Macrophage Infiltration Correlates with Genomic Instability in Classic Hodgkin Lymphoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tissue Microarray
2.3. Immunohistochemical Staining
2.4. Fluorescent In Situ Hybridization
2.5. In Situ Hybridization
2.6. Statistical Analysis
3. Results
3.1. Patients’ Clinical Data
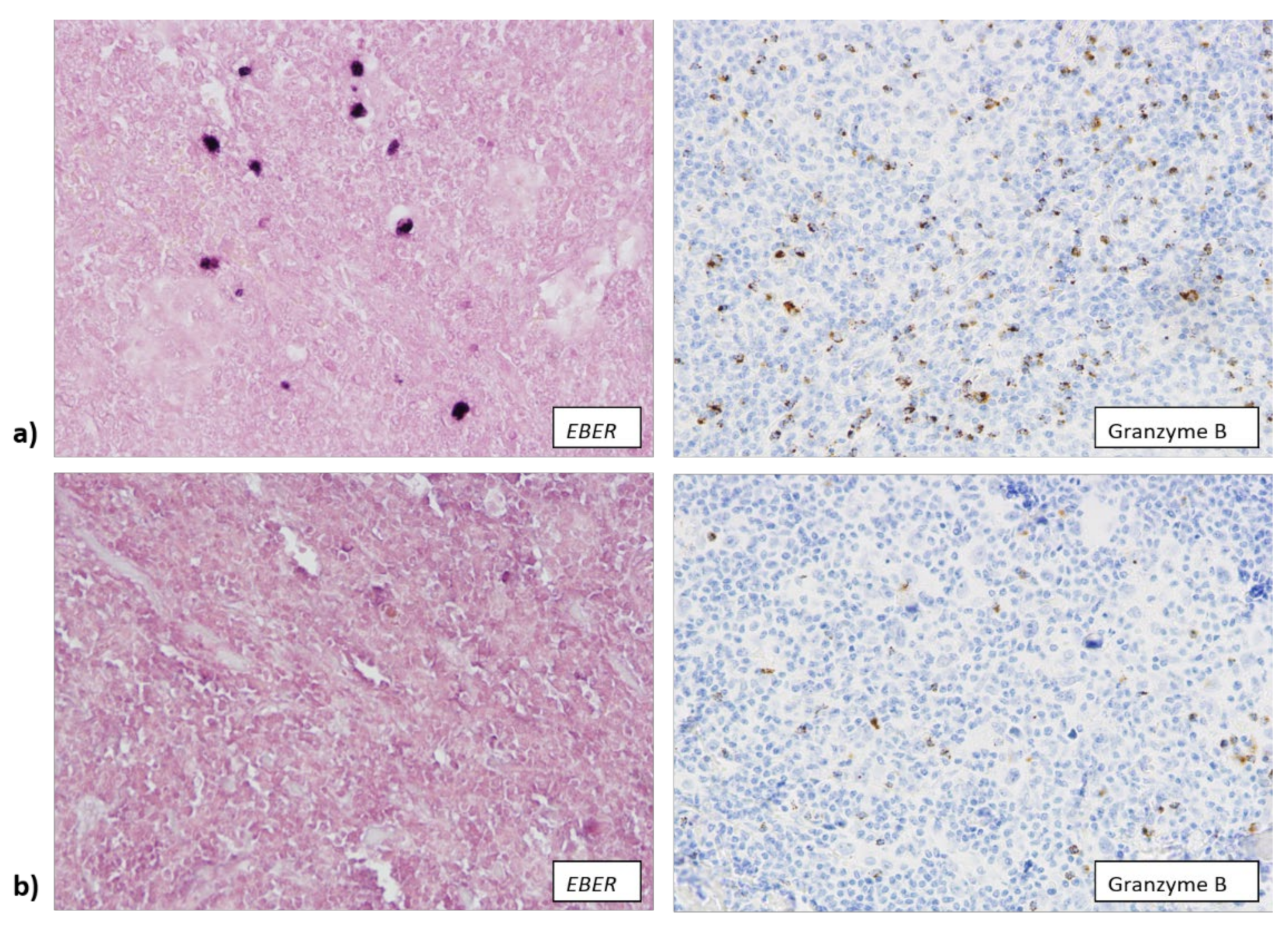
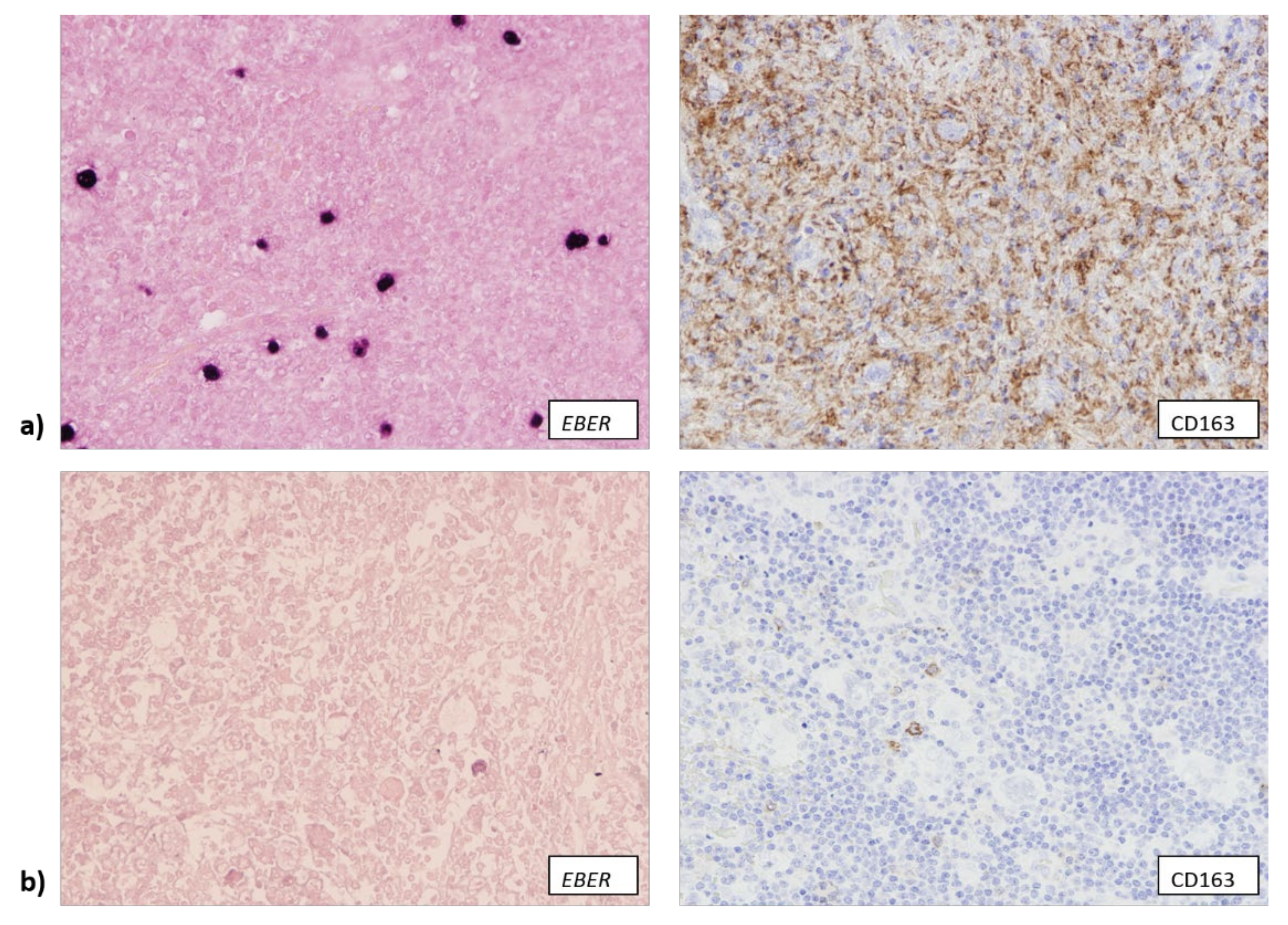
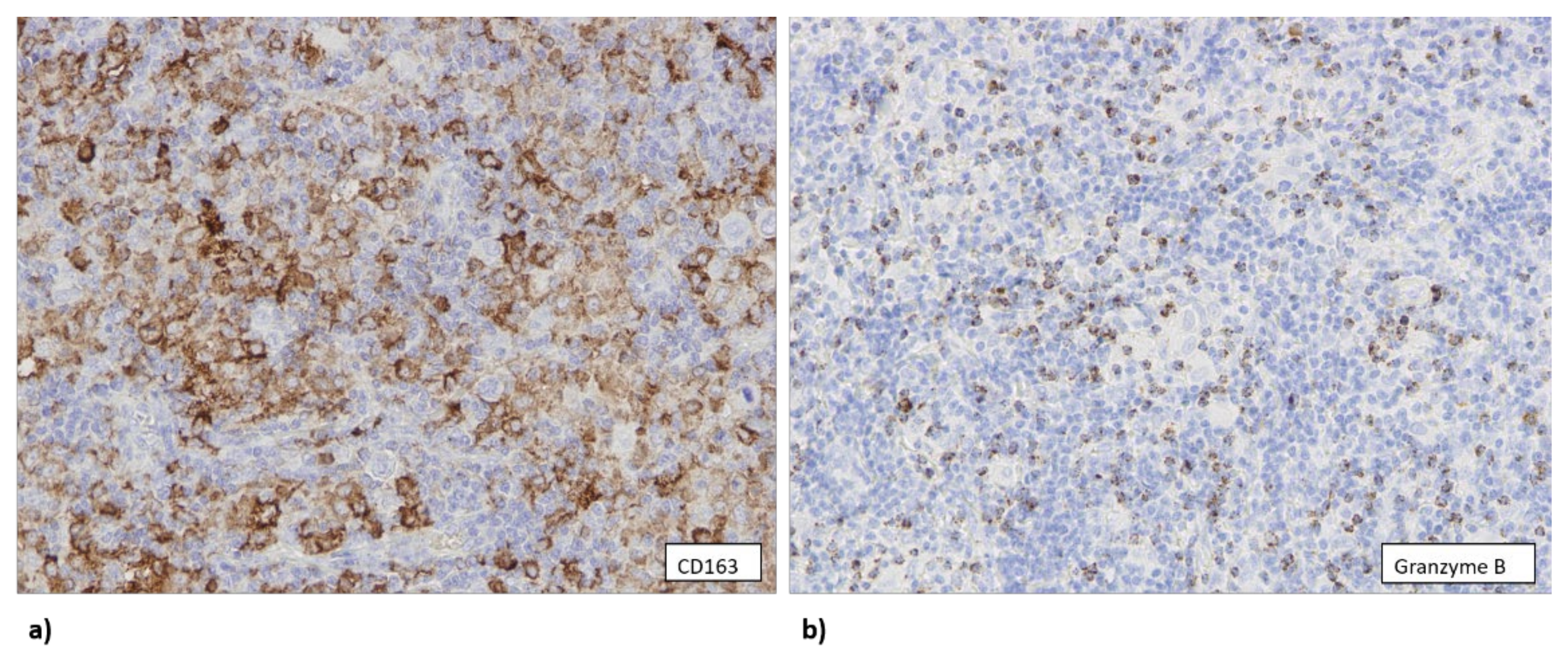
3.2. EBV Infection Attracts Cytotoxic T Lymphocytes and Macrophages to TME
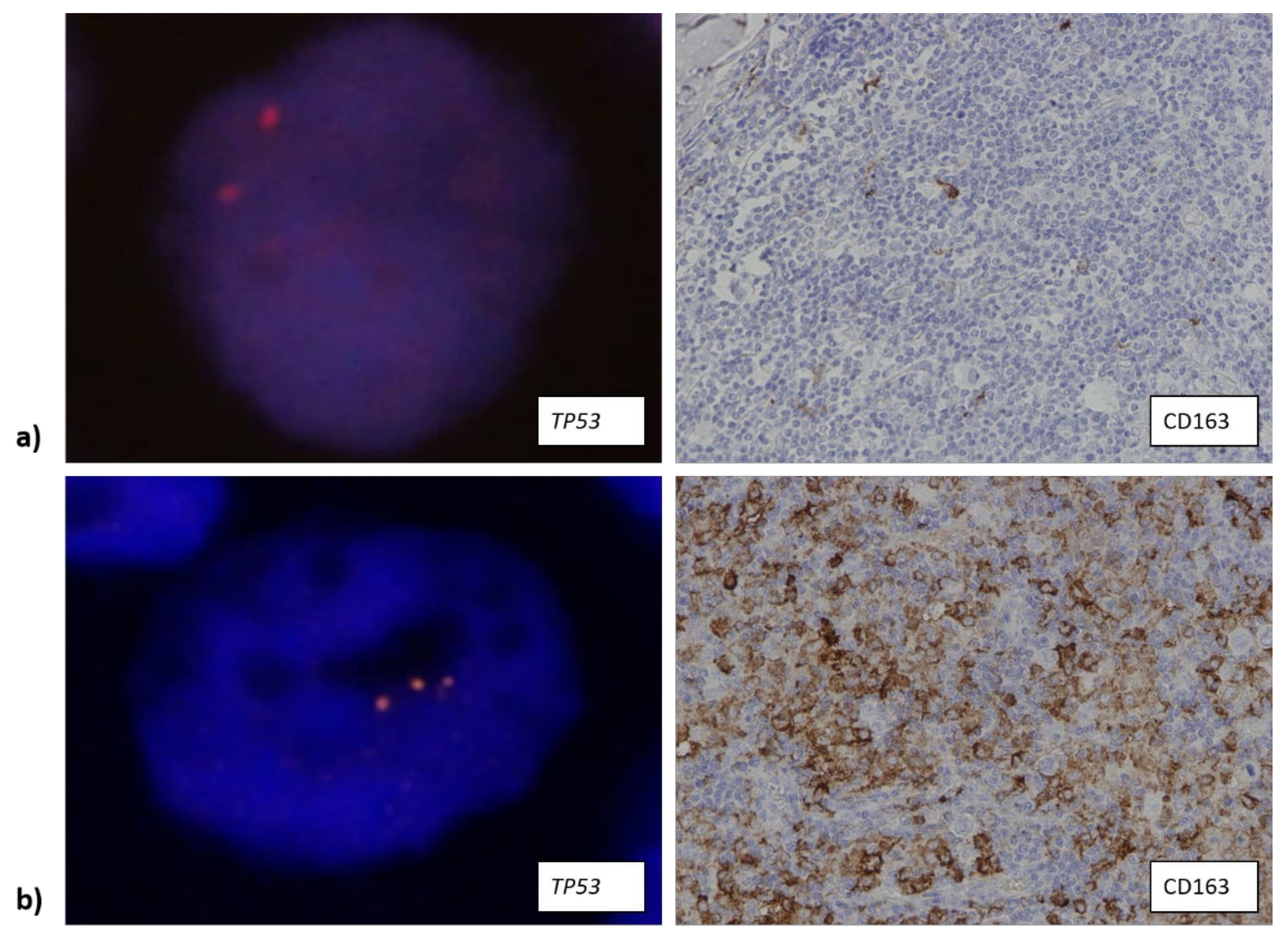
3.3. The Number of CD163+ Cells Is Associated with Higher Copy Number of TP53 Gene
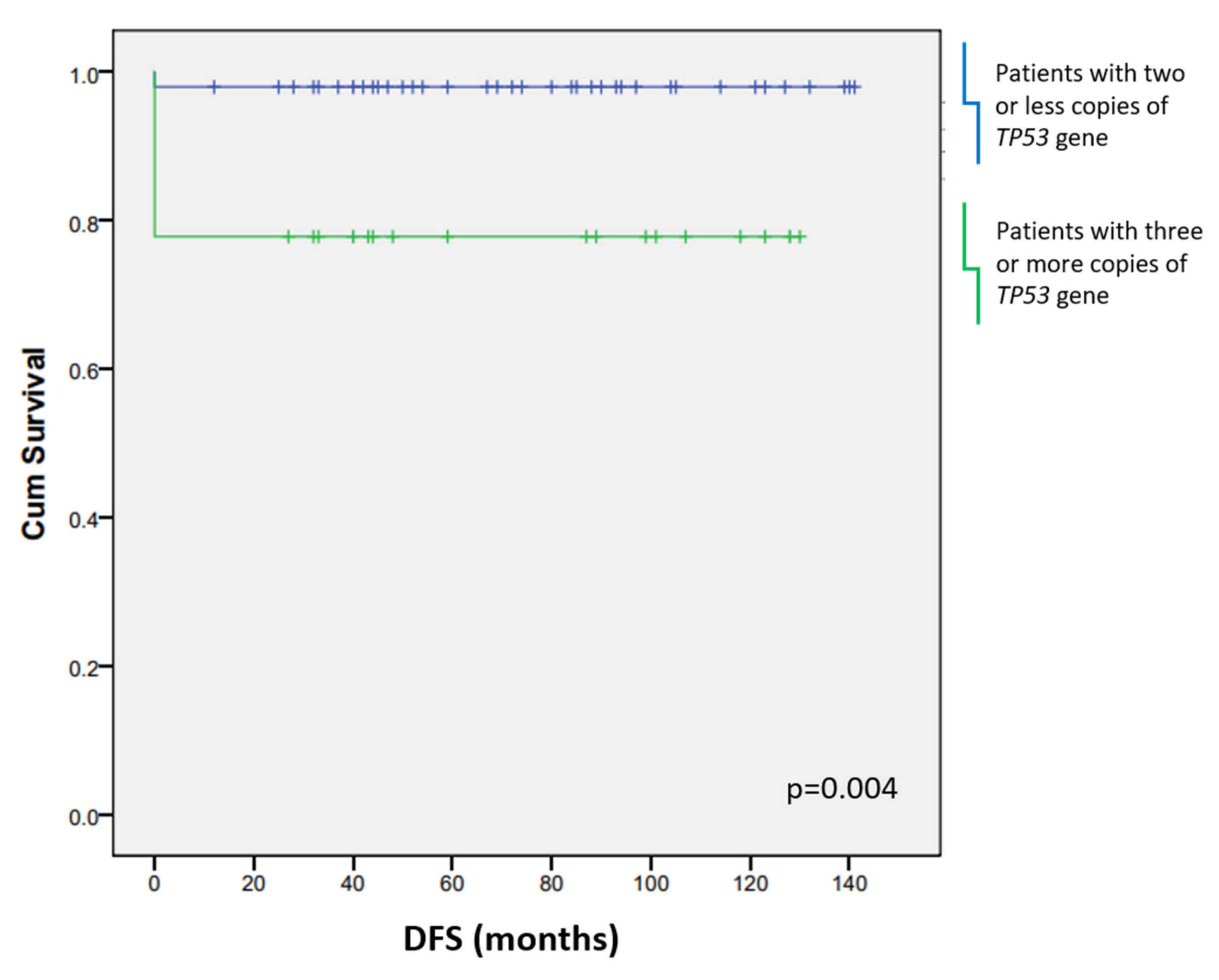
3.4. CD163+ Macrophages and Additional TP53 Copies Affect the Patients’ Survival
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stein, H.; Pileri, S.A.; Weiss, L.M.; Opppema, S.; Gascoyne, R.D.; Jaffe, E.S. Hodgkin lymphomas. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2017; pp. 423–442. [Google Scholar]
- Küppers, R. Molecular biology of Hodgkin’s lymphoma. Adv. Cancer Res. 2002, 44, 277–312. [Google Scholar]
- Weiss, L.M.; Chan, J.K.C.; MacLennan, K.; Warnke, R.A. Pathology of classical Hodgkin’s disease. In Hodgkin’s Disease; Mauch, P.M., Armitage, J.O., Diehl, V., Hoppe, R.T., Weiss, L.M., Eds.; Lippencott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 101–120. [Google Scholar]
- Hsu, S.M.; Jaffe, E.S. Leu M1 and peanut agglutinin stain the neoplastic cells of Hodgkin’s disease. Am. J. Clin. Pathol. 1984, 82, 29–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oudejans, J.J.; Kummer, J.A.; Jiwa, M.; van der Valk, P.; Ossenkoppele, G.J.; Kluin, P.M.; Kluin-Nelemans, J.C.; Meijer, C.J. Granzyme B expression in Reed-Sternberg cells of Hodgkin’s disease. Am. J. Pathol. 1996, 148, 233–240. [Google Scholar] [PubMed]
- Van den Berg, A.; Visser, L.; Poppema, S. High expression of the CC chemokine TARC in Reed-Sternberg cells. A possible explanation for the characteristic T-cell infiltration Hodgkin’s lymphoma. Am. J. Pathol. 1999, 154, 1685–1691. [Google Scholar] [CrossRef]
- Küppers, R.; Klein, U.; Schwering, I.; Distler, V.; Bräuninger, A.; Cattoretti, G.; Tu, Y.; Stolovitzky, G.A.; Califano, A.; Hansmann, M.; et al. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J. Clin. Investig. 2003, 111, 529–537. [Google Scholar] [CrossRef] [Green Version]
- Piccaluga, P.P.; Agostinelli, C.; Gazzola, A.; Tripodo, C.; Bacci, F.; Sabattini, E.; Sista, M.T.; Mannu, C.; Sapienza, M.R.; Rossi, M.; et al. Pathobiology of Hodgkin Lymphoma. Adv. Hematol. 2011, 2011, 920898. [Google Scholar] [CrossRef] [PubMed]
- Ann Arbor Staging System|Radiology Reference Article. Available online: https://radiopaedia.org/articles/ann-arbor-staging-system (accessed on 14 January 2022).
- German Hodgkin Study Group: Home–GHSG. Available online: https://en.ghsg.org/ (accessed on 14 January 2022).
- ECOG Performance Status Scale. Available online: https://ecog-acrin.org/resources/ecog-performance-status (accessed on 14 January 2022).
- Opinto, G.; Agostinelli, C.; Ciavarella, S.; Guarini, A.; Maiorano, E.; Ingravallo, G. Hodgkin Lymphoma: A Special Microenvironment. J. Clin. Med. 2021, 10, 4665. [Google Scholar] [CrossRef]
- Skinnider, B.F.; Mak, T.W. The role of cytokines in classical Hodgkin lymphoma. Blood 2002, 99, 4283–4297. [Google Scholar] [CrossRef] [Green Version]
- Aldinucci, D.; Gloghini, A.; Pinto, A.; De Filippi, R.; Carbone, A. The classical Hodgkin’s lymphoma microenvironment and its role in promoting tumour growth and immune escape. J. Pathol. 2010, 221, 248–263. [Google Scholar] [CrossRef]
- Vardhana, S.; Younes, A. The immune microenvironment in Hodgkin lymphoma: T cells, B cells, and immune checkpoints. Haematologica 2016, 101, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Visser, L.; Blokzijl, T.; Harms, G.; Atayar, C.; Poppema, S.; van den Berg, A. The CD4+CD26- T-cell population in classical Hodgkin’s lymphoma displays a distinctive regulatory T-cell profile. Lab. Investig. 2008, 88, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.K.; Lambley, E.; Duraiswamy, J.; Dua, U.; Smith, C.; Elliott, S.; Gill, D.; Marlton, P.; Seymour, J.; Khanna, R. Expression of LAG-3 by tumor-infiltrating lymphocytes is coincident with the suppression of latent membrane antigen-specific CD8+ T-cell function in Hodgkin lymphoma patients. Blood 2006, 108, 2280–2289. [Google Scholar] [CrossRef] [PubMed]
- Re, D.; Kuppers, R.; Diehl, V. Molecular pathogenesis of Hodgkin’s lymphoma. J. Clin. Oncol. 2005, 23, 6379–6386. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Barbi, J.; Pan, F. The regulation of immune tolerance by FOXP3. Nat. Rev. Immunol. 2017, 17, 703–717. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Connors, J.M.; Gascoyne, R.D. Molecular pathogenesis of Hodgkin’s lymphoma: Increasing evidence of the importance of the microenvironment. J. Clin. Oncol. 2011, 29, 1812–1826. [Google Scholar] [CrossRef]
- Teichmann, M.; Meyer, B.; Beck, A.; Niedobitek, G. Expression of the interferon-inducible chemokine IP-10 (CXCL10), a chemokine with proposed anti-neoplastic functions, in Hodgkin lymphoma and nasopharyngeal carcinoma. J. Pathol. 2005, 206, 68–75. [Google Scholar] [CrossRef]
- Massini, G.; Siemer, D.; Hohaus, S. EBV in Hodgkin Lymphoma. Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009013. [Google Scholar] [CrossRef]
- Qian, B.; Pollard, J.W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, O.; El Bastawisy, A.; Allahlobi, N.; Abdellateif, M.S.; Zekri, A.R.N.; Shaarawy, S.; Korany, Z.; Mohanad, M.; Bahnassy, A.A. The role of CD68+ macrophage in classical Hodgkin lymphoma patients from Egypt. Diagn. Pathol. 2020, 15, 10. [Google Scholar] [CrossRef]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N. Engl. J. Med. 2010, 362, 875–885. [Google Scholar] [CrossRef] [Green Version]
- Werner, L.; Dreyer, J.H.; Hartmann, D.; Barros, M.H.M.; Büttner-Herold, M.; Grittner, U.; Niedobitek, G. Tumor-associated macrophages in classical Hodgkin lymphoma: Hormetic relationship to outcome. Sci. Rep. 2020, 10, 9410. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.P.; Hopman, A.H.; Haesevoets, A.M.; Gennotte, I.A.; Bot, F.J.; Arends, J.W.; Ramaekers, F.C.; Schouten, H.C. Chromosomal abnormalities in Hodgkin’s disease are not restricted to Hodgkin/Reed-Sternberg cells. J. Pathol. 1998, 185, 145–152. [Google Scholar] [CrossRef]
- Re, D.; Zander, T.; Diehl, V.; Wolf, J. Genetic instability in Hodgkin’s lymphoma. Ann. Oncol. 2002, 13, 19–22. [Google Scholar] [CrossRef]
- Cuceu, C.; Hempel, W.M.; Sabatier, L.; Bosq, J.; Carde, P.; M’kacher, R. Chromosomal Instability in Hodgkin Lymphoma: An In-Depth Review and Perspectives. Cancers 2018, 10, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Re, D.; Benenson, L.; Wickenhauser, C.; Starostik, P.; Staratschek-Jox, A.; Müller-Hermelink, H.K.; Diehl, V.; Wolf, J. Proficient expression of mismatch repair genes in Hodgkin-Reed Sternberg cells. Int. J. Cancer 2002, 97, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Park, K.; Kim, J.; Kim, H.S.; Shin, H.S. Isolated human germinal center centroblasts have an intact mismatch repair system. J. Immunol. 1998, 161, 6128–6132. [Google Scholar]
- Starczynski, J.; Simmons, W.; Flavell, J.R.; Byrd, P.J.; Stewart, G.S.; Kullar, H.S.; Groom, A.; Crocker, J.; Moss, P.A.H.; Reynolds, G.M.; et al. Variations in ATM protein expression during normal lymphoid differentiation and among B-cell-derived neoplasias. Am. J. Pathol. 2003, 163, 423–432. [Google Scholar] [CrossRef] [Green Version]
- Weber-Matthiesen, K.; Deerberg, J.; Poetsch, M.; Grote, W.; Schlegelberger, B. Numerical chromosome aberrations are present within the CD30+ Hodgkin and Reed–Sternberg cells in 100% of analyzed cases of Hodgkin’s disease. Blood 1995, 86, 1464–1468. [Google Scholar] [CrossRef] [Green Version]
- Knecht, H.; Righolt, C.; Mai, S. Genomic instability: The driving force behind refractory/relapsing Hodgkin’s lymphoma. Cancers 2013, 5, 714–725. [Google Scholar] [CrossRef] [Green Version]
- Guffei, A.; Sarkar, R.; Klewes, L.; Righolt, C.; Knecht, H.; Mai, S. Dynamic chromosomal rearrangements in Hodgkin’s lymphoma are due to ongoing three-dimensional nuclear remodeling and breakage-bridge-fusion cycles. Haematologica 2010, 95, 2038–2046. [Google Scholar] [CrossRef]
- Küppers, R.; Schmitz, R.; Distler, V.; Renné, C.; Bräuninger, A.; Hansmann, M. Pathogenesis of Hodgkin’s lymphoma. Eur. J. Haematol. Suppl. 2005, 66, 26–33. [Google Scholar] [CrossRef]
- Baumforth, K.R.N.; Birgersdotter, A.; Reynolds, G.M.; Wei, W.; Kapatai, G.; Flavell, J.R.; Kalk, E.; Piper, K.; Lee, S.; Machado, L.; et al. Expression of the Epstein-Barr virus-encoded Epstein-Barr virus nuclear antigen 1 in Hodgkin’s lymphoma cells mediates Up-regulation of CCL20 and the migration of regulatory T cells. Am. J. Pathol. 2008, 173, 195–204. [Google Scholar] [CrossRef] [Green Version]
- Vockerodt, M.; Cader, F.Z.; Shannon-Lowe, C.; Murray, P. Epstein-Barr virus and the origin of Hodgkin lymphoma. Chin. J. Cancer 2014, 33, 591–597. [Google Scholar] [CrossRef]
- Glaser, S.L.; Lin, R.J.; Stewart, S.L.; Ambinder, R.F.; Jarrett, R.F.; Brousset, P.; Pallesen, G.; Gulley, M.L.; Khan, G.; O’Grady, J.; et al. Epstein-Barr virus-associated Hodgkin’s disease: Epidemiologic characteristics in international data. Int. J. Cancer 1997, 70, 375–382. [Google Scholar] [CrossRef]
- Grywalska, E.; Rolinski, J. Epstein-Barr virus-associated lymphomas. Semin. Oncol. 2015, 42, 291–303. [Google Scholar] [CrossRef] [Green Version]
- Oudejans, J.J.; Jiwa, N.W.; Kummer, J.A.; Ossenkoppele, G.J.; van Heerde, P.; Baars, J.W.; Kluin, P.M.; Kluin-Nelemans, J.C.; van Diest, P.J.; Middeldorp, J.M.; et al. Activated Cytotoxic T Cells as Prognostic Marker in Hodgkin’s Disease. Blood 1997, 89, 1376–1382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100 Pt B, 1–441. [Google Scholar]
- Hislop, A.D.; Kuo, M.; Drake-Lee, A.B.; Akbar, A.N.; Bergler, W.; Hammerschmitt, N.; Khan, N.; Palendira, U.; Leese, A.M.; Timms, J.M.; et al. Tonsillar homing of Epstein-Barr virus-specific CD8+ T cells and the virus-host balance. J. Clin. Investig. 2005, 115, 2546–2555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, J.; Oh, E.J.; Yang, W.I.; Kim, S.J.; Yoon, S.O. Implications of infiltrating immune cells within bone marrow of patients with diffuse large B-cell lymphoma. Hum. Pathol. 2017, 64, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Jakovic, L.R.; Mihaljevic, B.S.; Andjelic, B.M.; Bogdanovic, A.D.; Perunicic Jovanovic, M.D.; Babic, D.D.; Bumbasirevic, V.Z. Prognostic value of lymphocyte/monocyte ratio in advanced Hodgkin lymphoma: Correlation with international prognostic score and tumor associated macrophages. Leuk. Lymphoma 2016, 57, 1839–1847. [Google Scholar] [CrossRef]
- Newman, A.M.; Zaka, M.; Zhou, P.; Blain, A.E.; Erhorn, A.; Barnard, A.; Crossland, R.E.; Wilkinson, S.; Enshaei, A.; De Zordi, J.; et al. Genomic abnormalities of TP53 define distinct risk groups of paediatric B-cell non-Hodgkin lymphoma. Leukemia 2021, 36, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Maggio, E.M.; Stekelenburg, E.; Van den Berg, A.; Poppema, S. TP53 gene mutations in Hodgkin lymphoma are infrequent and not associated with absence of Epstein-Barr virus. Int. J. Cancer 2001, 94, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Dedon, P.C.; Tannenbaum, S.R. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004, 423, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Tamma, R.; Ruggieri, S.; Annese, T.; Simone, G.; Mangia, A.; Rega, S.; Zito, F.A.; Nico, B.; Ribatti, D. Bcl6/p53 expression, macrophages/mast cells infiltration and microvascular density in invasive breast carcinoma. Oncotarget 2018, 9, 22727–22740. [Google Scholar] [CrossRef] [Green Version]
- Al Sayed Ahmed, H.; Raslan, W.F.; Deifalla, A.H.S.; Fathallah, M.D. CD163 is a predictive biomarker for prognosis of classical Hodgkin’s lymphoma in Saudi patients. Mol. Clin. Oncol. 2019, 11, 67–76. [Google Scholar] [CrossRef]
- Jakovic, L.R.; Mihaljevic, B.S.; Perunicic Jovanovic, M.D.; Bogdanovic, A.D.; Andjelic, B.M.; Bumbasirevic, V.Z. The prognostic relevance of tumor associated macrophages in advanced stage classical Hodgkin lymphoma. Leuk. Lymphoma 2011, 52, 1913–1919. [Google Scholar] [CrossRef]
- Tudor, C.S.; Bruns, H.; Daniel, C.; Distel, L.V.; Hartmann, A.; Gerbitz, A.; Buettne, M.J. Macrophages and dendritic cells as actors in the immune reaction of classical Hodgkin lymphoma. PLoS ONE 2014, 9, e114345. [Google Scholar] [CrossRef] [Green Version]
- Kamper, P.; Benedix, K.; Hamilton-Dutoit, S.; Honoré, B.; Nyengaard, J.R.; d’Amore, F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin’s lymphoma. Haematologica 2011, 96, 269–276. [Google Scholar] [CrossRef]
- Minami, K.; Hiwatashi, K.; Ueno, S.; Sakoda, M.; Iino, S.; Okumura, H.; Hashiguchi, M.; Kawasaki, Y.; Kurahara, H.; Mataki, Y.; et al. Prognostic significance of CD68, CD163 and Folate receptor-β positive macrophages in hepatocellular carcinoma. Exp. Ther. Med. 2018, 15, 4465–4476. [Google Scholar] [CrossRef] [Green Version]


| Marker | Antibody Clone/Manufacturer | Type of Cells | Cut-Off Value |
|---|---|---|---|
| Granzyme B | GrB-7/Dako/Agilent (Santa Clara, CA, USA) | Cytotoxic T cells and natural killer cells | 30 cell per high power field |
| FOXP3 | 236A/E7/Abcam (Cambridge, UK) | Regulatory T cells | 30% of the overall number of cells within the tumor tissue |
| CD68 | PG-M1/Dako/Agilent (Santa Clara, CA, USA) | M1 macrophages | 25% of the overall number of cells within the tumor tissue § |
| CD163 | MRQ-26/Cell Marque (Rocklin, CA, USA) | M2 macrophages | 25% of the overall number of cells within the tumor tissue § |
| Characteristic | Category | N | % |
|---|---|---|---|
| Sex | Male | 49 | 40.8 |
| Female | 71 | 59.2 | |
| Age | ≤40 | 93 | 77.5 |
| >40 | 27 | 22.5 | |
| Histology | NSCHL | 77 | 64.2 |
| MCCHL | 43 | 35.8 | |
| Ann Arbor | I | 12 | 11.3 |
| II | 69 | 65.1 | |
| III | 17 | 16 | |
| IV | 8 | 7.6 | |
| GHSG | Early stage | 24 | 22.7 |
| Intermediate stage | 49 | 46.2 | |
| Advanced stage | 33 | 31.1 | |
| ECOG | 1 | 103 | 97.2 |
| 2 | 2 | 1.9 | |
| 3 | 0 | 0 | |
| 4 | 1 | 0.9 | |
| B symptoms | + | 52 | 49.1 |
| − | 54 | 50.9 | |
| Bulky disease (>7.5 cm) | + | 35 | 33 |
| − | 71 | 67 | |
| Bone marrow infiltration | + | 4 | 3.8 |
| − | 102 | 96.2 | |
| First therapy | ABVD | 103 | 97.2 |
| Other | 3 | 2.8 | |
| Response to first therapy | Complete remission | 95 | 89.6 |
| Partial remission | 9 | 8.5 | |
| Disease progression | 2 | 1.9 | |
| Relapse | + | 12 | 11.3 |
| − | 94 | 88.7 | |
| Second therapy | Stem cell transplantation | 21 | 19.8 |
| Other | 2 | 1.9 | |
| None | 83 | 78.3 | |
| Response to Second therapy | Complete remission | 16 | 69.6 |
| Partial remission | 5 | 21.8 | |
| No response | 1 | 4.3 | |
| Unknown | 1 | 4.3 | |
| CD20 | + | 9 | 7.5 |
| − | 111 | 92.5 | |
| CD15 | + | 109 | 90.8 |
| − | 11 | 9.2 | |
| EBV-ISH | + | 20 | 16.7 |
| − | 100 | 83.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hančić, S.; Gršković, P.; Gašparov, S.; Ostojić Kolonić, S.; Dominis, M.; Korać, P. Macrophage Infiltration Correlates with Genomic Instability in Classic Hodgkin Lymphoma. Biomedicines 2022, 10, 579. https://doi.org/10.3390/biomedicines10030579
Hančić S, Gršković P, Gašparov S, Ostojić Kolonić S, Dominis M, Korać P. Macrophage Infiltration Correlates with Genomic Instability in Classic Hodgkin Lymphoma. Biomedicines. 2022; 10(3):579. https://doi.org/10.3390/biomedicines10030579
Chicago/Turabian StyleHančić, Suzana, Paula Gršković, Slavko Gašparov, Slobodanka Ostojić Kolonić, Mara Dominis, and Petra Korać. 2022. "Macrophage Infiltration Correlates with Genomic Instability in Classic Hodgkin Lymphoma" Biomedicines 10, no. 3: 579. https://doi.org/10.3390/biomedicines10030579
APA StyleHančić, S., Gršković, P., Gašparov, S., Ostojić Kolonić, S., Dominis, M., & Korać, P. (2022). Macrophage Infiltration Correlates with Genomic Instability in Classic Hodgkin Lymphoma. Biomedicines, 10(3), 579. https://doi.org/10.3390/biomedicines10030579






