A Non-Canonical Link between Non-Coding RNAs and Cardiovascular Diseases
Abstract
:1. Introduction
2. Biogenesis and Development of Atherosclerosis, the Principal CVD Worldwide
3. Non-Coding RNAs
4. Small ncRNAs
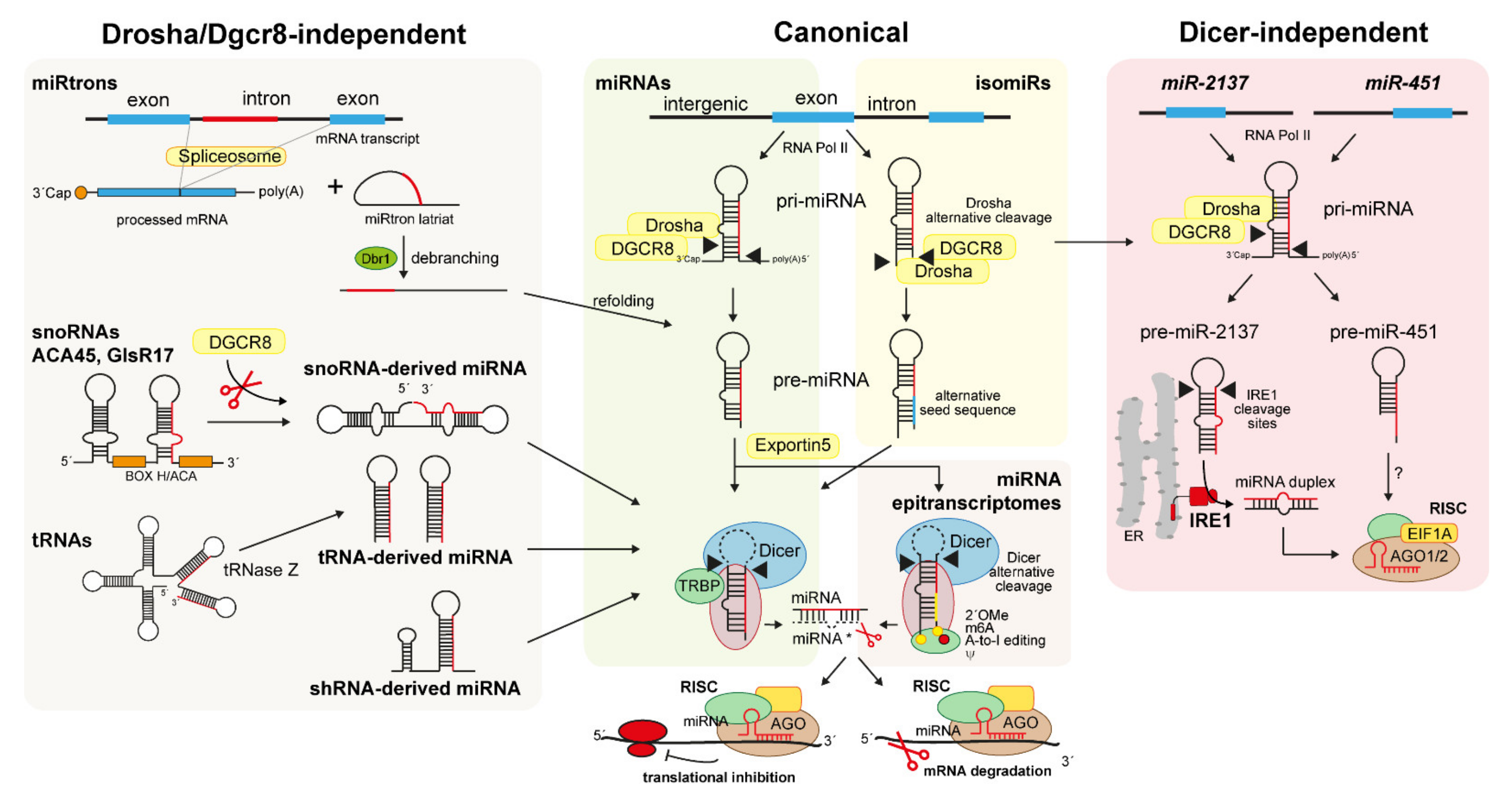
4.1. Canonical Biogenesis of miRNAs
4.2. Non-Canonical Biogenesis of Small RNAs
4.2.1. snoRNA-, shRNA-, and tRNA-Derived miRNAs
4.2.2. MiRtrons
4.2.3. IsomiRs
4.2.4. Dicer-Independent miRNAs and Simtrons
5. lncRNAs
5.1. Canonical Biogenesis of lncRNAs
5.2. Non-Canonical Biogenesis of lncRNAs and circRNAs
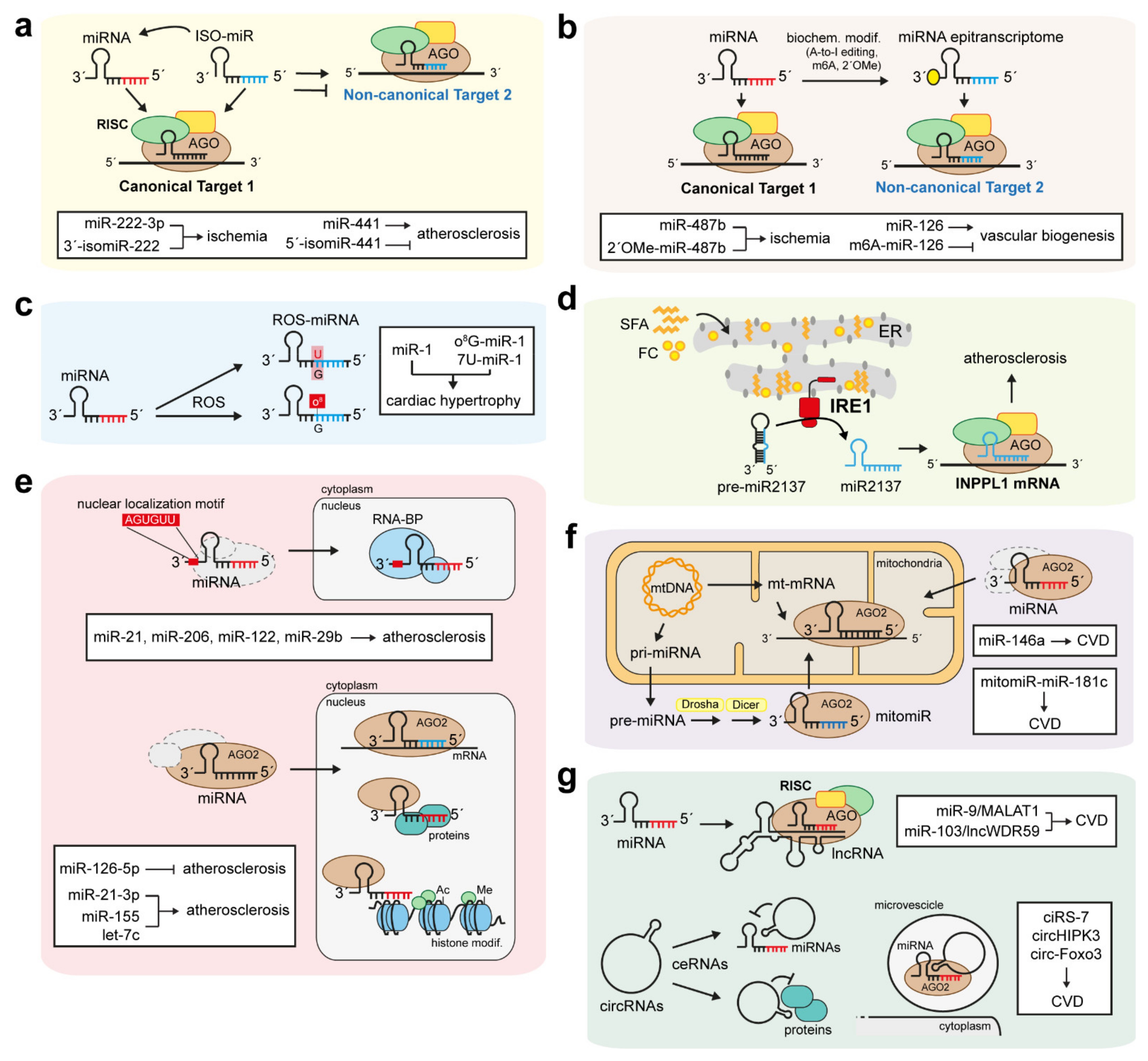
6. Non-Canonical Function of ncRNAs in CVD
6.1. IsomiRs and miRNA Epitranscriptomes
6.2. Small-Derived miRNAs and MiRtrons
6.3. Oxidative and ER Stress-Related Non-Canonical miRNAs
6.4. Non-Canonical Function of Nuclear and Mitochondrial miRNAs
6.5. Antagonistic Functions of miRNAs and lncRNAs
6.6. circRNAs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 7 February 2022).
- World Health Organization. World Health Statistics 2021: A Visual Summary. Available online: https://www.who.int/data/stories/world-health-statistics-2021-a-visual-summary (accessed on 7 February 2022).
- World Health Organization. Global Health Estimates: Life Expectancy and Leading Causes of Death and Disability. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 7 February 2022).
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Health Statistics: Monitoring Health for the SDGs 2021. Available online: https://apps.who.int/iris/bitstream/handle/10665/342703/9789240027053-eng.pdf (accessed on 10 January 2022).
- Jayasuriya, R.; Ganesan, K.; Xu, B.; Ramkumar, K.M. Emerging role of long non-coding RNAs in endothelial dysfunction and their molecular mechanisms. Biomed. Pharmacother. 2022, 145, 112421. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimata, H.; Sugimoto, N. Roles of non-canonical structures of nucleic acids in cancer and neurodegenerative diseases. Nucleic Acids Res. 2021, 49, 7839–7855. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Ni, M.; Song, B.; Cao, L. Circulating Circular RNAs: Novel Biomarkers for Heart Failure. Front Pharm. 2020, 11, 560537. [Google Scholar] [CrossRef]
- Jaé, N.; Dimmeler, S. Noncoding RNAs in Vascular Diseases. Circ. Res. 2020, 126, 1127–1145. [Google Scholar] [CrossRef]
- Abdelfattah, A.M.; Park, C.; Choi, M.Y. Update on non-canonical microRNAs. Biomol. Concepts 2014, 5, 275–287. [Google Scholar] [CrossRef] [Green Version]
- Correction to: Heart Disease and Stroke Statistics—2019 Update: A Report from the American Heart Association. Circulation 2020, 141, e33. [CrossRef]
- Pearson-Stuttard, J.; Guzman-Castillo, M.; Penalvo, J.L.; Rehm, C.D.; Afshin, A.; Danaei, G.; Kypridemos, C.; Gaziano, T.; Mozaffarian, D.; Capewell, S.; et al. Modeling Future Cardiovascular Disease Mortality in the United States: National Trends and Racial and Ethnic Disparities. Circulation 2016, 133, 967–978. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Huth, J.K. Time-series study of cardiovascular rates in India: A systematic analysis between 1990 and 2017. Indian Heart J. 2020, 72, 194–196. [Google Scholar] [CrossRef]
- Yusuf, S.; Hawken, S.; Ounpuu, S.; Dans, T.; Avezum, A.; Lanas, F.; McQueen, M.; Budaj, A.; Pais, P.; Varigos, J.; et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet 2004, 364, 937–952. [Google Scholar] [CrossRef]
- Ross, R. Atherosclerosis--an inflammatory disease. N. Engl. J. Med. 1999, 340, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Natarelli, L.; Schober, A. MicroRNAs and the response to injury in atherosclerosis. Hamostaseologie 2015, 35, 142–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Effects of shear stress on endothelial cells: Go with the flow. Acta Physiol. 2017, 219, 382–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Sheng, L.; He, T.; Nin, X.; Xue, A.; Zhang, H.; Liu, Z. High fluid shear stress prevents atherosclerotic plaque formation by promoting endothelium denudation and synthetic phenotype of vascular smooth muscle cells. Mol. Med. Rep. 2021, 24, 577. [Google Scholar] [CrossRef]
- Fuster, J.J.; Fernández, P.; González-Navarro, H.; Silvestre, C.; Nabah, Y.N.; Andrés, V. Control of cell proliferation in atherosclerosis: Insights from animal models and human studies. Cardiovasc. Res. 2010, 86, 254–264. [Google Scholar] [CrossRef] [Green Version]
- Robbins, C.S.; Hilgendorf, I.; Weber, G.F.; Theurl, I.; Iwamoto, Y.; Figueiredo, J.L.; Gorbatov, R.; Sukhova, G.K.; Gerhardt, L.M.; Smyth, D.; et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat. Med. 2013, 19, 1166–1172. [Google Scholar] [CrossRef]
- Yamamoto, E.; Siasos, G.; Zaromytidou, M.; Coskun, A.U.; Xing, L.; Bryniarski, K.; Zanchin, T.; Sugiyama, T.; Lee, H.; Stone, P.H.; et al. Low Endothelial Shear Stress Predicts Evolution to High-Risk Coronary Plaque Phenotype in the Future. Circ. Cardiovasc. Interv. 2017, 10, e005455. [Google Scholar] [CrossRef]
- Schober, A.; Nazari-Jahantigh, M.; Wei, Y.; Bidzhekov, K.; Gremse, F.; Grommes, J.; Megens, R.T.; Heyll, K.; Noels, H.; Hristov, M.; et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat. Med. 2014, 20, 368–376. [Google Scholar] [CrossRef] [Green Version]
- Tabas, I.; Williams, K.J.; Borén, J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: Update and therapeutic implications. Circulation 2007, 116, 1832–1844. [Google Scholar] [CrossRef]
- Bekkering, S.; Quintin, J.; Joosten, L.A.; van der Meer, J.W.; Netea, M.G.; Riksen, N.P. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arter. Thromb. Vasc. Biol. 2014, 34, 1731–1738. [Google Scholar] [CrossRef]
- Nielsen, L.B. Transfer of low density lipoprotein into the arterial wall and risk of atherosclerosis. Atherosclerosis 1996, 123, 1–15. [Google Scholar] [CrossRef]
- Wolf, D.; Ley, K. Immunity and Inflammation in Atherosclerosis. Circ. Res. 2019, 124, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Reglero-Real, N.; Colom, B.; Bodkin, J.V.; Nourshargh, S. Endothelial Cell Junctional Adhesion Molecules. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2048–2057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doran, A.C.; Yurdagul, A., Jr.; Tabas, I. Efferocytosis in health and disease. Nat. Rev. Immunol. 2020, 20, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Bentzon, J.F.; Otsuka, F.; Virmani, R.; Falk, E. Mechanisms of plaque formation and rupture. Circ Res 2014, 109, 1852–1866. [Google Scholar] [CrossRef] [PubMed]
- Panning, B.; Jaenisch, R. RNA and the Epigenetic Regulation of X Chromosome Inactivation. Cell 1998, 93, 305–308. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.; Guttman, M. RNA and dynamic nuclear organization. Science 2014, 345, 1240–1241. [Google Scholar] [CrossRef] [Green Version]
- Stark, B.C.; Kole, R.; Bowman, E.J.; Altman, S. Ribonuclease P: An enzyme with an essential RNA component. Proc. Natl. Acad. Sci. USA 1978, 75, 3717–3721. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulos, G.L.; Reczko, M.; Simossis, V.A.; Sethupathy, P.; Hatzigeorgiou, A.G. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2008, 37, D155–D158. [Google Scholar] [CrossRef] [Green Version]
- Bentwich, I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005, 579, 5904–5910. [Google Scholar] [CrossRef] [Green Version]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doolittle, W.F.; Brunet, T.D.P.; Linquist, S.; Gregory, T.R. Distinguishing between “Function” and “Effect” in Genome Biology. Genome Biol. Evol. 2014, 6, 1234–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, N.; McGrath, A.; Chen, Y.-P.P. Evaluation of deep learning in non-coding RNA classification. Nat. Mach. Intell. 2019, 1, 246–256. [Google Scholar] [CrossRef]
- Park, S.; Kang, I.; Shin, C. MicroRNA clustering on the biogenesis of suboptimal microRNAs. Appl. Biol. Chem. 2021, 64, 51. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- López-Urrutia, E.; Bustamante Montes, L.P.; Ladrón de Guevara Cervantes, D.; Pérez-Plasencia, C.; Campos-Parra, A.D. Crosstalk Between Long Non-coding RNAs, Micro-RNAs and mRNAs: Deciphering Molecular Mechanisms of Master Regulators in Cancer. Front. Oncol. 2019, 9, 669. [Google Scholar] [CrossRef]
- Natarelli, L.; Geißler, C.; Csaba, G.; Wei, Y.; Zhu, M.; di Francesco, A.; Hartmann, P.; Zimmer, R.; Schober, A. miR-103 promotes endothelial maladaptation by targeting lncWDR59. Nat. Commun. 2018, 9, 2645. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Czech, B.; Zhou, R.; Erlich, Y.; Brennecke, J.; Binari, R.; Villalta, C.; Gordon, A.; Perrimon, N.; Hannon, G.J. Hierarchical rules for Argonaute loading in Drosophila. Mol. Cell 2009, 36, 445–456. [Google Scholar] [CrossRef] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Okamura, K.; Liu, N.; Lai, E.C. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol. Cell 2009, 36, 431–444. [Google Scholar] [CrossRef] [Green Version]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011, 12, R126. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Fan, J.; Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 4034–4039. [Google Scholar] [CrossRef] [Green Version]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dika, E.; Broseghini, E.; Porcellini, E.; Lambertini, M.; Riefolo, M.; Durante, G.; Loher, P.; Roncarati, R.; Bassi, C.; Misciali, C.; et al. Unraveling the role of microRNA/isomiR network in multiple primary melanoma pathogenesis. Cell Death Dis. 2021, 12, 473. [Google Scholar] [CrossRef] [PubMed]
- Hoernes, T.P.; Erlacher, M.D. Translating the epitranscriptome. Wiley Interdiscip. Rev. RNA 2017, 8, e1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.S.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. Rna 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [Green Version]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Liu, B.; Cao, J.; Wang, X.; Guo, C.; Liu, Y.; Wang, T. Deciphering the tRNA-derived small RNAs: Origin, development, and future. Cell Death Dis. 2021, 13, 24. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [Green Version]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Rorbach, G.; Unold, O.; Konopka, B.M. Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods. Sci. Rep. 2018, 8, 7560. [Google Scholar] [CrossRef] [Green Version]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA Family Gets Wider: The IsomiRs Classification and Role. Front. Cell Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef]
- Van der Kwast, R.; Quax, P.H.A.; Nossent, A.Y. An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization. Cells 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 Mediates RNA Cleavage Targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Lührmann, R.; Tuschl, T. Single-Stranded Antisense siRNAs Guide Target RNA Cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.-S.; Maurin, T.; Lai, E.C. Functional parameters of Dicer-independent microRNA biogenesis. Rna 2012, 18, 945–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, T.; Arthanari, H.; Akabayov, B.; Song, H.; Papadopoulos, E.; Qi, H.H.; Jedrychowski, M.; Güttler, T.; Guo, C.; Luna, R.E.; et al. eIF1A augments Ago2-mediated Dicer-independent miRNA biogenesis and RNA interference. Nat. Commun. 2015, 6, 7194. [Google Scholar] [CrossRef] [Green Version]
- Havens, M.; Reich, A.; Duelli, D.; Hastings, M. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef] [Green Version]
- Hamid, S.M.; Citir, M.; Terzi, E.M.; Cimen, I.; Yildirim, Z.; Dogan, A.E.; Kocaturk, B.; Onat, U.I.; Arditi, M.; Weber, C.; et al. Inositol-requiring enzyme-1 regulates phosphoinositide signaling lipids and macrophage growth. EMBO Rep. 2020, 21, e51462. [Google Scholar] [CrossRef]
- Avril, T.; Chevet, E. IRE1-mediated miRNA maturation in macrophage phosphoinositide signaling. EMBO Rep. 2020, 21, e51929. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 107, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Schlackow, M.; Nojima, T.; Gomes, T.; Dhir, A.; Carmo-Fonseca, M.; Proudfoot, N.J. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol. Cell 2017, 65, 25–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, C.; Hamada, M. Identifying sequence features that drive ribosomal association for lncRNA. BMC Genom. 2018, 19, 906. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Hirose, T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013, 10, 456–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, X.; Jia, Y.; Zhang, Y.; Shi, L.; Li, Q.; Zang, A.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 2020, 12, 267–283. [Google Scholar] [CrossRef]
- Van der Kwast, R.; Woudenberg, T.; Quax, P.H.A.; Nossent, A.Y. MicroRNA-411 and Its 5′-IsomiR Have Distinct Targets and Functions and Are Differentially Regulated in the Vasculature under Ischemia. Mol. Ther. J. Am. Soc. Gene Ther. 2020, 28, 157–170. [Google Scholar] [CrossRef]
- Raval, Z.; Losordo, D.W. Cell Therapy of Peripheral Arterial Disease. Circ. Res. 2013, 112, 1288–1302. [Google Scholar] [CrossRef]
- Guduric-Fuchs, J.; O’Connor, A.; Cullen, A.; Harwood, L.; Medina, R.J.; O’Neill, C.L.; Stitt, A.W.; Curtis, T.M.; Simpson, D.A. Deep sequencing reveals predominant expression of miR-21 amongst the small non-coding RNAs in retinal microvascular endothelial cells. J. Cell. Biochem. 2012, 113, 2098–2111. [Google Scholar] [CrossRef] [Green Version]
- Voellenkle, C.; Rooij, J.; Guffanti, A.; Brini, E.; Fasanaro, P.; Isaia, E.; Croft, L.; David, M.; Capogrossi, M.C.; Moles, A.; et al. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. Rna 2012, 18, 472–484. [Google Scholar] [CrossRef] [Green Version]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. The role of miR-126 in embryonic angiogenesis, adult vascular homeostasis, and vascular repair and its alterations in atherosclerotic disease. J. Mol. Cell. Cardiol. 2016, 97, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Van der Kwast, R.V.; van Ingen, E.; Parma, L.; Peters, H.A.; Quax, P.H.; Nossent, A.Y. Adenosine-to-Inosine Editing of MicroRNA-487b Alters Target Gene Selection After Ischemia and Promotes Neovascularization. Circ. Res. 2018, 122, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.-Z.; Yang, F.; Zhou, C.-C.; Liu, F.; Yuan, J.-H.; Wang, F.; Wang, T.-T.; Xu, Q.-G.; Zhou, W.-P.; Sun, S.-H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef] [PubMed]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Bracken, C.P.; Pillman, K.A.; Lawrence, D.M.; Goodall, G.J.; Callen, D.F.; Neilsen, P.M. p53 Represses the Oncogenic Sno-MiR-28 Derived from a SnoRNA. PLoS ONE 2015, 10, e0129190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, D.G.; Roberts, J.T.; King, V.M.; Houserova, D.; Barnhill, E.C.; Crucello, A.; Polska, C.J.; Brantley, L.W.; Kaufman, G.C.; Nguyen, M.; et al. Human snoRNA-93 is processed into a microRNA-like RNA that promotes breast cancer cell invasion. NPJ Breast Cancer 2017, 3, 25. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, S.; Li, H.; Zhan, J.; Wang, F.; Fan, J.; Nie, X.; Wang, Y.; Wen, Z.; Chen, Y.; et al. The double face of miR-320: Cardiomyocytes-derived miR-320 deteriorated while fibroblasts-derived miR-320 protected against heart failure induced by transverse aortic constriction. Signal Transduct. Target. Ther. 2021, 6, 69. [Google Scholar] [CrossRef]
- Kuscu, C.; Kumar, P.; Kiran, M.; Su, Z.; Malik, A.; Dutta, A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. Rna 2018, 24, 1093–1105. [Google Scholar] [CrossRef] [Green Version]
- Seok, H.; Lee, H.; Lee, S.; Ahn, S.H.; Lee, H.-S.; Kim, G.-W.D.; Peak, J.; Park, J.; Cho, Y.K.; Jeong, Y.; et al. Position-specific oxidation of miR-1 encodes cardiac hypertrophy. Nature 2020, 584, 279–285. [Google Scholar] [CrossRef]
- Colpaert, R.M.W.; Calore, M. Epigenetics and microRNAs in cardiovascular diseases. Genomics 2021, 113, 540–551. [Google Scholar] [CrossRef]
- Çimen, I.; Kocatürk, B.; Koyuncu, S.; Tufanlı, Ö.; Onat, U.I.; Yıldırım, A.D.; Apaydın, O.; Demirsoy, Ş.; Aykut, Z.G.; Nguyen, U.T.; et al. Prevention of atherosclerosis by bioactive palmitoleate through suppression of organelle stress and inflammasome activation. Sci. Transl. Med. 2016, 8, 358ra126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Földes-Papp, Z.; König, K.; Studier, H.; Bückle, R.; Breunig, H.G.; Uchugonova, A.; Kostner, G.M. Trafficking of mature miRNA-122 into the nucleus of live liver cells. Curr. Pharm. Biotechnol. 2009, 10, 569–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegel, A.J.; Liu, Y.; Fang, Y.; Ding, X.; Liang, M. The miR-29 family: Genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genom. 2012, 44, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Sarshad, A.A.; Juan, A.H.; Muler, A.I.C.; Anastasakis, D.G.; Wang, X.; Genzor, P.; Feng, X.; Tsai, P.F.; Sun, H.W.; Haase, A.D.; et al. Argonaute-miRNA Complexes Silence Target mRNAs in the Nucleus of Mammalian Stem Cells. Mol. Cell 2018, 71, 1040–1050.e8. [Google Scholar] [CrossRef] [Green Version]
- Yan, M.; Chen, C.; Gong, W.; Yin, Z.; Zhou, L.; Chaugai, S.; Wang, D.W. miR-21-3p regulates cardiac hypertrophic response by targeting histone deacetylase-8. Cardiovasc. Res. 2014, 105, 340–352. [Google Scholar] [CrossRef]
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourão, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunßen, C.; Horckmans, M.; et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 2020, 12, eaaz2294. [Google Scholar] [CrossRef]
- Su, Q.; Xu, Y.; Cai, R.; Dai, R.; Yang, X.; Liu, Y.; Kong, B. miR-146a inhibits mitochondrial dysfunction and myocardial infarction by targeting cyclophilin D. Mol. Ther. -Nucleic Acids 2021, 23, 1258–1271. [Google Scholar] [CrossRef]
- Gao, J.; Hu, C.; Liang, H.; Liu, X.; Gao, X. miRNA143 Downregulated the Expression of Cyclophilin A to Inhibit the Progression of Atherosclerosis. J. Biomater. Tissue Eng. 2019, 9, 935–942. [Google Scholar] [CrossRef]
- Pinti, M.V.; Hathaway, Q.A.; Hollander, J.M. Role of microRNA in metabolic shift during heart failure. Am. J. Physiology. Heart Circ. Physiol. 2017, 312, H33–H45. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Song, C.; Zhou, X.; Han, X.; Li, J.; Wang, Z.; Shang, H.; Liu, Y.; Cao, H. Mitochondria Associated MicroRNA Expression Profiling of Heart Failure. BioMed Res. Int. 2017, 2017, 4042509. [Google Scholar] [CrossRef]
- Song, R.; Hu, X.-Q.; Zhang, L. Mitochondrial MiRNA in Cardiovascular Function and Disease. Cells 2019, 8, 1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, L.; Binder, P.; Chen, H.; Wang, X. Regulation of Long Non-coding RNAs and MicroRNAs in Heart Disease: Insight Into Mechanisms and Therapeutic Approaches. Front. Physiol. 2020, 11, 798. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef] [Green Version]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrøm, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013, 3, 2535. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019, 176, 831–843.e22. [Google Scholar] [CrossRef] [Green Version]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018, 9, 417. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Natarelli, L.; Weber, C. A Non-Canonical Link between Non-Coding RNAs and Cardiovascular Diseases. Biomedicines 2022, 10, 445. https://doi.org/10.3390/biomedicines10020445
Natarelli L, Weber C. A Non-Canonical Link between Non-Coding RNAs and Cardiovascular Diseases. Biomedicines. 2022; 10(2):445. https://doi.org/10.3390/biomedicines10020445
Chicago/Turabian StyleNatarelli, Lucia, and Christian Weber. 2022. "A Non-Canonical Link between Non-Coding RNAs and Cardiovascular Diseases" Biomedicines 10, no. 2: 445. https://doi.org/10.3390/biomedicines10020445
APA StyleNatarelli, L., & Weber, C. (2022). A Non-Canonical Link between Non-Coding RNAs and Cardiovascular Diseases. Biomedicines, 10(2), 445. https://doi.org/10.3390/biomedicines10020445





