Hyperspectral Imaging during Normothermic Machine Perfusion—A Functional Classification of Ex Vivo Kidneys Based on Convolutional Neural Networks
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Normothermic Machine Perfusion
2.3. Hyperspectral Imaging System
2.4. HSI Data Acquisition and Data Correction
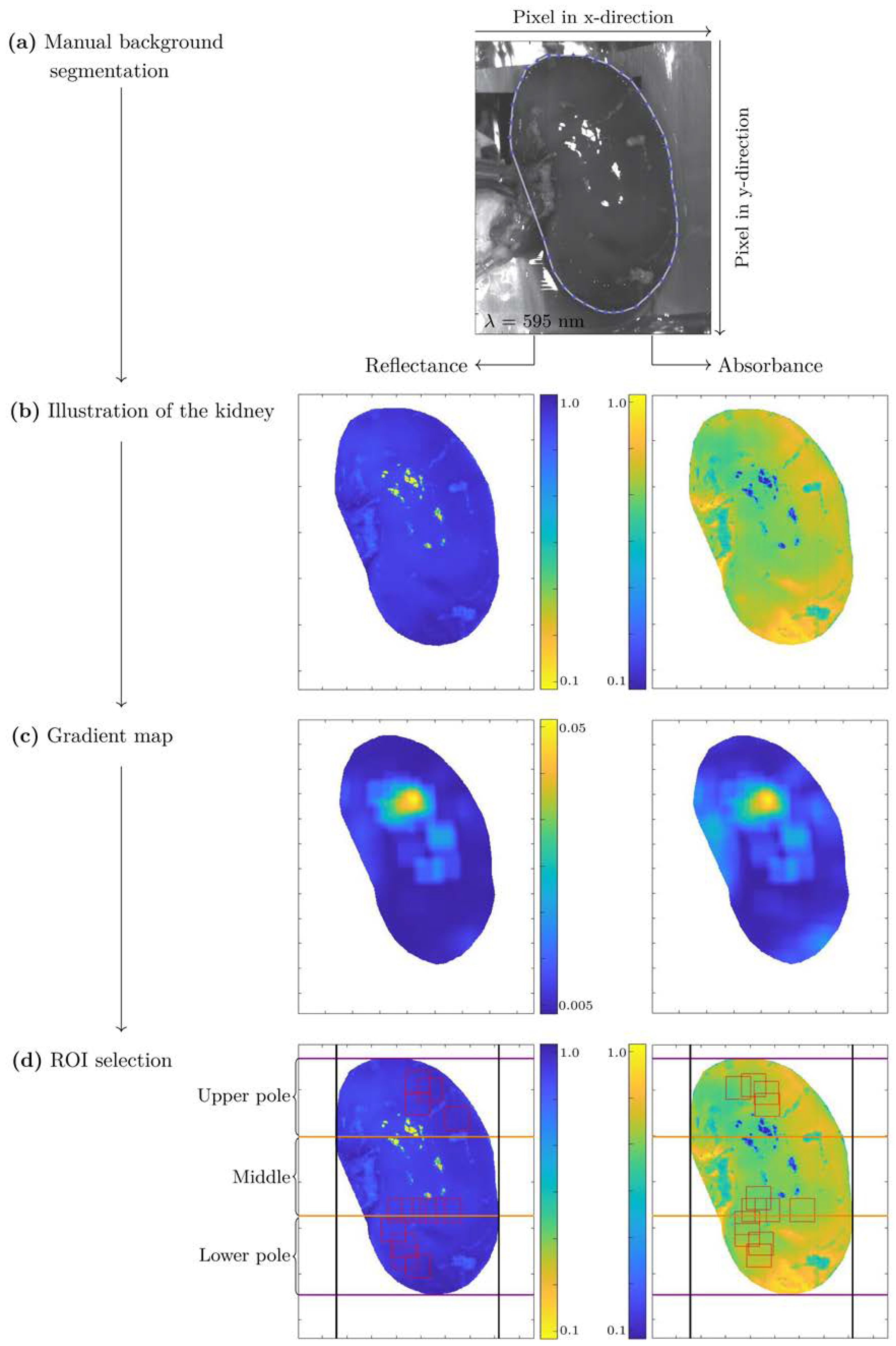
2.5. HSI Data Preprocessing
- (1)
- Manual background segmentation,
- (2)
- Wavelength range selection (550 nm–995 nm),
- (3)
- Automated region of interest (ROI) selection,
- (4)
- Vector normalization,
- (5)
- Savitzky-Golay smoothing.
2.6. Data Set
2.7. Normalized Cross Correlation
2.8. CNN Model Architecture and Optimization
2.9. Validation Strategy
2.10. KidneyResNet Model Evaluation
3. Results
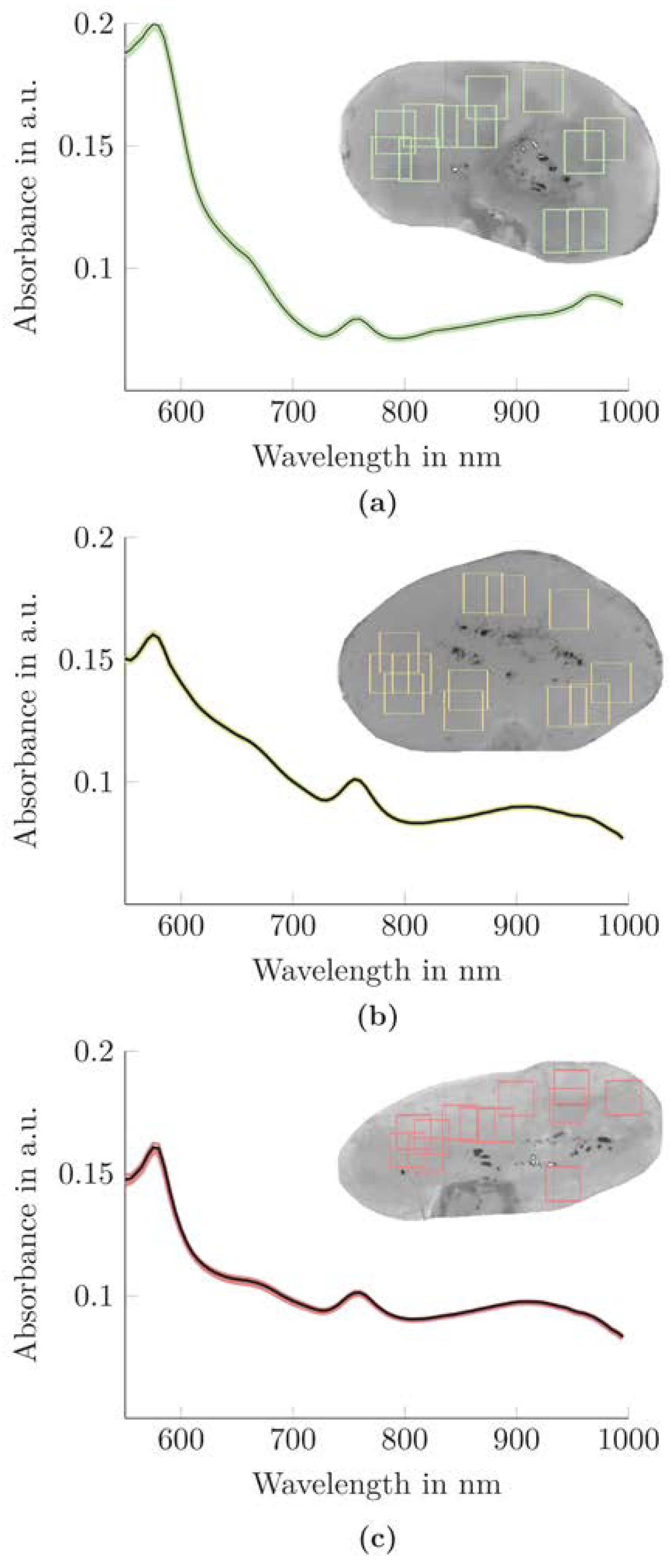
3.1. Spectra Comparison of the Kidneys According to the Pig Characteristics
3.2. Spectral Properties of the Kidneys According to the Functional Classes
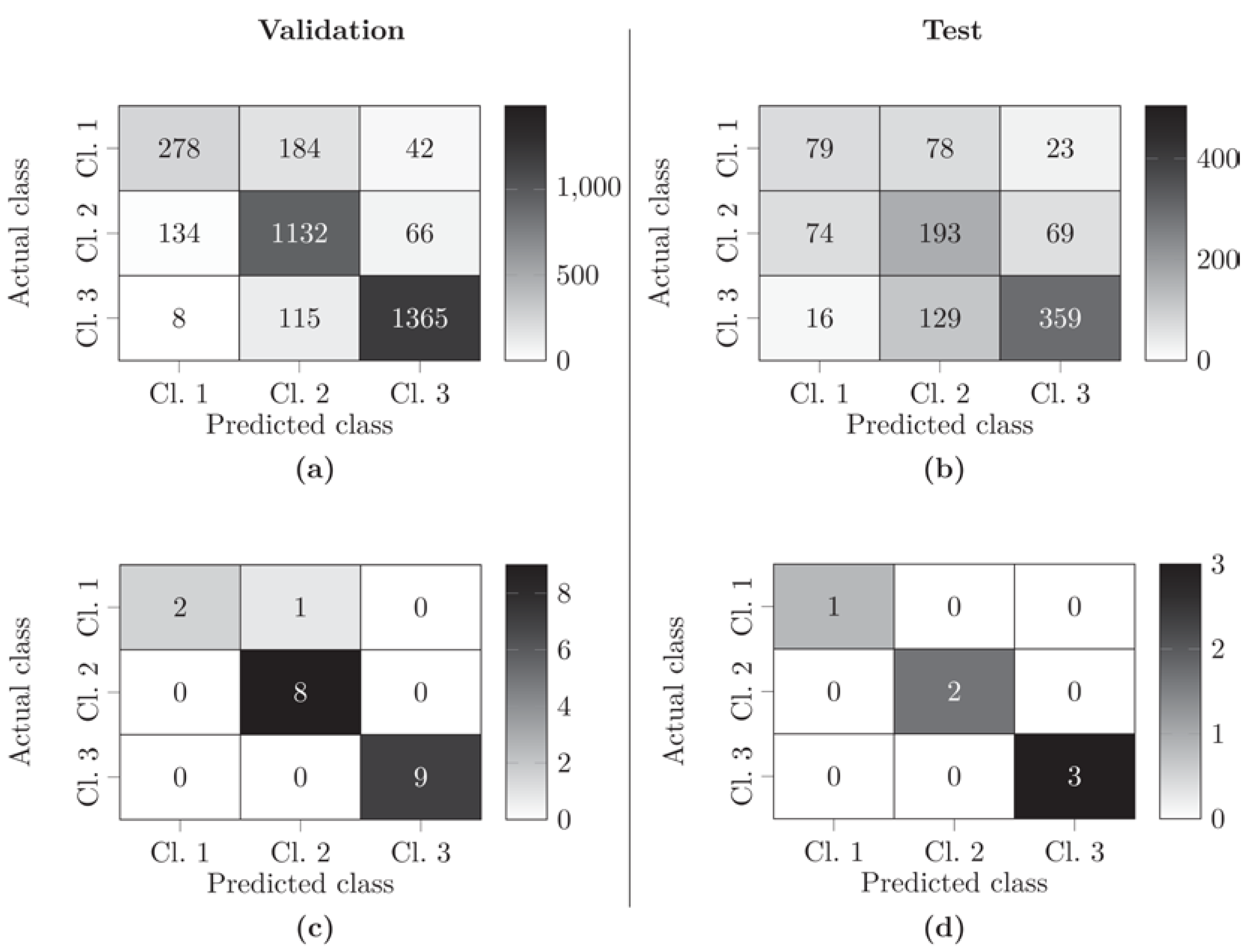
3.3. Optimization of Classification of ROIs Using the KidneyResNet Model
3.4. Classification of Kidneys Using the KidneyResNet Model
4. Discussion
4.1. Organ Classification Methods
4.2. ROI Selection for Preprocessing of HSI Data
4.3. CNN Model Architecture and Optimization Methods
4.4. Analysis of Exclusively Tissue-Specific Data Enables Functional Evaluation of Kidneys
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romagnani, P.; Remuzzi, G.; Glassock, R.; Levin, A.; Jager, K.J.; Tonelli, M.; Massy, Z.; Wanner, C.; Anders, H.-J. Chronic kidney disease. Nat. Rev. Dis. Primers 2017, 3, 17088. [Google Scholar] [CrossRef]
- Branger, P.; Vogelaar, S. Annual Report 2020; Eurotransplant International Foundation: Leiden, The Netherlands, 2021. [Google Scholar]
- Port, F.K.; Bragg-Gresham, J.L.; Metzger, R.A.; Dykstra, D.M.; Gillespie, B.W.; Young, E.W.; Delmonico, F.L.; Wynn, J.J.; Merion, R.M.; Wolfe, R.A.; et al. Donor characteristics associated with reduced graft survival: An approach to expanding the pool of donor kidneys. Transplantation 2002, 74, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Remuzzi, G.; Grinyò, J.; Ruggenenti, P.; Beatini, M.; Cole, E.H.; Milford, E.L.; Brenner, B.M. Early experience with dual kidney transplantation in adults using expanded donor criteria. JASN 1999, 10, 2591–2598. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.S.; Schaubel, D.E.; Guidinger, M.K.; Andreoni, K.A.; Wolfe, R.A.; Merion, R.M.; Port, F.K.; Sung, R.S. A comprehensive risk quantification score for deceased donor kidneys: The kidney donor risk index. Transplantation 2009, 88, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Stallone, G.; Grandaliano, G. To discard or not to discard: Transplantation and the art of scoring. CKJ 2019, 12, 564–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moeckli, B.; Sun, P.; Lazeyras, F.; Morel, P.; Moll, S.; Pascual, M.; Bühler, L.H. Evaluation of donor kidneys prior to transplantation: An update of current and emerging methods. Transpl. Int. 2019, 32, 459–469. [Google Scholar] [CrossRef]
- Kaths, J.M.; Paul, A.; Robinson, L.A.; Selzner, M. Ex vivo machine perfusion for renal graft preservation. Transplant. Rev. 2018, 32, 1–9. [Google Scholar] [CrossRef]
- Resch, T.; Cardini, B.; Oberhuber, R.; Weissenbacher, A.; Dumfarth, J.; Krapf, C.; Boesmueller, C.; Oefner, D.; Grimm, M.; Schneeberger, S. Transplanting Marginal Organs in the Era of Modern Machine Perfusion and Advanced Organ Monitoring. Front. Immunol. 2020, 11, 631. [Google Scholar] [CrossRef]
- Markgraf, W.; Janssen, M.W.W.; Lilienthal, J.; Feistel, P.; Thiele, C.; Stöckle, M.; Malberg, H. Hyperspectral imaging for ex-vivo organ characterization during normothermic machine perfusion. Eur. Urol. Suppl. 2018, 17, e767. [Google Scholar] [CrossRef]
- De Beule, J.; Jochmans, I. Kidney Perfusion as an Organ Quality Assessment Tool-Are We Counting Our Chickens Before They Have Hatched? J. Clin. Med. 2020, 9, 879. [Google Scholar] [CrossRef] [Green Version]
- Kaths, J.M.; Hamar, M.; Echeverri, J.; Linares, I.; Urbanellis, P.; Cen, J.Y.; Ganesh, S.; Dingwell, L.S.; Yip, P.; John, R.; et al. Normothermic ex vivo kidney perfusion for graft quality assessment prior to transplantation. Am. J. Transplant. 2018, 18, 580–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Thompson, E.; Bates, L.; Pither, T.L.; Hosgood, S.A.; Nicholson, M.L.; Watson, C.J.E.; Wilson, C.; Fisher, A.J.; Ali, S.; et al. Flavin mononucleotide as a biomarker of organ quality—A pilot study. Transplant. Direct 2020, 6, e600. [Google Scholar] [CrossRef] [PubMed]
- Woud, W.W.; Merino, A.; Hoogduijn, M.J.; Boer, K.; van den Hoogen, M.W.F.; Baan, C.C.; Minnee, R.C. Nanoparticle release by extended criteria donor kidneys during normothermic machine perfusion. Transplantation 2019, 103, e110–e111. [Google Scholar] [CrossRef] [PubMed]
- Hosgood, S.A.; Barlow, A.D.; Hunter, J.P.; Nicholson, M.L. Ex vivo normothermic perfusion for quality assessment of marginal donor kidney transplants. Br. J. Surg. 2015, 102, 1433–1440. [Google Scholar] [CrossRef]
- Hosgood, S.A.; Thompson, E.; Moore, T.; Wilson, C.H.; Nicholson, M.L. Normothermic machine perfusion for the assessment and transplantation of declined human kidneys from donation after circulatory death donors. Br. J. Surg. 2018, 105, 388–394. [Google Scholar] [CrossRef] [Green Version]
- Hosgood, S.A.; Saeb-Parsy, K.; Hamed, M.O.; Nicholson, M.L. Successful transplantation of human kidneys deemed untransplantable but resuscitated by ex vivo normothermic machine perfusion. Am. J. Transplant. 2016, 16, 3282–3285. [Google Scholar] [CrossRef] [Green Version]
- Markgraf, W.; Mühle, R.; Lilienthal, L.; Kromnik, S.; Thiele, C.; Malberg, H.; Janssen, M.; Putz, J. Inulin Clearance during ex vivo Normothermic Machine Perfusion as a Marker of Renal Function. ASAIO J. 2021. [Google Scholar] [CrossRef]
- Lu, G.; Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 2014, 19, 10901. [Google Scholar] [CrossRef]
- Goto, A.; Nishikawa, J.; Kiyotoki, S.; Nakamura, M.; Nishimura, J.; Okamoto, T.; Ogihara, H.; Fujita, Y.; Hamamoto, Y.; Sakaida, I. Use of hyperspectral imaging technology to develop a diagnostic support system for gastric cancer. J. Biomed. Opt. 2015, 20, 16017. [Google Scholar] [CrossRef]
- Fei, B.; Lu, G.; Wang, X.; Zhang, H.; Little, J.V.; Patel, M.R.; Griffith, C.C.; El-Diery, M.W.; Chen, A.Y. Label-free reflectance hyperspectral imaging for tumor margin assessment: A pilot study on surgical specimens of cancer patients. J. Biomed. Opt. 2017, 22, 86009. [Google Scholar] [CrossRef] [Green Version]
- Mühle, R.; Ernst, H.; Sobottka, S.; Morgenstern, U. Workflow and hardware for intraoperative hyperspectral data acquisition in neurosurgery. Biomed. Eng./Biomed. Tech. 2020, 66, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Bjorgan, A.; Randeberg, L.L. Estimation of skin optical parameters for real-time hyperspectral imaging applications. J. Biomed. Opt. 2014, 19, 066003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancio, L.C.; Batchinsky, A.I.; Mansfield, J.R.; Panasyuk, S.; Hetz, K.; Martini, D.; Jordan, B.S.; Tracey, B.; Freeman, J.E. Hyperspectral imaging: A new approach to the diagnosis of hemorrhagic shock. J. Trauma Acute Care Surg. 2006, 60, 1087–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markgraf, W.; Feistel, P.; Thiele, C.; Malberg, H. Algorithms for mapping kidney tissue oxygenation during normothermic machine perfusion using hyperspectral imaging. Biomed. Eng./Biomed. Tech. 2018, 63, 557–566. [Google Scholar] [CrossRef]
- Beach, J.; Ning, J.; Khoobehi, B. Oxygen saturation in optic nerve head structures by hyperspectral image analysis. Curr. Eye Res. 2007, 32, 161–170. [Google Scholar] [CrossRef]
- Markgraf, W.; Lilienthal, J.; Feistel, P.; Thiele, C.; Malberg, H. Algorithm for mapping kidney tissue water content during normothermic machine perfusion using hyperspectral imaging. Algorithms 2020, 13, 289. [Google Scholar] [CrossRef]
- Dietrich, M.; Marx, S.; von der Forst, M.; Bruckner, T.; Schmitt, F.C.F.; Fiedler, M.O.; Nickel, F.; Studier-Fischer, A.; Müller-Stich, B.P.; Hackert, T.; et al. Hyperspectral imaging for perioperative monitoring of microcirculatory tissue oxygenation and tissue water content in pancreatic surgery—An observational clinical pilot study. Perioper. Med. 2021, 10, 42. [Google Scholar] [CrossRef]
- Mühle, R.; Markgraf, W.; Hilsmann, A.; Malberg, H.; Eisert, P.; Wisotzky, E.L. Comparison of different spectral cameras for image-guided organ transplantation. J. Biomed. Opt. 2021, 26, 076007. [Google Scholar] [CrossRef]
- Sucher, R.; Wagner, T.; Köhler, H.; Sucher, E.; Guice, H.; Recknagel, S.; Lederer, A.; Hau, H.M.; Rademacher, S.; Schneeberger, S.; et al. Hyperspectral imaging (HSI) of human kidney allografts. Ann. Surg. 2020. In Print. [Google Scholar] [CrossRef]
- Holmer, A.; Tetschke, F.; Marotz, J.; Malberg, H.; Markgraf, W.; Thiele, C.; Kulcke, A. Oxygenation and perfusion monitoring with a new hyperspectral camera system for chemical based tissue analysis. Physiol. Meas. 2016, 37, 2064–2078. [Google Scholar] [CrossRef]
- Tetschke, F.; Markgraf, W.; Gransow, M.; Koch, S.; Thiele, C.; Kulcke, A.; Malberg, H. Hyperspectral imaging for monitoring oxygen saturation levels during normothermic kidney perfusion. J. Sens. Sens. Syst. 2016, 5, 313–318. [Google Scholar] [CrossRef] [Green Version]
- Markgraf, W.; Lilienthal, J.; Feistel, P.; Mühle, R.; Thiele, C.; Malberg, H.; Janssen, M. Hyperspectral imaging of porcine kidneys during normothermic ex vivo perfusion—An analysis of tissue-related renal ischemia injury. Transpl. Int. 2020, 33, 16–36. [Google Scholar] [CrossRef]
- Fodor, F.; Hofmann, J.; Lanser, L.; Otarashvili, G.; Pühringer, M.; Hautz, T.; Sucher, R.; Schneeberger, S. Hyperspectral Imaging and Machine Perfusion in Solid Organ Transplantation: Clinical Potentials of Combining Two Novel Technologies. J. Clin. Med. 2021, 10, 3838. [Google Scholar] [CrossRef] [PubMed]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep learning meets hyperspectral image analysis: A multidisciplinary review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Ortega, S.; Halicek, M.; Fabelo, H.; Guerra, R.; Lopez, C.; Lejaune, M.; Godtliebsen, F.; Callico, G.M.; Fei, B. Hyperspectral imaging and deep learning for the detection of breast cancer cells in digitized histological images. Proc. SPIE Int. Soc. Opt. Eng. 2020, 11320–113200V. [Google Scholar] [CrossRef]
- Hu, B.; Du, J.; Zhang, Z.; Wang, Q. Tumor tissue classification based on microhyperspectral technology and deep learning. Biomed. Opt. Express 2019, 10, 6370. [Google Scholar] [CrossRef]
- Halicek, M.; Little, J.V.; Wang, X.; Patel, M.; Griffith, C.C.; Chen, A.Y.; Fei, B. Tumor margin classification of head and neck cancer using hyperspectral imaging and convolutional neural networks. Proc. SPIE Int. Soc. Opt. Eng. 2018, 10576, 1057605. [Google Scholar] [CrossRef]
- Fabelo, H.; Halicek, M.; Ortega, S.; Szolna, A.; Morera, J.; Sarmiento, R.; Callico, G.M.; Fei, B. Surgical aid visualization system for glioblastoma tumor identification based on deep learning and in-vivo hyperspectral images of human patients. Proc. SPIE Int. Soc. Opt. Eng. 2019, 10951, 1095110. [Google Scholar] [CrossRef]
- Baumann, K.; Clerc, J.T. Computer-assisted IR spectra prediction-linked similarity searches for structures and spectra. Anal. Chim. Acta 1997, 348, 327–343. [Google Scholar] [CrossRef]
- Hinkle, D.E.; Wiersma, W.; Jurs, S.G. Applied Statistics for the Behavioral Sciences, 5th ed.; Houghton Mifflin: Boston, MA, USA, 2003. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef] [Green Version]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G. 2016. Available online: https://github.com/pytorch (accessed on 20 December 2021).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization, in: ICLR’15, San Diego. arXiv 2015, arXiv:1412.6980. [Google Scholar]
- Perk, S.; Izamis, M.-L.; Tolboom, H.; Uygun, B.; Yarmush, M.L.; Uygun, K. A fitness index for transplantation of perfused DCD rat livers. BMC Res. Notes 2012, 5, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruinsma, B.G.; Sridharan, G.V.; Weeder, P.D.; Avruch, J.H.; Saeidi, N.; Özer, S.; Geerts, S.; Porte, R.J.; Heger, M.; van Gulik, T.M.; et al. Metabolic profiling during ex vivo machine perfusion of the human liver. Sci. Rep. 2016, 6, 22415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Vekemans, K.; Iania, L.; Komuta, M.; Parkkinen, J.; Heedfeld, V.; Wylin, T.; Monbaliu, D.; Pirenne, J.; van Pelt, J. Assessing warm ischemic injury of pig livers at hypothermic machine perfusion. J. Surg. Res. 2014, 186, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Qureshi, S.A. A review of the medical hyperspectral imaging systems and unmixing algorithms’ in biological tissues. Photodiagnosis Photodyn. Ther. 2021, 33, 102165. [Google Scholar] [CrossRef]
- Halicek, M.; Fabelo, H.; Ortega, S.; Little, J.V.; Wang, X.; Chen, A.Y.; Callico, G.M.; Myers, L.; Sumer, B.D.; Fei, B. Hyperspectral imaging for head and neck cancer detection: Specular glare and variance of the tumor margin in surgical specimens. J. Med. Imaging 2019, 6, 035004. [Google Scholar] [CrossRef]
- Kho, E.; Dashtbozorg, B.; de Boer, L.L.; Van de Vijver, K.K.; Sterenborg, H.J.C.M.; Ruers, T.J.M. Broadband hyperspectral imaging for breast tumor detection using spectral and spatial information. Biomed. Opt. Express 2019, 10, 4496–4515. [Google Scholar] [CrossRef] [Green Version]
- Jeyaraj, P.R.; Samuel Nadar, E.R. Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. J. Cancer Res. Clin. Oncol. 2019, 145, 829–837. [Google Scholar] [CrossRef]
- Grigoroiu, A.; Yoon, J.; Bohndiek, S.E. Deep learning applied to hyperspectral endoscopy for online spectral classification. Sci. Rep. 2020, 10, 3947. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; He, Y.; Yao, C.; Wang, S.; Xue, Y.; Zhang, Z.; Wang, J.; Liu, X. Classification and Segmentation of Hyperspectral Data of Hepatocellular Carcinoma Samples Using 1-D Convolutional Neural Network. Cytom. Part A 2020, 97A, 31–38. [Google Scholar] [CrossRef]
- Huang, Q.; Li, W.; Zhang, B.; Li, Q.; Tao, R.; Lovell, N.H. Blood Cell Classification Based on Hyperspectral Imaging with Modulated Gabor and CNN. IEEE J. Biomed. Health Inform. 2020, 24, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Lasch, P.; Stämmler, M.; Zhang, M.; Baranska, M.; Bosch, A.; Majzner, K. FT-IR Hyperspectral Imaging and Artificial Neural Network Analysis for Identification of Pathogenic Bacteria. Anal. Chem. 2018, 90, 8896–8904. [Google Scholar] [CrossRef]
- Noor, S.S.M.; Michael, K.; Marshall, S.; Ren, J. Hyperspectral image enhancement and mixture deep-learning classification of corneal epithelium injuries. Sensors 2017, 17, 2644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhang, F.; Yu, J.; Chen, X.; Liu, B.; Meng, X. A rapid and non-destructive detection of Escherichia coli on the surface of fresh-cut potato slices and application using hyperspectral imaging. Postharvest Biol. Technol. 2021, 171, 111352. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, J.; Ma, L.; Jiang, H.; Zhao, H. Deep residual networks for hyperspectral image classification. IGARSS 2017, 1824–1827. [Google Scholar] [CrossRef] [Green Version]
| Pig and Kidney Characteristics | Class 1 | Class 2 | Class 3 |
|---|---|---|---|
| Overall | n = 4 | n = 10 | n = 12 |
| Female:Male | 3:1 | 4:6 | 4:8 |
| German Landrace:Swabian Hall | 3:1 | 4:6 | 4:8 |
| Warm ischemia time in min | 122 ± 52 | 67 ± 30 | 12 ± 7 |
| Cold ischemia time in min | 404 ± 246 | 402 ± 201 | 312 ± 170 |
| GFR in mL/min/100 g | 1.3 ± 0.6 | 3.0 ± 0.9 | 14.8 ± 10.1 |
| Ie,total in % | 45 ± 1 | 72 ± 9 | 97 ± 3 |
| Training and validation | n = 3 | n = 8 | n = 9 |
| Female:Male | 2:1 | 3:5 | 2:7 |
| German Landrace:Swabian Hall | 2:1 | 3:5 | 2:7 |
| Warm ischemia time in min | 114 ± 61 | 66 ± 34 | 12 ± 7 |
| Cold ischemia time in min | 341 ± 259 | 397 ± 233 | 305 ± 125 |
| GFR in mL/min/100 g | 1.5 ± 0.8 | 2.9 ± 0.8 | 16.4 ± 10.4 |
| Ie,total in % | 45 ± 1 | 72 ± 9 | 97 ± 3 |
| Test | n = 1 | n = 2 | n = 3 |
| Female:Male | 1:0 | 1:1 | 2:1 |
| German Landrace:Swabian Hall | 1:0 | 1:1 | 2:1 |
| Warm ischemia time in min | 145 | 73 ± 10 | 13 ± 8 |
| Cold ischemia time in min | 594 | 420 ± 121 | 333 ± 309 |
| GFR in mL/min/100 g | 0.8 | 3.4 ± 1.4 | 10.0 ± 9.3 |
| Ie,total in % | 44 | 75 ± 12 | 98 ± 1 |
| Data Origin/ | Code | Variant | ||
|---|---|---|---|---|
| Training Parameter | I | II | III | |
| Data origin | A | Absorbance | Reflectance | |
| Dropout rate in % | B | 0 | 25 | 50 |
| Weight decay | C | 0 | 0.0005 | |
| Learning rate | D | 0 | 0.11 |
| Data Augmentation | Code | Variant | |||
|---|---|---|---|---|---|
| Method | I | II | III | IV | |
| Rotation | A | 0° | 90° | 180° | 270° |
| Gaussian noise, 3σ | B | 0 | 0.00625 | ||
| Random occlusion in % | C | 0 | 25 | ||
| Nr. | Race | Sex | Body Weight in kg | Warm Ischemia Time in min | Cold Ischemia Time in min |
|---|---|---|---|---|---|
| 1 | Swabian Hall | Male | 40 ± 5 | 20 | 136 |
| 2 | German Landrace | Female | 40 ± 5 | 20 | 221 |
| 3 | German Landrace | Female | 80 ± 3 | 25 | 343 |
| 4 | Swabian Hall | Male | 40 ± 5 | 60 | 277 |
| 5 | German Landrace | Female | 40 ± 5 | 118 | 463 |
| 6 | German Landrace | Female | 80 ± 3 | 80 | 334 |
| Comparison | Pearson Correlation Coefficient | Comparison | Pearson Correlation Coefficient |
|---|---|---|---|
| 1 vs. 2 | 0.991 | 4 vs. 5 | 0.997 |
| 1 vs. 3 | 0.998 | 4 vs. 6 | 0.995 |
| 2 vs. 3 | 0.993 | 5 vs. 6 | 0.998 |
| Model No. | Parameter | Median Early Stopping Epoch | Validation Mean Accuracy | |||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| 1 | I | I | I | I | 9 | 0.80 |
| 2 | I | I | I | II | 2 | 0.72 |
| 3 | I | I | II | I | 9 | 0.85 |
| 4 | I | I | II | II | 5 | 0.77 |
| 5 | I | II | I | I | 13 | 0.82 |
| 6 | I | II | I | II | 4 | 0.69 |
| 7 | I | II | II | I | 12 | 0.81 |
| 8 | I | II | II | II | 3 | 0.79 |
| 9 | I | III | I | I | 11 | 0.84 |
| 10 | I | III | I | II | 7 | 0.84 |
| 11 | I | III | II | I | 6 | 0.75 |
| 12 | I | III | II | II | 6 | 0.78 |
| 13 | II | I | I | I | 8 | 0.83 |
| 14 | II | I | I | II | 5 | 0.69 |
| 15 | II | I | II | I | 15 | 0.82 |
| 16 | II | I | II | II | 4 | 0.72 |
| 17 | II | II | I | I | 9 | 0.79 |
| 18 | II | II | I | II | 4 | 0.71 |
| 19 | II | II | II | I | 12 | 0.81 |
| 20 | II | II | II | II | 3 | 0.74 |
| 21 | II | III | I | I | 10 | 0.79 |
| 22 | II | III | I | II | 3 | 0.69 |
| 23 | II | III | II | I | 12 | 0.81 |
| 24 | II | III | II | II | 4 | 0.73 |
| Model No. | Parameter | Median Early Stopping Epoch | Validation Mean Accuracy | ||
|---|---|---|---|---|---|
| A | B | C | |||
| Input KidneyResNet model configuration: model No. 3, Table 6 | |||||
| 1 | I–IV | I | I | 7 | 0.85 |
| 2 | I | II | I | 5 | 0.73 |
| 3 | I | I | II | 6 | 0.83 |
| Input KidneyResNet model configuration: model No. 9, Table 6 | |||||
| 4 | I–IV | I | I | 10 | 0.81 |
| 5 | I | II | I | 10 | 0.84 |
| 6 | I | I | II | 10 | 0.79 |
| Input KidneyResNet model configuration: model No. 10, Table 6 | |||||
| 7 | I–IV | I | I | 3 | 0.65 |
| 8 | I | II | I | 2 | 0.78 |
| 9 | I | I | II | 4 | 0.67 |
| Model No. | Test | ||
|---|---|---|---|
| Accuracy | Recall | Precision | |
| Table 6 | |||
| 3 | 0.28 | 0.25 | 0.29 |
| 9 | 0.41 | 0.32 | 0.32 |
| 10 | 0.62 | 0.58 | 0.58 |
| Table 7 | |||
| 1 | 0.55 | 0.49 | 0.56 |
| 5 | 0.59 | 0.48 | 0.49 |
| Kidney No. | Actual Class | Predicted Class | Classification Reliability | |
|---|---|---|---|---|
| 1 | 3 | 3 | 99% |  |
| 2 | 3 | 3 | 99% | |
| 3 | 3 | 3 | 98% | |
| 4 | 3 | 3 | 98% | |
| 5 | 2 | 2 | 98% | |
| 6 | 2 | 2 | 97% | |
| 7 | 3 | 3 | 96% | |
| 8 | 2 | 2 | 94% | |
| 9 | 3 | 3 | 93% | |
| 10 | 3 | 3 | 92% | |
| 11 * | 3 | 3 | 91% | |
| 12 | 2 | 2 | 88% | |
| 13 | 2 | 2 | 86% | |
| 14 | 3 | 3 | 84% | |
| 15 | 2 | 2 | 81% | |
| 16 | 2 | 2 | 80% | |
| 17 | 1 | 1 | 71% | |
| 18 * | 2 | 2 | 70% | |
| 19 * | 3 | 3 | 69% | |
| 20 | 3 | 3 | 66% | |
| 21 | 1 |  | 64% | |
| 22 | 1 | 1 | 61% | |
| 23 | 2 | 2 | 57% | |
| 24 * | 3 | 3 | 54% | |
| 25 * | 1 | 1 | 44% | |
| 26 * | 2 | 2 | 43% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sommer, F.; Sun, B.; Fischer, J.; Goldammer, M.; Thiele, C.; Malberg, H.; Markgraf, W. Hyperspectral Imaging during Normothermic Machine Perfusion—A Functional Classification of Ex Vivo Kidneys Based on Convolutional Neural Networks. Biomedicines 2022, 10, 397. https://doi.org/10.3390/biomedicines10020397
Sommer F, Sun B, Fischer J, Goldammer M, Thiele C, Malberg H, Markgraf W. Hyperspectral Imaging during Normothermic Machine Perfusion—A Functional Classification of Ex Vivo Kidneys Based on Convolutional Neural Networks. Biomedicines. 2022; 10(2):397. https://doi.org/10.3390/biomedicines10020397
Chicago/Turabian StyleSommer, Florian, Bingrui Sun, Julian Fischer, Miriam Goldammer, Christine Thiele, Hagen Malberg, and Wenke Markgraf. 2022. "Hyperspectral Imaging during Normothermic Machine Perfusion—A Functional Classification of Ex Vivo Kidneys Based on Convolutional Neural Networks" Biomedicines 10, no. 2: 397. https://doi.org/10.3390/biomedicines10020397
APA StyleSommer, F., Sun, B., Fischer, J., Goldammer, M., Thiele, C., Malberg, H., & Markgraf, W. (2022). Hyperspectral Imaging during Normothermic Machine Perfusion—A Functional Classification of Ex Vivo Kidneys Based on Convolutional Neural Networks. Biomedicines, 10(2), 397. https://doi.org/10.3390/biomedicines10020397






