial. Ann. Oncol. 2019, 30, 1613–1621. [Google Scholar] [CrossRef]
- Johnson, S.M.; Torrice, C.D.; Bell, J.F.; Monahan, K.B.; Jiang, Q.; Wang, Y.; Ramsey, M.R.; Jin, J.; Wong, K.-K.; Su, L.; et al. Mitigation of hematologic radiation toxicity in mice through pharmacological quiescence induced by CDK4/6 inhibition. J. Clin. Investig. 2010, 120, 2528–2536. [Google Scholar] [CrossRef]
- Wei, L.; Leibowitz, B.J.; Wang, X.; Epperly, M.; Greenberger, J.; Zhang, L.; Yu, J. Inhibition of CDK4/6 protects against radiation-induced intestinal injury in mice. J. Clin. Investig. 2016, 126, 4076–4087. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, R.; Zhang, A.; Mueller, S.; Prados, M.D.; Lulla, R.R.; Goldman, S.; Saratsis, A.M.; Mazar, A.P.; Stegh, A.H.; Cheng, S.-Y.; et al. Inhibition of DNA damage repair by the CDK4/6 inhibitor palbociclib delays irradiated intracranial atypical teratoid rhabdoid tumor and glioblastoma xenograft regrowth. Neuro-Oncology 2016, 18, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; Sowers, A.; Choudhuri, R.; Wissler, M.; Gamson, J.; Mathias, A.; Cook, J.A.; Mitchell, J.B. Abemaciclib, a Selective CDK4/6 Inhibitor, Enhances the Radiosensitivity of Non-Small Cell Lung Cancer In Vitro and In Vivo. Clin. Cancer Res. 2018, 24, 3994–4005. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Barbero, B.; Alvarez-Fernandez, M.; Zapatero-Solana, E.; El Bakkali, A.; del Camino Menendez, M.; Lopez-Casas, P.P.; Di Domenico, T.; Xie, T.; VanArsdale, T.; Shields, D.J.; et al. CDK4/6 Inhibitors Impair Recovery from Cytotoxic Chemotherapy in Pancreatic Adenocarcinoma. Cancer Cell 2020, 37, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.V.R.; Akula, B.; Cosenza, S.C.; Athuluridivakar, S.; Mallireddigari, M.R.; Pallela, V.R.; Billa, V.K.; Subbaiah, D.R.C.V.; Bharathi, E.V.; Vasquez-Del Carpio, R.; et al. Discovery of 8-Cyclopentyl-2- 4-(4-methyl-piperazin-1-yl)-phenylamino -7-oxo-7,8-dihy dro-pyrido 2,3-d pyrimidine-6-carbonitrile (7x) as a Potent Inhibitor of Cyclin-Dependent Kinase 4 (CDK4) and AMPK-Related Kinase 5 (ARKS). J. Med. Chem. 2014, 57, 578–599. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Borland, M.; Brain, C.; Chen, C.H.T.; Cheng, H.; Chopra, R.; Chung, K.; Groarke, J.; He, G.; Hou, Y.; et al. 4-(Pyrazol-4-yl)-pyrimidines as Selective Inhibitors of Cyclin-Dependent Kinase 4/6. J. Med. Chem. 2010, 53, 7938–7957. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Eksterowicz, J.; Gribble, M.W., Jr.; Alba, G.Q.; Ayres, M.; Carlson, T.J.; Chen, A.; Chen, X.; Cho, R.; et al. Discovery of AMG 925, a FLT3 and CDK4 Dual Kinase Inhibitor with Preferential Affinity for the Activated State of FLT3. J. Med. Chem. 2014, 57, 3430–3449. [Google Scholar] [CrossRef]
- Li, C.; Liu, L.; Liang, L.; Xia, Z.; Li, Z.; Wang, X.; McGee, L.R.; Newhall, K.; Sinclair, A.; Kamb, A.; et al. AMG 925 Is a Dual FLT3/CDK4 Inhibitor with the Potential to Overcome FLT3 Inhibitor Resistance in Acute Myeloid Leukemia. Mol. Cancer Ther. 2015, 14, 375–383. [Google Scholar] [CrossRef]
- Wang, Y.; Zhi, Y.; Jin, Q.; Lu, S.; Lin, G.; Yuan, H.; Yang, T.; Wang, Z.; Yao, C.; Ling, J.; et al. Discovery of 4-((7H-Pyrrolo 2,3-d pyrimidin-4-yl)amino)-N-(4-((4-methylpiperazin-1-yl )methyl)phenyl)-1H-pyrazole-3-carboxamide (FN-1501), an FLT3-and CDK-Kinase Inhibitor with Potentially High Efficiency against Acute Myelocytic Leukemia. J. Med. Chem. 2018, 61, 1499–1518. [Google Scholar] [CrossRef]
- Konstantinidou, M.; Li, J.; Zhang, B.; Wang, Z.; Shaabani, S.; Ter Brake, F.; Essa, K.; Domling, A. PROTACs- a game-changing technology. Expert Opin. Drug Discov. 2019, 14, 1255–1268. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Feng, F.; Liu, W.; Sun, H. Degradation of proteins by PROTACs and other strategies. Acta Pharm. Sin. B 2020, 10, 207–238. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019, 4, 64. [Google Scholar] [CrossRef]
- Sun, X.; Rao, Y. PROTACs as Potential Therapeutic Agents for Cancer Drug Resistance. Biochemistry 2020, 59, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Cai, M.; Shao, L.; Zhang, J. Targeting Protein Kinases Degradation by PROTACs. Front. Chem. 2021, 9, 679120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Burgess, K. PROTACs suppression of CDK4/6, crucial kinases for cell cycle regulation in cancer. Chem. Commun. 2019, 55, 2704–2707. [Google Scholar] [CrossRef] [PubMed]
- Puyol, M.; Martin, A.; Dubus, P.; Mulero, F.; Pizcueta, P.; Khan, G.; Guerra, C.; Santamaria, D.; Barbacid, M. A Synthetic Lethal Interaction between K-Ras Oncogenes and Cdk4 Unveils a Therapeutic Strategy for Non-small Cell Lung Carcinoma. Cancer Cell 2010, 18, 63–73. [Google Scholar] [CrossRef]
- Handschick, K.; Beuerlein, K.; Jurida, L.; Bartkuhn, M.; Mueller, H.; Soelch, J.; Weber, A.; Dittrich-Breiholz, O.; Schneider, H.; Scharfe, M.; et al. Cyclin-Dependent Kinase 6 Is a Chromatin-Bound Cofactor for NF-kappa B-Dependent Gene Expression. Mol. Cell 2014, 53, 193–208. [Google Scholar] [CrossRef]
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.-H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017, 13, 514–521. [Google Scholar] [CrossRef]
- Nowak, R.P.; DeAngelo, S.L.; Buckley, D.; He, Z.; Donovan, K.A.; An, J.; Safaee, N.; Jedrychowski, M.P.; Ponthier, C.M.; Ishoey, M.; et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 2018, 14, 706–714. [Google Scholar] [CrossRef]
- Rana, S.; Bendjennat, M.; Kour, S.; King, H.M.; Kizhake, S.; Zahid, M.; Natarajan, A. Selective degradation of CDK6 by a palbociclib based PROTAC. Bioorganic Med. Chem. Lett. 2019, 29, 1375–1379. [Google Scholar] [CrossRef]
- Brand, M.; Jiang, B.; Bauer, S.; Donovan, K.A.; Liang, Y.; Wang, E.S.; Nowak, R.P.; Yuan, J.C.; Zhang, T.; Kwiatkowski, N.; et al. Homolog-Selective Degradation as a Strategy to Probe the Function of CDK6 in AML. Cell Chem. Biol. 2019, 26, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Steinebach, C.; Ng, Y.L.D.; Sosic, I.; Lee, C.-S.; Chen, S.; Lindner, S.; Lan Phuong, V.; Bricelj, A.; Haschemi, R.; Monschke, M.; et al. Systematic exploration of different E3 ubiquitin ligases: An approach towards potent and selective CDK6 degraders. Chem. Sci. 2020, 11, 3474–3486. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.A.; Cryan, J.; Ahmed, A.; Dai, H.; McGonagle, G.A.; Rozier, C.; Benowitz, A.B. Selective CDK6 degradation mediated by cereblon, VHL, and novel IAP-recruiting PROTACs. Bioorganic Med. Chem. Lett. 2020, 30, 127106. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Yang, Z.; Gao, H.; Yang, H.; Zhu, S.; An, Z.; Wang, J.; Li, Q.; Chandarlapaty, S.; Deng, H.; et al. Potent and Preferential Degradation of CDK6 via Proteolysis Targeting Chimera Degraders. J. Med. Chem. 2019, 62, 7575–7582. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, I.; Denissova, N.G.; Wang, G.N.; He, D.M.; Long, J.Y.; Liu, F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 2004, 430, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Zhao, R.; Cao, Y.; Wei, Y.; Li, M.; Dong, Z.; Liu, Y.; Ruan, H.; Li, Y.; Cao, S.; et al. First orally bioavailable prodrug of proteolysis targeting chimera (PROTAC) degrades cyclin-dependent kinases 2/4/6 in vivo. Eur. J. Med. Chem. 2021, 209, 112903. [Google Scholar] [CrossRef]
- Johansson, M.; Persson, J.L. Cancer Therapy: Targeting Cell Cycle Regulators. Anti-Cancer Agents Med. Chem. 2008, 8, 723–731. [Google Scholar] [CrossRef]
- Heptinstall, A.B.; Adiyasa, I.W.S.; Cano, C.; Hardcastle, I.R. Recent advances in CDK inhibitors for cancer therapy. Future Med. Chem. 2018, 10, 1369–1388. [Google Scholar] [CrossRef]
- Cicenas, J.; Kalyan, K.; Sorokinas, A.; Jatulyte, A.; Valiunas, D.; Kaupinis, A.; Valius, M. Highlights of the Latest Advances in Research on CDK Inhibitors. Cancers 2014, 6, 2224–2242. [Google Scholar] [CrossRef]
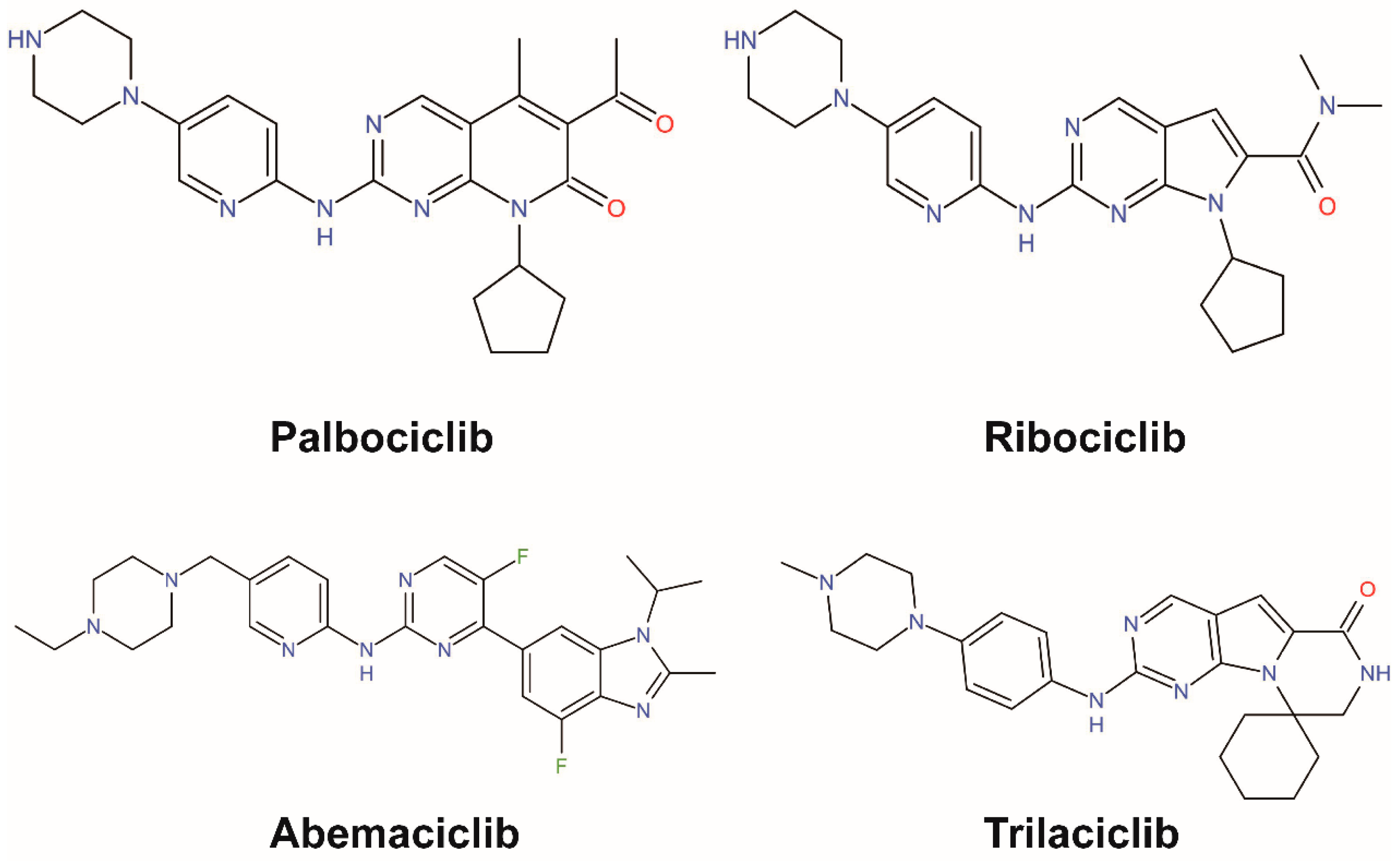

| Drug | Palbociclib (Pfizer) (PD0332991, lbrance) | Ribociclib (Novartis) (LEE011) | Abemaciclib (Eli Lilly) (LY2835219) |
|---|---|---|---|
| IC50 (in vitro kinase assay, recombinant proteins) | CDK4: 9-11 nM CDK6: 15 nM | CDK4: 10 nM CDK6: 39 nM | CDK4: 2 nM CDK6: 5 nM CDK9: 57 nM |
| Cell cycle arrest | G1 | G1 | G1, G2 |
| Off-targets | TTK, TRK, ULK2-3, PIK3R4, PIP5K2C, PIK3CD, PIP4K2A/C CAMK2δ, FLT3, PIP4K2B, HIPK1-3, MPSK1, CDK16/17, CLK1-2, CSNK2α | TTK, GAK, QIK, CAMK2δ, CAMK2α-γ | CDK1/2/5/7-9/12-17/19, CAMK2δ, CAMK2α-γ, FLT3, PIP4K2B, HIPK1-3, MPSK1, CLK1-2, CSNK2α1-2, ERK8, GSK3α/β, PIM1/3, AAK1, IRAK1, DYRK1-3, CSNK2α3 |
| CDK4/6i | Cancer Type | Combination Therapy | Trial Name |
|---|---|---|---|
| Palbociclib | Mantle cell lymphoma | ibrutinib | NCT03478514 |
| Metastatic colorectal cancer | cetuximab | NCT03446157 | |
| Multiple myeloma | bortezomib; dexamethasone | NCT00555906 | |
| Ewing sarcoma | ganitumab | NCT04129151 | |
| Advanced KRAS mutant non-small cell lung cancer | binimetinib | NCT03170206 | |
| Acute myeloid leukemia | vyxeos | NCT03844997 | |
| Squamous cell carcinoma of the head and neck | carboplatin | NCT03194373 | |
| Breast cancer | Letrozole | NCT01740427 (PALOMA-2) | |
| Breast cancer | fulvestrant | NCT01942135(PALOMA-3) | |
| Breast cancer | anastrozole | NCT01723774 | |
| Breast cancer | paclitaxel | NCT01320592 | |
| Ribociclib | Recurrent platinum sensitive ovarian cancer | paclitaxel; carboplatin | NCT03056833 |
| ALK-positive non-small cell lung cancer | ceritinib | NCT02292550 | |
| Liposarcoma | siremadlin | NCT02343172 | |
| Metastatic pancreatic adenocarcinoma | everolimus | NCT02985125 | |
| Triple-negative breast cancer | bicalutamide | NCT03090165 | |
| Squamous cell carcinoma of the head and neck | cetuximab | NCT02429089 | |
| Acute lymphoblastic leukemia | everolimus; dexamethasone | NCT03740334 | |
| Prostate cancer | enzalutamide | NCT02555189 | |
| Breast cancer | letrozole | NCT01958021 | |
| Breast cancer | fulvestrant | NCT02422615 (MONALEESA-3) | |
| Breast cancer | tamoxifen, NSAI | NCT2278120 (MONALEESA-7) | |
| Abemaciclib | Renal cell carcinoma metastatic | sunitinib | NCT03905889 |
| Recurrent glioblastoma | bevacizumab | NCT04074785 | |
| Hepatocellular carcinoma | nivolumab | NCT03781960 | |
| Non-small cell lung cancer | necitumumab | NCT02411591 | |
| Gastroesophageal cancer Adenocarcinoma | pembrolizumab | NCT03997448 | |
| Non-small cell lung cancer | erlotinib | NCT02152631 | |
| Brain metastases | GDC-0084 entrectinib | NCT03994796 | |
| Breast cancer | anastrozole | NCT02441946 (NeoMONARCH) | |
| Breast cancer | fulvestrant | NCT02107703 (MONARCH-2) | |
| Trilaciclib | Metastatic triple-negative breast cancer | gemcitabine plus carbo-platin | NCT02978716 |
| Extensive-stage small cell lung cancer | etoposide, carboplatin | NCT02499770 | |
| Extensive-stage small cell lung cancer | etoposide, carboplatin, atezolizumab | NCT03041311 | |
| Extensive-stage small cell lung cancer | topotecan | NCT02514447 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, J.; Ouyang, Z. Targeting CDK4/6 for Anticancer Therapy. Biomedicines 2022, 10, 685. https://doi.org/10.3390/biomedicines10030685
Qi J, Ouyang Z. Targeting CDK4/6 for Anticancer Therapy. Biomedicines. 2022; 10(3):685. https://doi.org/10.3390/biomedicines10030685
Chicago/Turabian StyleQi, Jiating, and Zhuqing Ouyang. 2022. "Targeting CDK4/6 for Anticancer Therapy" Biomedicines 10, no. 3: 685. https://doi.org/10.3390/biomedicines10030685
APA StyleQi, J., & Ouyang, Z. (2022). Targeting CDK4/6 for Anticancer Therapy. Biomedicines, 10(3), 685. https://doi.org/10.3390/biomedicines10030685






