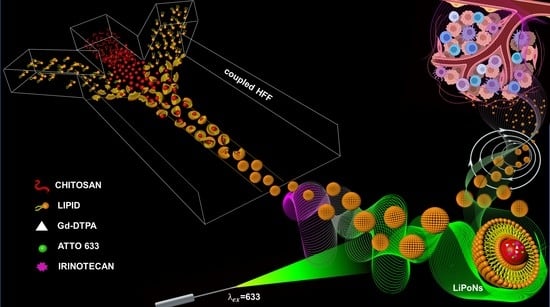
coupled Hydrodynamic Flow Focusing (cHFF) to Engineer Lipid–Polymer Nanoparticles (LiPoNs) for Multimodal Imaging and Theranostic Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Microfluidic Set-Up for coupled Flow Focusing Approach
2.3. One Step HFF for Lipid Polymer Nanoparticles (LiPoNs) Production
2.4. Purification and Concentration of NPs
2.5. Characterisation of LiPoNs Nanoparticles
2.5.1. Physicochemical Characterisation
2.5.2. LiPoNs Imaging
2.5.3. In Vitro MRI
2.6. Evaluation of Co-Encapsulation Efficiency
2.7. Cell Viability by MTT Assay
2.8. Flow Cytometry Analysis
2.9. Cell Uptake Study by Confocal Imaging
3. Results
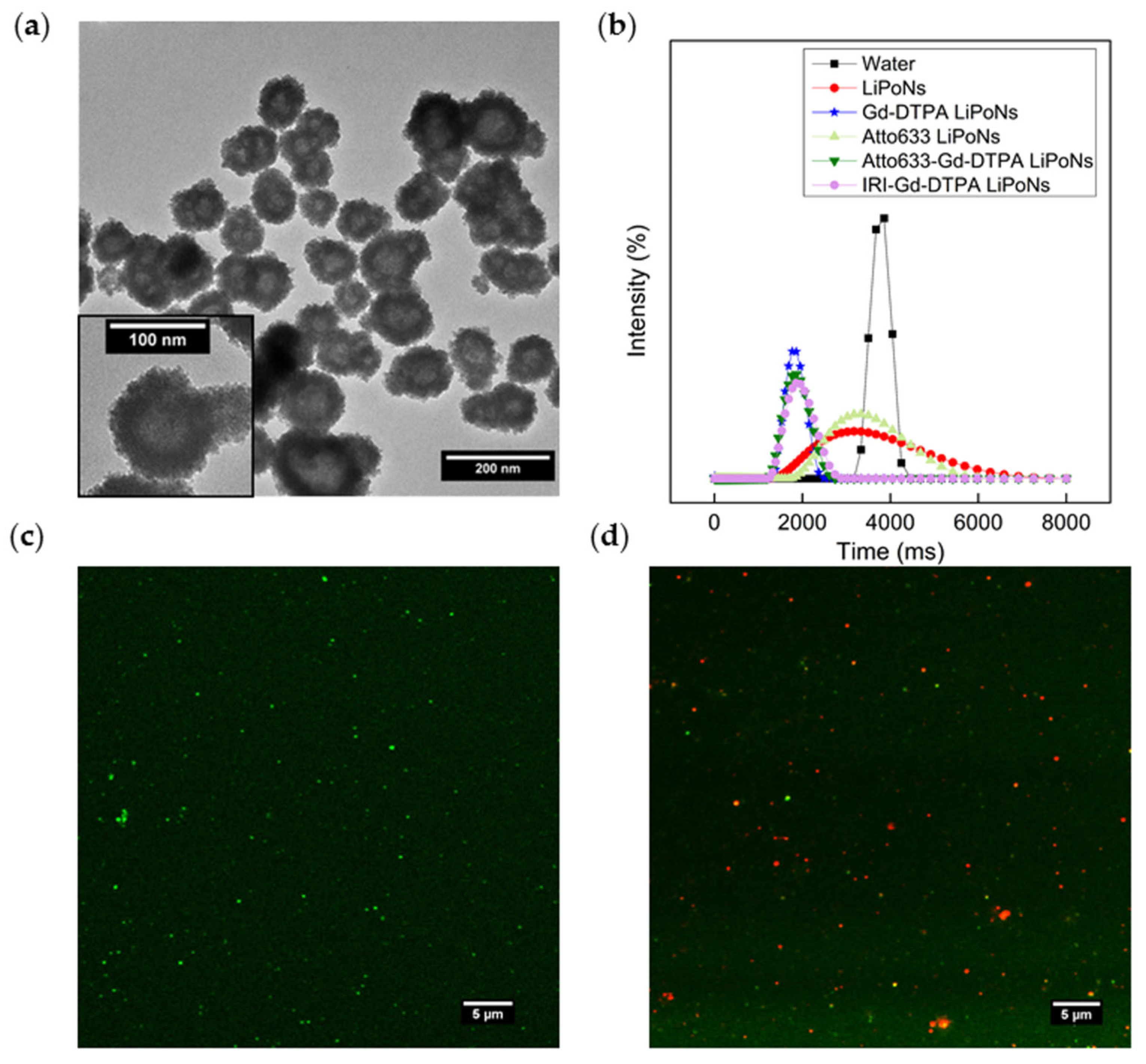
3.1. coupled Hydrodynamic Flow Focusing (cHFF) for LiPoNs Production
3.2. Microfluidic Platform to Obtain coupled Hydrodynamic Flow Focusing (cHFF) and the Optimisation of Process Parameters to Control the LiPoNs’ Nanostructures
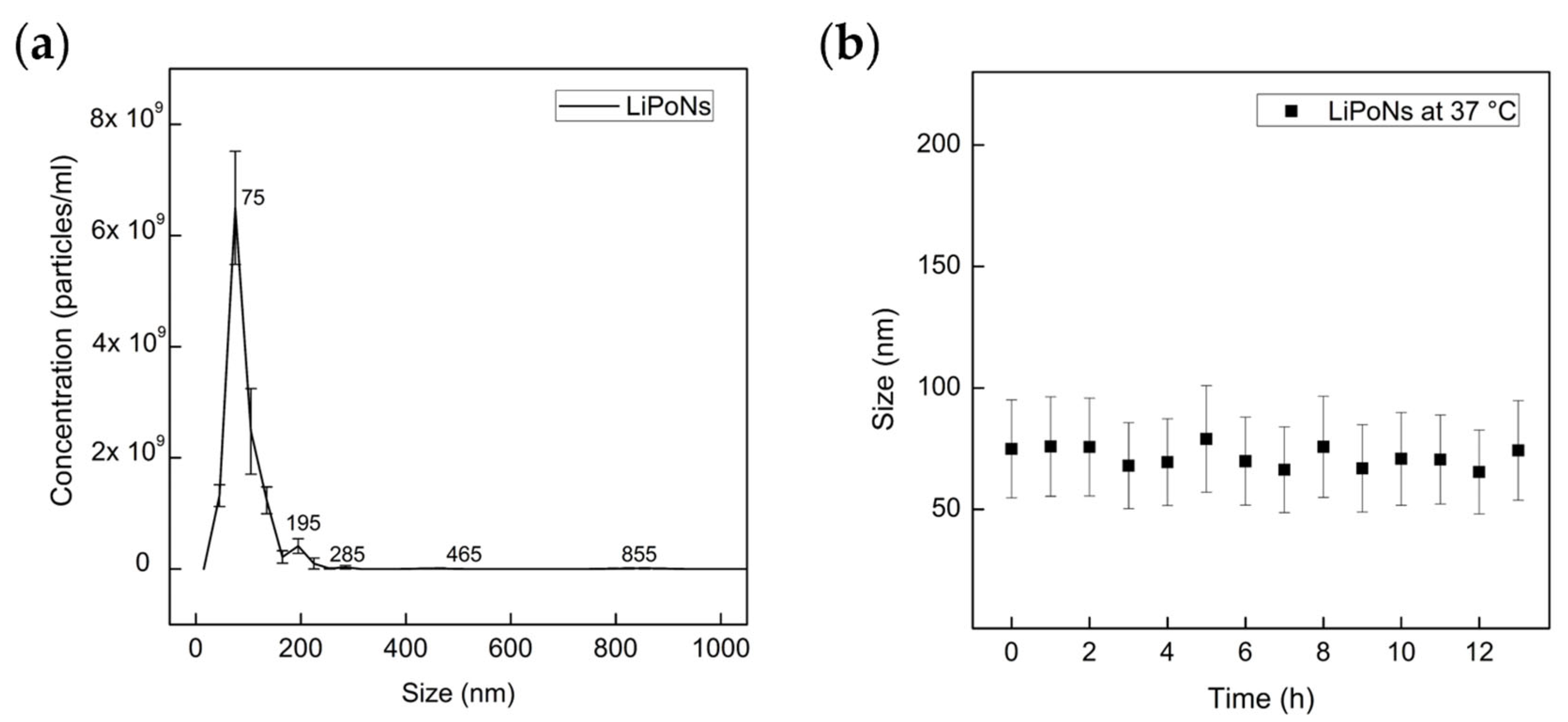
3.3. Stability Study of LiPoNs Nanoparticles
3.4. Co-Encapsulation Efficacy of Multifunctional LiPoNs
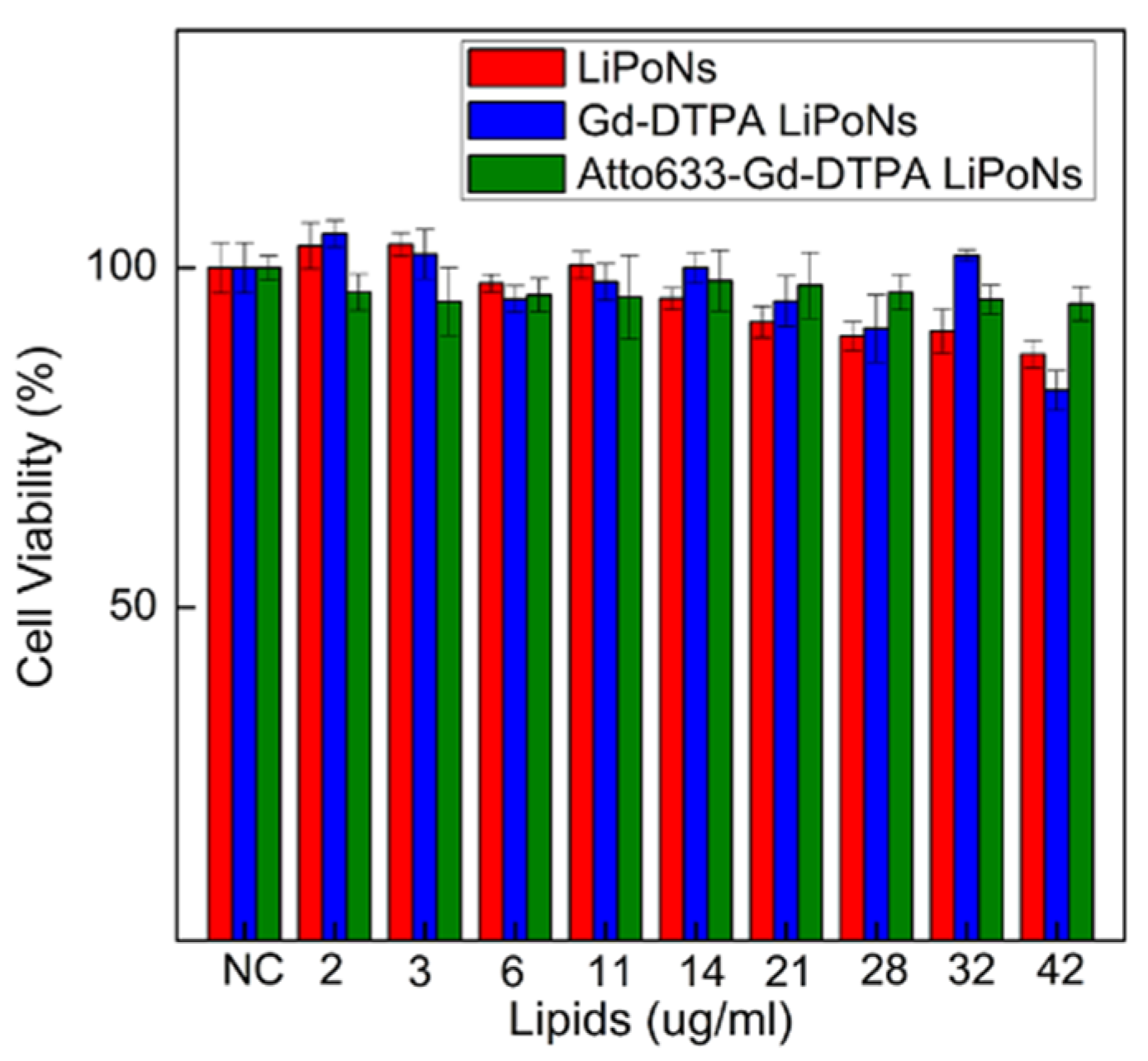
3.5. In Vitro Cytotoxicity Study
3.6. Evaluation of Cellular Uptake of Multimodal Imaging LiPoNs
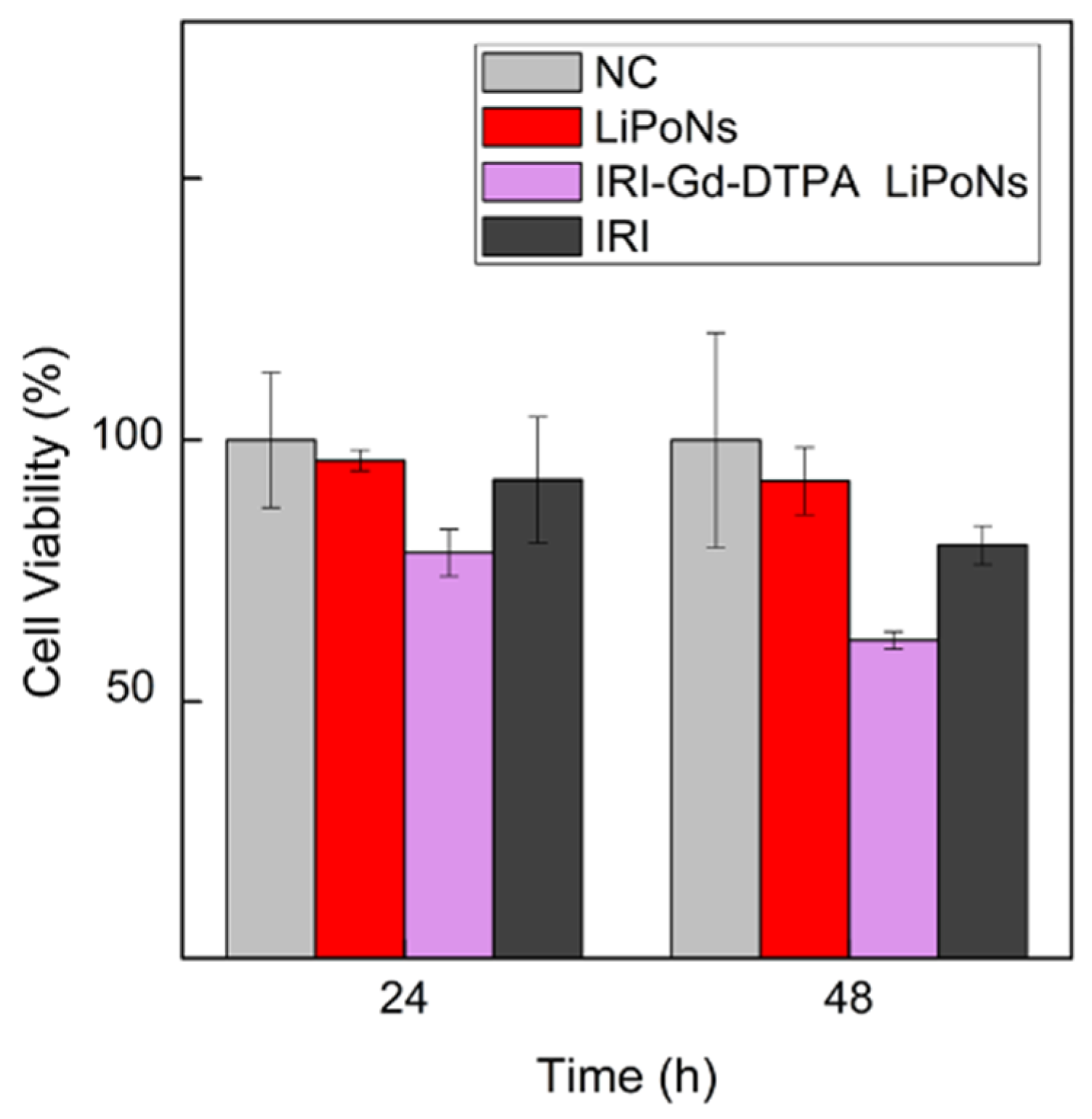
3.7. In Vitro Assessment of Cytotoxicity Activity of Theranostic IRI Gd-DTPA-Loaded LiPoNs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elvira, K.S.; Solvas, X.C.I.; Wootton, R.C.R.; deMello, A.J. The past, present and potential for microfluidic reactor technology in chemical synthesis. Nat. Chem. 2013, 5, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Q.; Ozcelik, A.; Grigsby, C.L.; Zhao, Y.H.; Guo, F.; Leong, K.W.; Huang, T.J. Microfluidic hydrodynamic focusing for synthesis of nanomaterials. Nano Today 2016, 11, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Amoyav, B.; Benny, O. Controlled and tunable polymer particles’ production using a single microfluidic device. Appl. Nanosci. 2018, 8, 905–914. [Google Scholar] [CrossRef]
- Majedi, F.S.; Hasani-Sadrabadi, M.M.; Emami, S.H.; Shokrgozar, M.A.; VanDersarl, J.J.; Dashtimoghadam, E.; Bertsch, A.; Renaud, P. Microfluidic assisted self-assembly of chitosan-based nanoparticles as drug delivery agents. Lab Chip 2013, 13, 204–207. [Google Scholar] [CrossRef]
- Russo, M.; Bevilacqua, P.; Netti, P.A.; Torino, E. A Microfluidic Platform to design crosslinked Hyaluronic Acid Nanoparticles (cHANPs) for enhanced MRI. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Smeraldo, A.; Ponsiglione, A.M.; Netti, P.A.; Torino, E. Tuning of Hydrogel Architectures by Ionotropic Gelation in Microfluidics: Beyond Batch Processing to Multimodal Diagnostics. Biomedicines 2021, 9, 1551. [Google Scholar] [CrossRef]
- Karnik, R.; Gu, F.; Basto, P.; Cannizzaro, C.; Dean, L.; Kyei-Manu, W.; Langer, R.; Farokhzad, O.C. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Lett. 2008, 8, 2906–2912. [Google Scholar] [CrossRef]
- Jahn, A.; Vreeland, W.N.; Gaitan, M.; Locascio, L.E. Controlled vesicle self-assembly in microfluidic channels with hydrodynamic focusing. J. Am. Chem. Soc. 2004, 126, 2674–2675. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Q.H.; Ma, Y.; Sun, J.S. Microfluidic Methods for Fabrication and Engineering of Nanoparticle Drug Delivery Systems. ACS Appl. Bio Mater. 2020, 3, 107–120. [Google Scholar] [CrossRef]
- Mohanty, A.; Uthaman, S.; Park, I.K. Utilization of Polymer-Lipid Hybrid Nanoparticles for Targeted Anti-Cancer Therapy. Molecules 2020, 25, 4377. [Google Scholar] [CrossRef]
- Mandal, B.; Bhattacharjee, H.; Mittal, N.; Sah, H.; Balabathula, P.; Thoma, L.A.; Wood, G.C. Core-shell-type lipid-polymer hybrid nanoparticles as a drug delivery platform. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 474–491. [Google Scholar] [CrossRef] [PubMed]
- Poon, W.; Kingston, B.R.; Ouyang, B.; Ngo, W.; Chan, W.C.W. A framework for designing delivery systems. Nat. Nanotechnol. 2020, 15, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Stavis, S.M.; Lacerda, S.H.D.; Locascio, L.E.; Raghavan, S.R.; Gaitan, M. Microfluidic Directed Self-Assembly of Liposome-Hydrogel Hybrid Nanoparticles. Langmuir 2010, 26, 11581–11588. [Google Scholar] [CrossRef] [PubMed]
- Valencia, P.M.; Basto, P.A.; Zhang, L.F.; Rhee, M.; Langer, R.; Farokhzad, O.C.; Karnik, R. Single-Step Assembly of Homogenous Lipid—Polymeric and Lipid—Quantum Dot Nanoparticles Enabled by Microfluidic Rapid Mixing. Acs Nano 2010, 4, 1671–1679. [Google Scholar] [CrossRef]
- Kim, Y.; Fay, F.; Cormode, D.P.; Sanchez-Gaytan, B.L.; Tang, J.; Hennessy, E.J.; Ma, M.M.; Moore, K.; Farokhzad, O.C.; Fisher, E.A.; et al. Single Step Reconstitution of Multifunctional High-Density Lipoprotein-Derived Nanomaterials Using Microfluidics. ACS Nano 2013, 7, 9975–9983. [Google Scholar] [CrossRef]
- Mieszawska, A.J.; Kim, Y.; Gianella, A.; van Rooy, I.; Priem, B.; Labarre, M.P.; Ozcan, C.; Cormode, D.P.; Petrov, A.; Langer, R.; et al. Synthesis of Polymer-Lipid Nanoparticles for Image-Guided Delivery of Dual Modality Therapy. Bioconjugate Chem. 2013, 24, 1429–1434. [Google Scholar] [CrossRef]
- Hadinoto, K.; Sundaresan, A.; Cheow, W.S. Lipid-polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013, 85, 427–443. [Google Scholar] [CrossRef]
- Mahzabin, A.; Das, B. A Review of Lipid-Polymer Hybrid Nanoparticles as A New Generation Drug Delivery System. Int. J. Pharm. Sci. Res. 2021, 12, 65–75. [Google Scholar] [CrossRef]
- Guimaraes, D.; Noro, J.; Loureiro, A.; Lager, F.; Renault, G.; Cavaco-Paulo, A.; Nogueira, E. Increased Encapsulation Efficiency of Methotrexate in Liposomes for Rheumatoid Arthritis Therapy. Biomedicines 2020, 8, 630. [Google Scholar] [CrossRef]
- Younis, M.A.; Tawfeek, H.M.; Abdellatif, A.A.H.; Abdel-Aleem, J.A.; Harashima, H. Clinical translation of nanomedicines: Challenges, opportunities, and keys. Adv. Drug Deliv. Rev. 2022, 181, 114083. [Google Scholar] [CrossRef]
- Hua, S.; de Matos, M.B.C.; Metselaar, J.M.; Storm, G. Current Trends and Challenges in the Clinical Translation of Nanoparticulate Nanomedicines: Pathways for Translational Development and Commercialization. Front. Pharmacol. 2018, 9, 790. [Google Scholar] [CrossRef] [PubMed]
- Tabeling, P. Introduction to Microfluidics; OUP Oxford: Oxford, UK, 2005. [Google Scholar]
- Valencia, P.M.; Farokhzad, O.C.; Karnik, R.; Langer, R. Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nat. Nanotechnol. 2012, 7, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Capretto, L.; Carugo, D.; Mazzitelli, S.; Nastruzzi, C.; Zhang, X.L. Microfluidic and lab-on-a-chip preparation routes for organic nanoparticles and vesicular systems for nanomedicine applications. Adv. Drug Deliv. Rev. 2013, 65, 1496–1532. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Yu, M.R.; Miao, Y.Q.; He, S.F.; Dai, Z.; Song, W.Y.; Liu, Y.; Song, S.; Ahmad, E.; Wang, D.K.; et al. Cholesterol-tuned liposomal membrane rigidity directs tumor penetration and anti-tumor effect. Acta Pharm. Sin. B 2019, 9, 858–870. [Google Scholar] [CrossRef]
- Coderch, L.; Fonollosa, J.; Estelrich, J.; De La Maza, A.; Parra, J.L. Influence of cholesterol on liposome fluidity by EPR—Relationship with percutaneous absorption. J. Control. Release 2000, 68, 85–95. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.N.; Li, X.D.; Chang, J. Multimodality imaging in nanomedicine and nanotheranostics. Cancer Biol. Med. 2016, 13, 339–348. [Google Scholar] [CrossRef]
- Rogosnitzky, M.; Branch, S. Gadolinium-based contrast agent toxicity: A review of known and proposed mechanisms. Biometals 2016, 29, 365–376. [Google Scholar] [CrossRef]
- Smeraldo, A.; Netti, P.A.; Torino, E. New Strategies in the Design of Paramagnetic CAs. Contrast Med. Mol. Imaging 2020, 2020, 4327479. [Google Scholar] [CrossRef]
- Marasini, R.; Nguyen, T.D.T.; Aryal, S. Integration of gadolinium in nanostructure for contrast enhanced-magnetic resonance imaging. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1580. [Google Scholar] [CrossRef]
- Cruz, L.J.; Que, I.; Aswendt, M.; Chan, A.; Hoehn, M.; Lowik, C. Targeted nanoparticles for the non-invasive detection of traumatic brain injury by optical imaging and fluorine magnetic resonance imaging. Nano Res. 2016, 9, 1276–1289. [Google Scholar] [CrossRef]
- Food and Drug Administration. FDA approves irinotecan as first-line therapy for colorectal cancer. Oncol. N. Y. 2000, 14, 652–654. [Google Scholar]
- Taghizadehghalehjoughi, A.; Hacimuftuoglu, A.; Cetin, M.; Ugur, A.B.; Galateanu, B.; Mezhuev, Y.; Okkay, U.; Taspinar, N.; Taspinar, M.; Uyanik, A.; et al. Effect of metformin/irinotecan-loaded poly-lactic-co-glycolic acid nanoparticles on glioblastoma: In vitro and in vivo studies. Nanomedicine 2018, 13, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Di Polidoro, A.C.; Zambito, G.; Haeck, J.; Mezzanotte, L.; Lamfers, M.; Netti, P.A.; Torino, E. Theranostic Design of Angiopep-2 Conjugated Hyaluronic Acid Nanoparticles (Thera-ANG-cHANPs) for Dual Targeting and Boosted Imaging of Glioma Cells. Cancers 2021, 13, 503. [Google Scholar] [CrossRef]
- Reeves, J.P.; Dowben, R.M. Formation and properties of thin-walled phospholipid vesicles. J. Cell. Physiol. 1969, 73, 49–60. [Google Scholar] [CrossRef]
- Capretto, L.; Cheng, W.; Carugo, D.; Katsamenis, O.L.; Hill, M.; Zhang, X.L. Mechanism of co-nanoprecipitation of organic actives and block copolymers in a microfluidic environment. Nanotechnology 2012, 23, 375602. [Google Scholar] [CrossRef] [PubMed]
- Jahn, A.; Vreeland, W.N.; DeVoe, D.L.; Locascio, L.E.; Gaitan, M. Microfluidic directed formation of liposomes of controlled size. Langmuir 2007, 23, 6289–6293. [Google Scholar] [CrossRef] [PubMed]
- Jahn, A.; Stavis, S.M.; Hong, J.S.; Vreeland, W.N.; Devoe, D.L.; Gaitan, M. Microfluidic Mixing and the Formation of Nanoscale Lipid Vesicles. ACS Nano 2010, 4, 2077–2087. [Google Scholar] [CrossRef]
- Abdelkarim, M.; Abd Ellah, N.H.; Elsabahy, M.; Abdelgawad, M.; Abouelmagd, S.A. Microchannel geometry vs flow parameters for controlling nanoprecipitation of polymeric nanoparticles. Colloids Surf. Physicochem. Eng. Asp. 2021, 611, 125774. [Google Scholar] [CrossRef]
- Lallana, E.; Donno, R.; Magri, D.; Barker, K.; Nazir, Z.; Treacher, K.; Lawrence, M.J.; Ashford, M.; Tirelli, N. Microfluidic-assisted nanoprecipitation of (PEGylated) poly (D,L-lactic acid-co-caprolactone): Effect of macromolecular and microfluidic parameters on particle size and paclitaxel encapsulation. Int. J. Pharm. 2018, 548, 530–539. [Google Scholar] [CrossRef]
- Donno, R.; Gennari, A.; Lallana, E.; De La Rosa, J.M.R.; d’Arcy, R.; Treacher, K.; Hill, K.; Ashford, M.; Tirelli, N. Nanomanufacturing through microfluidic-assisted nanoprecipitation: Advanced analytics and structure-activity relationships. Int. J. Pharm. 2017, 534, 97–107. [Google Scholar] [CrossRef]
- Wright, M. Nanoparticle tracking analysis for the multiparameter characterization and counting of nanoparticle suspensions. Methods Mol. Biol. 2012, 906, 511–524. [Google Scholar] [CrossRef] [PubMed]
- FDA. Liposome Drug Products Chemistry, Manufacturing, and Controls; Human Pharmacokinetics and Bioavailability; and Labeling Documentation. Guidance for Industry. 2018. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/liposome-drug-products-chemistry-manufacturing-and-controls-human-pharmacokinetics-and (accessed on 14 January 2022).
- Inglut, C.T.; Sorrin, A.J.; Kuruppu, T.; Vig, S.; Cicalo, J.; Ahmad, H.; Huang, H.C. Immunological and Toxicological Considerations for the Design of Liposomes. Nanomaterials 2020, 10, 190. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Z.; Gao, J.Q.; Lang, W.Q.; Nakagawa, S. Preparation and characterization of liposomes encapsulating chitosan nanoparticles. Biol. Pharm. Bull. 2005, 28, 387–390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hasan, M.; Ben Messaoud, G.; Michaux, F.; Tamayol, A.; Kahn, C.J.F.; Belhaj, N.; Linder, M.; Arab-Tehrany, E. Chitosan-coated liposomes encapsulating curcumin: Study of lipid-polysaccharide interactions and nanovesicle behavior. RSC Adv. 2016, 6, 45290–45304. [Google Scholar] [CrossRef]
- Yandrapati, R.K. Effect of Lipid Composition on the Physical Properties of Liposomes: A Light Scattering Study. Master’s Thesis, Missouri University of Science and Technology, Rolla, MO, USA, 2012. [Google Scholar]
- Shi, M.H.; Anantha, M.; Wehbe, M.; Bally, M.B.; Fortin, D.; Roy, L.O.; Charest, G.; Richer, M.; Paquette, B.; Sanche, L. Liposomal formulations of carboplatin injected by convection-enhanced delivery increases the median survival time of F98 glioma bearing rats. J. Nanobiotechnol. 2018, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.C.; Sahdev, P.; Ochyl, L.J.; Akerberg, J.J.; Moon, J.J. Cationic liposome-hyaluronic acid hybrid nanoparticles for intranasal vaccination with subunit antigens. J. Control. Release 2015, 208, 121–129. [Google Scholar] [CrossRef]
- Witika, B.A.; Makoni, P.A.; Matafwali, S.K.; Chabalenge, B.; Mwila, C.; Kalungia, A.C.; Nkanga, C.I.; Bapolisi, A.M.; Walker, R.B. Biocompatibility of Biomaterials for Nanoencapsulation: Current Approaches. Nanomaterials 2020, 10, 1649. [Google Scholar] [CrossRef]
- Parnham, M.J.; Wetzig, H. Toxicity screening of liposomes. Chem. Phys. Lipids 1993, 64, 263–274. [Google Scholar] [CrossRef]
- Drasler, B.; Sayre, P.; Steinhauser, K.G.; Petri-Fink, A.; Rothen-Rutishauser, B. In vitro approaches to assess the hazard of nanomaterials (vol 8, pg 99, 2017). Nanoimpact 2018, 9, 51. [Google Scholar] [CrossRef]
- Nguyen, T.L.; Nguyen, T.H.; Nguyen, D.H. Development and In Vitro Evaluation of Liposomes Using Soy Lecithin to Encapsulate Paclitaxel. Int. J. Biomater. 2017, 2017, 1–7. [Google Scholar] [CrossRef]
- Huang, M.; Khor, E.; Lim, L.Y. Uptake and cytotoxicity of chitosan molecules and nanoparticles: Effects of molecular weight and degree of deacetylation. Pharm. Res. 2004, 21, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Yang, N.; Jeon, S.K.; Yoon, T.H. Semi-quantitative Estimation of Cellular SiO2 Nanoparticles Using Flow Cytometry Combined with X-ray Fluorescence Measurements. Cytom. Part A 2014, 85A, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Salvati, A.; Aberg, C.; dos Santos, T.; Varela, J.; Pinto, P.; Lynch, I.; Dawson, K.A. Experimental and theoretical comparison of intracellular import of polymeric nanoparticles and small molecules: Toward models of uptake kinetics. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Montizaan, D.; Yang, K.N.; Reker-Smit, C.; Salvati, A. Comparison of the uptake mechanisms of zwitterionic and negatively charged liposomes by HeLa cells’. Nanomed. Nanotechnol. Biol. Med. 2020, 30, 102300. [Google Scholar] [CrossRef]
- Ducat, E.; Evrard, B.; Peulen, O.; Piel, G. Cellular uptake of liposomes monitored by confocal microscopy and flow cytometry. J. Drug Deliv. Sci. Technol. 2011, 21, 469–477. [Google Scholar] [CrossRef]
- Lichtenberg, S.S.; Tsyusko, O.V.; Palli, S.R.; Unrine, J.M. Uptake and Bioactivity of Chitosan/Double-Stranded RNA Polyplex Nanoparticles in Caenorhabditis elegans. Environ. Sci. Technol. 2019, 53, 3832–3840. [Google Scholar] [CrossRef]
- Manzanares, D.; Cena, V. Endocytosis: The Nanoparticle and Submicron Nanocompounds Gateway into the Cell. Pharmaceutics 2020, 12, 371. [Google Scholar] [CrossRef]
- Guo, P.; Liu, D.X.; Subramanyam, K.; Wang, B.R.; Yang, J.; Huang, J.; Auguste, D.T.; Moses, M.A. Nanoparticle elasticity directs tumor uptake. Nat. Commun. 2018, 9, 130. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef]
- Vredenburgh, J.J.; Desjardins, A.; Reardon, D.A.; Friedman, H.S. Experience with irinotecan for the treatment of malignant glioma. Neuro Oncol. 2009, 11, 80–91. [Google Scholar] [CrossRef]
- Casado, A.; Mora, M.; Sagrista, M.L.; Rello-Varona, S.; Acedo, P.; Stockert, J.C.; Canete, M.; Villanueva, A. Improved selectivity and cytotoxic effects of irinotecan via liposomal delivery: A comparative study on Hs68 and HeLa cells. Eur. J. Pharm. Sci. 2017, 109, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Burke, T.G.; Bom, D. Campthotecin Design and Delivery Approaches for Elevating Anti-Topoisomerase I Activities in Vivo. Ann. N. Y. Acad. Sci. 2000, 922, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Cai, R.; Chen, C.Y. The Nano-Bio Interactions of Nanomedicines: Understanding the Biochemical Driving Forces and Redox Reactions. Acc. Chem. Res. 2019, 52, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 1–12. [Google Scholar] [CrossRef]
- Anselmo, A.C.; Mitragotri, S. Impact of particle elasticity on particle-based drug delivery systems. Adv. Drug Deliv. Rev. 2017, 108, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Let’s talk about lipid nanoparticles. Nat. Rev. Mater. 2021, 6, 99. [Google Scholar] [CrossRef]
- Ouranidis, A.; Vavilis, T.; Mandala, E.; Davidopoulou, C.; Stamoula, E.; Markopoulou, C.K.; Karagianni, A.; Kachrimanis, K. mRNA Therapeutic Modalities Design, Formulation and Manufacturing under Pharma 4.0 Principles. Biomedicines 2022, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Colombo, S.; Beck-Broichsitter, M.; Becker, J.P.; Malmsten, M.; Rantanen, J.; Bohr, A. Transforming nanomedicine manufacturing toward Quality by Design and microfluidics. Adv. Drug Deliv. Rev. 2018, 128, 115–131. [Google Scholar] [CrossRef]
- Evers, M.J.W.; Kulkarni, J.A.; van der Meel, R.; Cullis, P.R.; Vader, P.; Schiffelers, R.M. State-of-the-Art Design and Rapid-Mixing Production Techniques of Lipid Nanoparticles for Nucleic Acid Delivery. Small Methods 2018, 2, 1700375. [Google Scholar] [CrossRef]
- D’Addio, S.M.; Prud’homme, R.K. Controlling drug nanoparticle formation by rapid precipitation. Adv. Drug Deliv. Rev. 2011, 63, 417–426. [Google Scholar] [CrossRef]
- Marsh, D. Thermodynamics of Phospholipid Self-Assembly. Biophys. J. 2012, 102, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.S.; Chow, S.F.; Zheng, Y. Application of flash nanoprecipitation to fabricate poorly water-soluble drug nanoparticles. Acta Pharm. Sin. B 2019, 9, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, D.; Grimaldi, A.M.; Forte, E.; Bevilacqua, P.; Netti, P.A.; Torino, E. Hybrid Core-Shell (HyCoS) Nanoparticles produced by Complex Coacervation for Multimodal Applications. Sci. Rep. 2017, 7, 45121. [Google Scholar] [CrossRef] [PubMed]
- Patil, Y.P.; Jadhav, S. Novel methods for liposome preparation. Chem. Phys. Lipids 2014, 177, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Antonietti, M.; Forster, S. Vesicles and liposomes: A self-assembly principle beyond lipids. Adv. Mater. 2003, 15, 1323–1333. [Google Scholar] [CrossRef]
- Derek, M. Handbook of Lipid Bilayers; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Israelachvili, J.N. Intermolecular and Surface Forces, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 1–674. [Google Scholar]
- Nagarajan, R. Molecular packing parameter and surfactant self-assembly: The neglected role of the surfactant tail. Langmuir 2002, 18, 31–38. [Google Scholar] [CrossRef]
- Lasic, D.D. The Mechanism of Vesicle Formation. Biochem. J. 1988, 256, 1–11. [Google Scholar] [CrossRef]
- Leng, J.; Egelhaaf, S.U.; Cates, M.E. Kinetic pathway of spontaneous vesicle formation. Europhys. Lett. 2002, 59, 311–317. [Google Scholar] [CrossRef]
- Kotoucek, J.; Hubatka, F.; Masek, J.; Kulich, P.; Velinska, K.; Bezdekova, J.; Fojtikova, M.; Bartheldyova, E.; Tomeckova, A.; Straska, J.; et al. Preparation of nanoliposomes by microfluidic mixing in herring-bone channel and the role of membrane fluidity in liposomes formation. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Jahn, A.; Lucas, F.; Wepf, R.A.; Dittrich, P.S. Freezing Continuous-Flow Self-Assembly in a Microfluidic Device: Toward Imaging of Liposome Formation. Langmuir 2013, 29, 1717–1723. [Google Scholar] [CrossRef]
- Tazina, E.V.; Kostin, K.V.; Oborotova, N.A. Specific Features of Drug Encapsulation in Liposomes (A Review). Pharm. Chem. J. 2011, 45, 481–490. [Google Scholar] [CrossRef]
- Gabba, M.; Frallicciardi, J.; van Klooster, J.; Henderson, R.; Syga, L.; Mans, R.; van Maris, A.J.A.; Poolman, B. Weak Acid Permeation in Synthetic Lipid Vesicles and Across the Yeast Plasma Membrane. Biophys. J. 2020, 118, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, N.; Martins, A.; Reis, R.L.; Neves, N.M. Liposomes in tissue engineering and regenerative medicine. J. R. Soc. Interface 2014, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.; Blume, A. A Comparative-Study of Diffusive and Osmotic Water Permeation Across Bilayers Composed of Phospholipids with Different Head Groups and Fatty Acyl Chains. Biophys. J. 1995, 68, 997–1008. [Google Scholar] [CrossRef]




| FR2 | Average Size (nm) | PDI | Zeta Potential (mV) | EE Gd-DTPA (%) | Co-EE Active Agent (%) | |
|---|---|---|---|---|---|---|
| LiPoNs | 0.073 | 77.4 | 0.22 | −17.4 | / | / |
| Gd-DTPA LiPoNs | 95.3 | 0.30 | −11 | 78% | / | |
| Atto633 LiPoNs | / | / | −3.7 | / | 57% | |
| Atto633-Gd-DTPA LiPoNs | / | / | −10.9 | 67% | 55% | |
| IRI-Gd-DTPA LiPoNs | 112.8 | 0.28 | −14.2 | 79% | 64% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roffo, F.; Ponsiglione, A.M.; Netti, P.A.; Torino, E. coupled Hydrodynamic Flow Focusing (cHFF) to Engineer Lipid–Polymer Nanoparticles (LiPoNs) for Multimodal Imaging and Theranostic Applications. Biomedicines 2022, 10, 438. https://doi.org/10.3390/biomedicines10020438
Roffo F, Ponsiglione AM, Netti PA, Torino E. coupled Hydrodynamic Flow Focusing (cHFF) to Engineer Lipid–Polymer Nanoparticles (LiPoNs) for Multimodal Imaging and Theranostic Applications. Biomedicines. 2022; 10(2):438. https://doi.org/10.3390/biomedicines10020438
Chicago/Turabian StyleRoffo, Felicia, Alfonso Maria Ponsiglione, Paolo Antonio Netti, and Enza Torino. 2022. "coupled Hydrodynamic Flow Focusing (cHFF) to Engineer Lipid–Polymer Nanoparticles (LiPoNs) for Multimodal Imaging and Theranostic Applications" Biomedicines 10, no. 2: 438. https://doi.org/10.3390/biomedicines10020438
APA StyleRoffo, F., Ponsiglione, A. M., Netti, P. A., & Torino, E. (2022). coupled Hydrodynamic Flow Focusing (cHFF) to Engineer Lipid–Polymer Nanoparticles (LiPoNs) for Multimodal Imaging and Theranostic Applications. Biomedicines, 10(2), 438. https://doi.org/10.3390/biomedicines10020438