Dynamic Interplay between Cockayne Syndrome Protein B and Poly(ADP-Ribose) Polymerase 1 during Oxidative DNA Damage Repair
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Menadione Treatment
2.2. Protein Fractionation and Western Blotting
2.3. shRNA Knockdown
2.4. Menadione Sensitivity Assays
2.5. ChIP-qPCR Analyses
2.6. Antibodies
3. Results
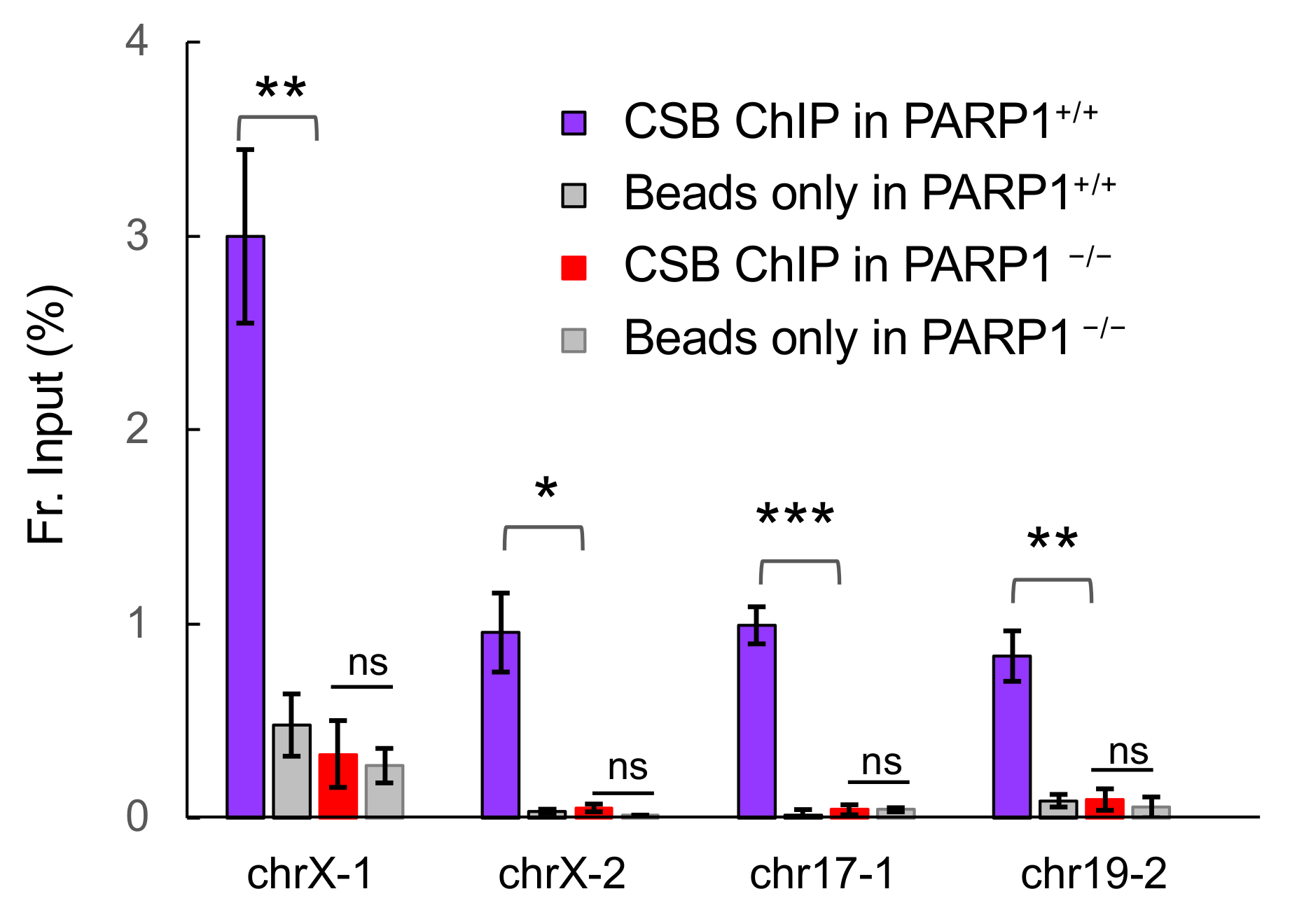
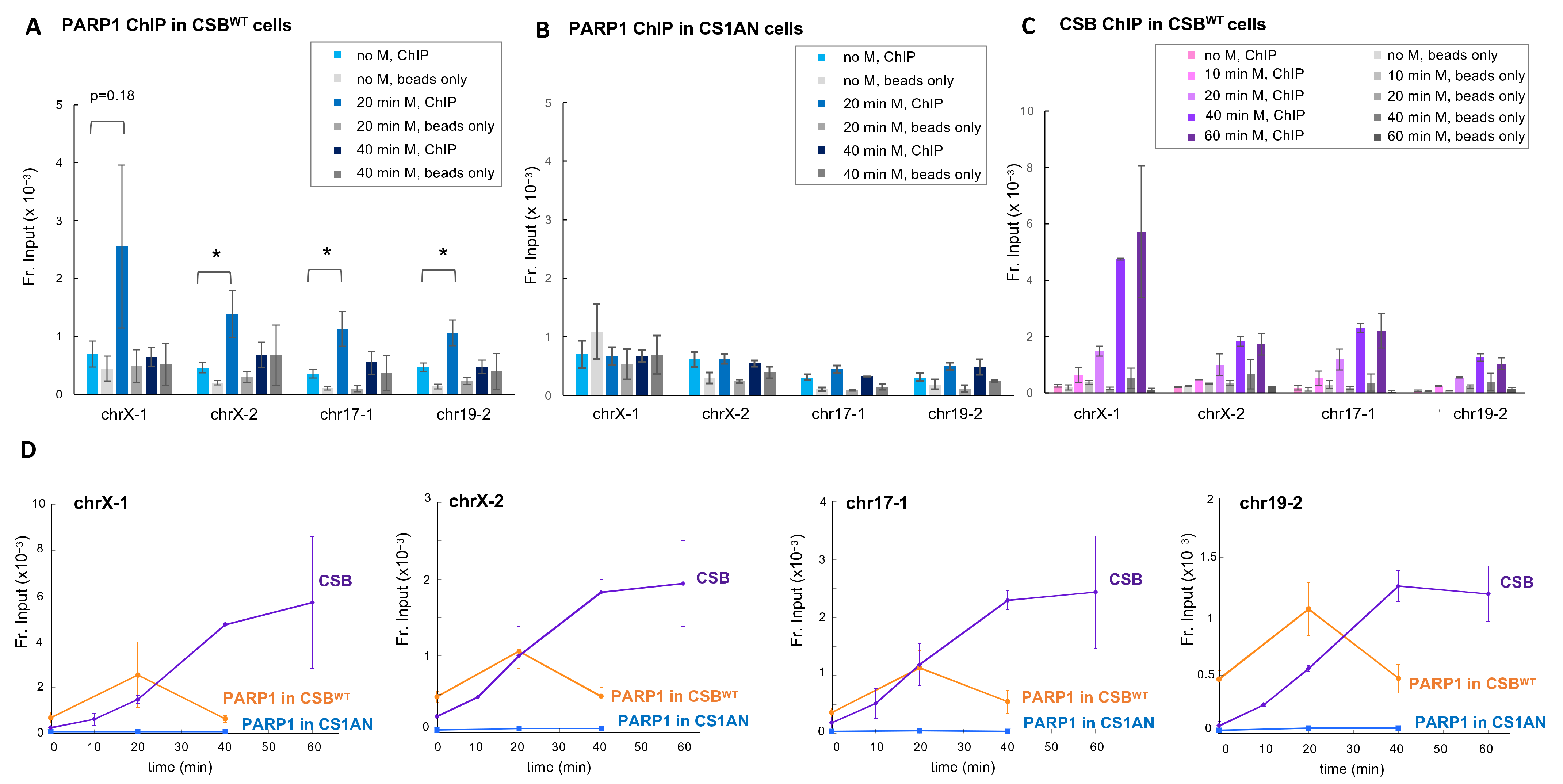
3.1. PARP1 Is Essential for CSB Recruitment to the Top Four Menadione-Induced CSB Binding Sites
3.2. CSB Regulates the Association of PARP1 with Chromatin upon Oxidative Stress
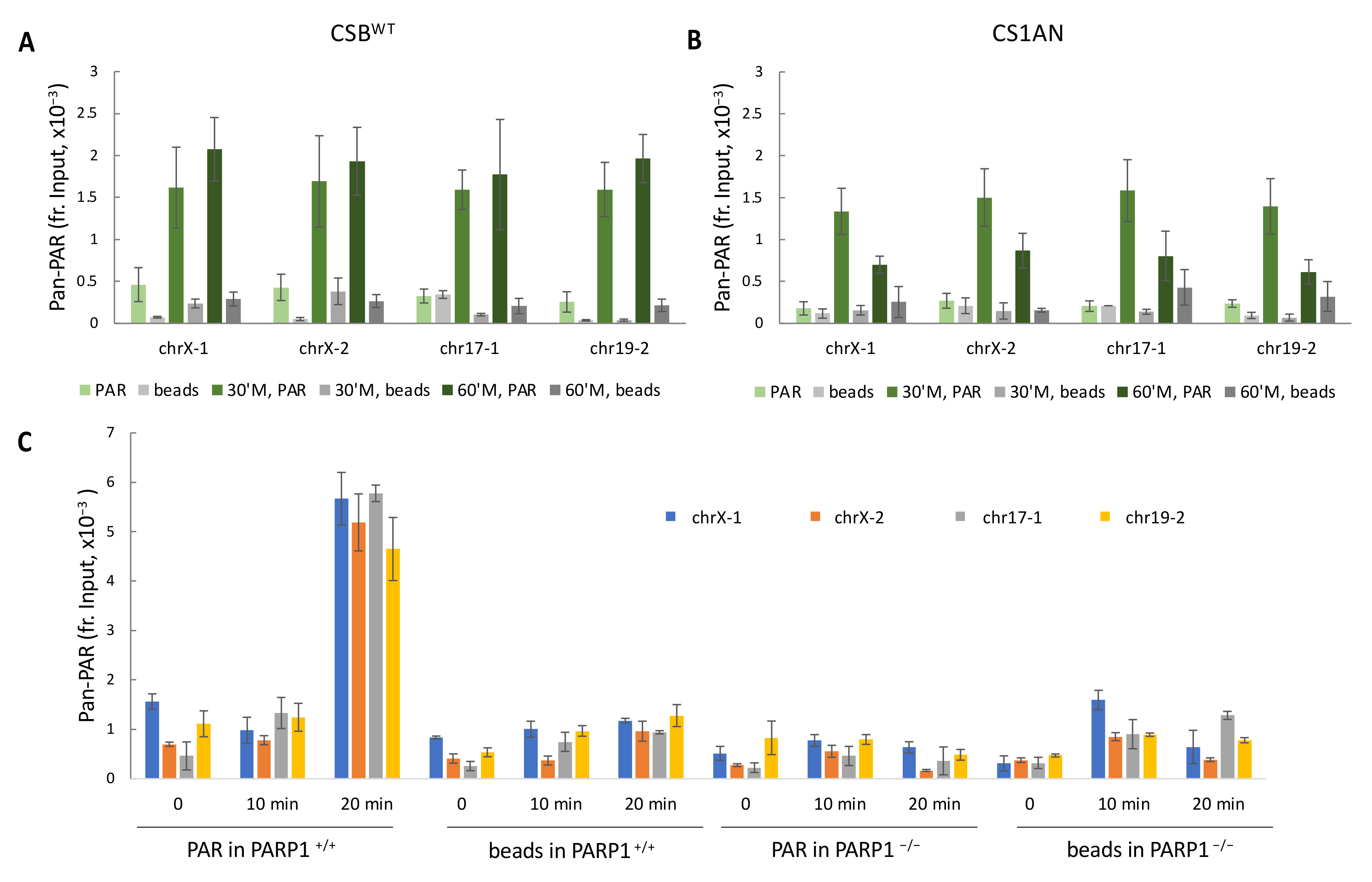
3.3. CSB Regulates PAR Levels on Chromatin in Oxidatively-Stressed Cells
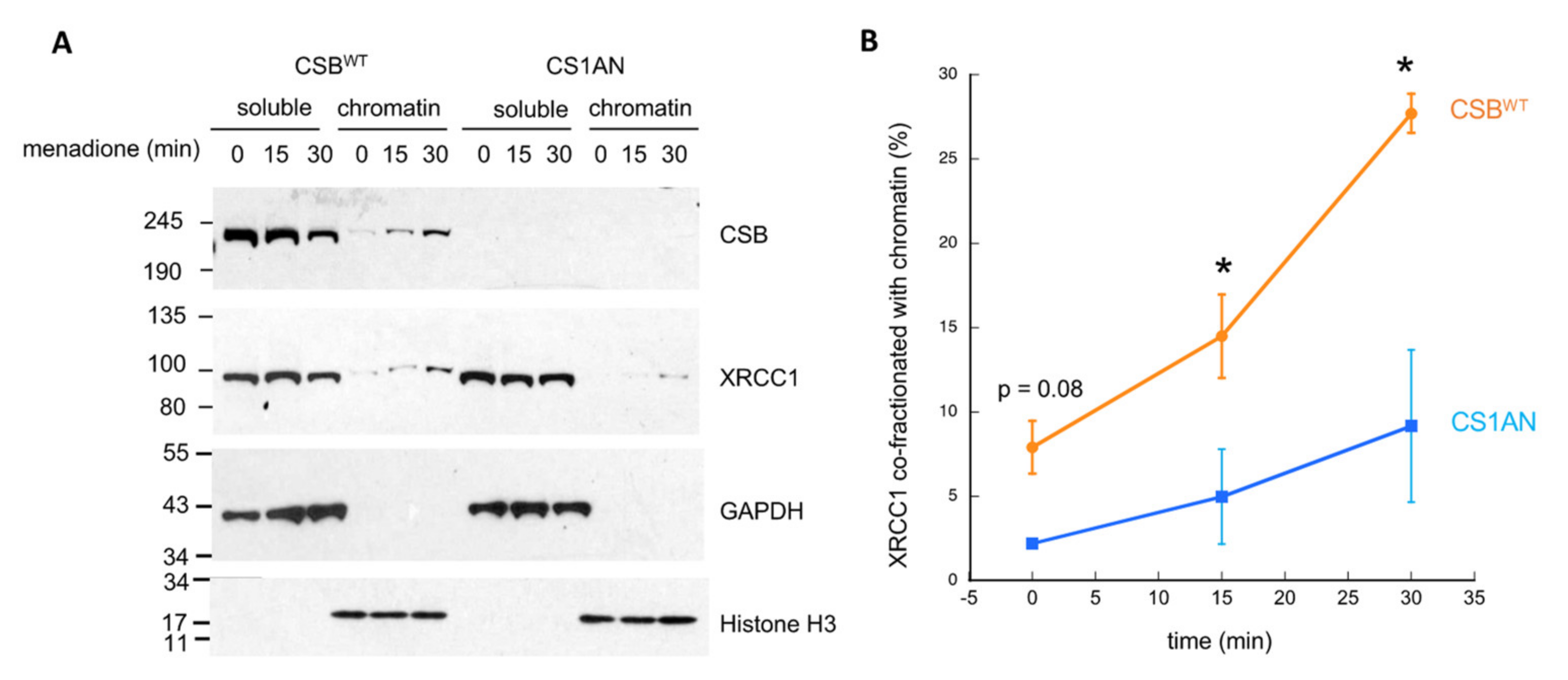
3.4. CSB Facilitates the Recruitment of XRCC1 to Oxidatively Damaged Chromatin
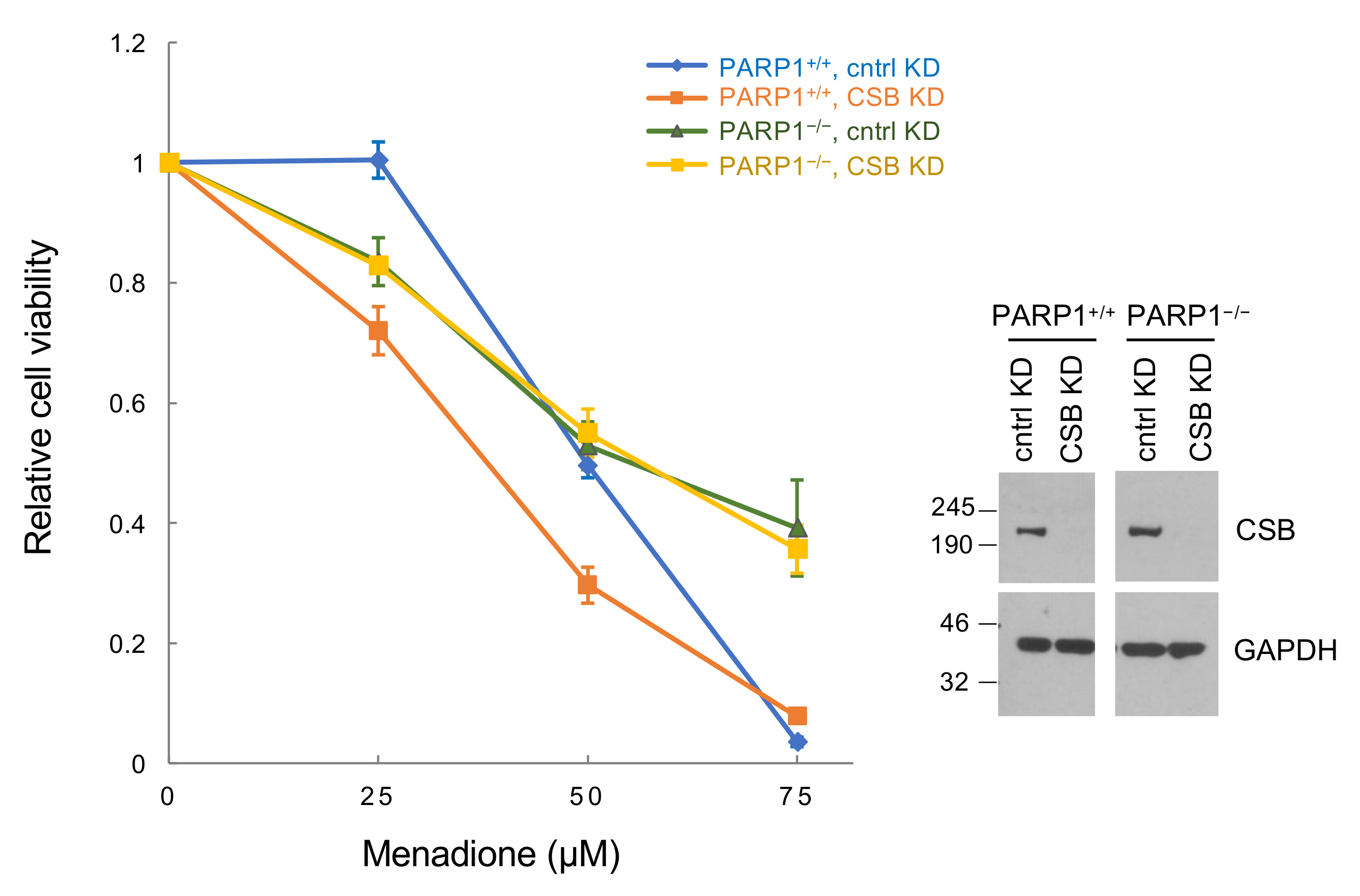
3.5. Regulation of PARP1 Activity Is a Major CSB Function in Oxidatively-Stressed Cells
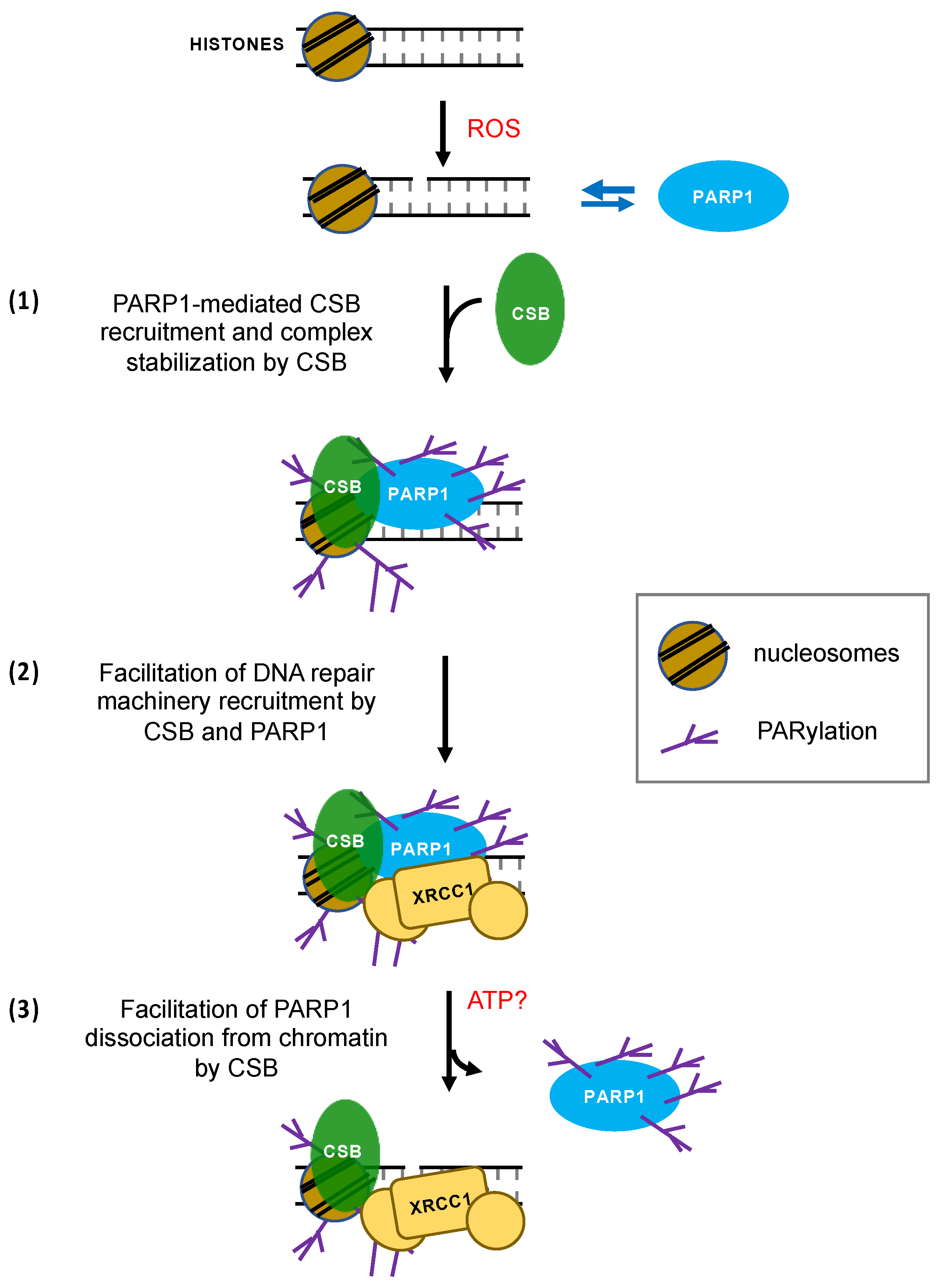
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ames, B.N.; Shigenaga, M.K.; Hagen, T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA 1993, 90, 7915–7922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindahl, T.; Satoh, M.S.; Poirier, G.G.; Klungland, A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995, 20, 405–411. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. Mammalian single-strand break repair: Mechanisms and links with chromatin. DNA Repair 2007, 6, 443–453. [Google Scholar] [CrossRef]
- Strom, C.E.; Johansson, F.; Uhlen, M.; Szigyarto, C.A.; Erixon, K.; Helleday, T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011, 39, 3166–3175. [Google Scholar] [CrossRef]
- Caldecott, K.W. XRCC1 protein; Form and function. DNA Repair 2019, 81, 102664. [Google Scholar] [CrossRef]
- Pandey, N.; Black, B.E. Rapid Detection and Signaling of DNA Damage by PARP-1. Trends Biochem. Sci. 2021, 46, 744–757. [Google Scholar] [CrossRef]
- Curtin, N.J.; Szabo, C. Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 2020, 19, 711–736. [Google Scholar] [CrossRef]
- Lake, R.J.; Fan, H.Y. Structure, function and regulation of CSB: A multi-talented gymnast. Mech. Ageing Dev. 2013, 134, 202–211. [Google Scholar] [CrossRef] [Green Version]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Blessing, C.; Mandemaker, I.K.; Gonzalez-Leal, C.; Preisser, J.; Schomburg, A.; Ladurner, A.G. The Oncogenic Helicase ALC1 Regulates PARP Inhibitor Potency by Trapping PARP2 at DNA Breaks. Mol. Cell 2020, 80, 862–875.e6. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Williams, R.T.; Calarco, J.P.; Miller, E.L.; Weber, C.M.; Braun, S.M.; Pulice, J.L.; Chory, E.J.; Crabtree, G.R. Crabtree, Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2017, 49, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Auble, D.T.; Wang, D.; Post, K.W.; Hahn, S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol. Cell. Biol. 1997, 17, 4842–4851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, A.R. Three complementation groups in Cockayne syndrome. Mutat. Res. 1982, 106, 347–356. [Google Scholar] [CrossRef]
- Nance, M.A.; Berry, S.A. Cockayne syndrome: Review of 140 cases. Am. J. Med. Genet. 1992, 42, 68–84. [Google Scholar] [CrossRef]
- Lake, R.J.; Geyko, A.; Hemashettar, G.; Zhao, Y.; Fan, H.Y. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell 2010, 37, 235–246. [Google Scholar] [CrossRef] [Green Version]
- Lake, R.J.; Boetefuer, E.L.; Won, K.J.; Fan, H.Y. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 2016, 44, 2125–2135. [Google Scholar] [CrossRef] [Green Version]
- Van Gool, A.J.; Citterio, E.; Rademakers, S.; van Os, R.; Vermeulen, W.; Constantinou, A.; Egly, J.M.; Bootsma, D.; Hoeijmakers, J.H. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997, 16, 5955–5965. [Google Scholar] [CrossRef]
- Troelstra, C.; van Gool, A.; de Wit, J.; Vermeulen, W.; Bootsma, D.; Hoeijmakers, J.H. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 1992, 71, 939–953. [Google Scholar] [CrossRef] [Green Version]
- Mellon, I.; Spivak, G.; Hanawalt, P.C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 1987, 51, 241–249. [Google Scholar] [CrossRef]
- Cho, I.; Tsai, P.F.; Lake, R.J.; Basheer, A.; Fan, H.Y. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013, 9, e1003407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boetefuer, E.L.; Lake, R.J.; Dreval, K.; Fan, H.Y. Poly(ADP-ribose) polymerase 1 (PARP1) promotes oxidative stress-induced association of Cockayne syndrome group B protein with chromatin. J. Biol. Chem. 2018, 293, 17863–17874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, H.K.; Muftuoglu, M.; Beck, G.; Imam, S.Z.; Bohr, V.A.; Wilson, D.M., 3rd. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007, 35, 4103–4113. [Google Scholar] [CrossRef]
- Menoni, H.; Hoeijmakers, J.H.; Vermeulen, W. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J. Cell Biol. 2012, 199, 1037–1046. [Google Scholar] [CrossRef]
- Tuo, J.; Chen, C.; Zeng, X.; Christiansen, M.; Bohr, V.A. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair 2002, 1, 913–927. [Google Scholar] [CrossRef]
- Tuo, J.; Jaruga, P.; Rodriguez, H.; Bohr, V.A.; Dizdaroglu, M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003, 17, 668–674. [Google Scholar] [CrossRef]
- Dianov, G.; Bischoff, C.; Sunesen, M.; Bohr, V.A. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 1999, 27, 1365–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menoni, H.; Wienholz, F.; Theil, A.F.; Janssens, R.C.; Lans, H.; Campalans, A.; Radicella, J.P.; Marteijn, J.A.; Vermeulen, W. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 2018, 46, 7747–7756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorslund, T.; von Kobbe, C.; Harrigan, J.A.; Indig, F.E.; Christiansen, M.; Stevnsner, T.; Bohr, V.A. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell. Biol. 2005, 25, 7625–7636. [Google Scholar] [CrossRef] [Green Version]
- Scheibye-Knudsen, M.; Mitchell, S.J.; Fang, E.F.; Iyama, T.; Ward, T.; Wang, J.; Dunn, C.A.; Singh, N.; Veith, S.; Hasan-Olive, M.M.; et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014, 20, 840–855. [Google Scholar] [CrossRef] [Green Version]
- Lake, R.J.; Boetefuer, E.L.; Tsai, P.F.; Jeong, J.; Choi, I.; Won, K.J.; Fan, H.Y. The sequence-specific transcription factor c-Jun targets Cockayne syndrome protein B to regulate transcription and chromatin structure. PLoS Genet. 2014, 10, e1004284. [Google Scholar] [CrossRef] [PubMed]
- Hanzlikova, H.; Gittens, W.; Krejcikova, K.; Zeng, Z.; Caldecott, K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017, 45, 2546–2557. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Wu, W.; Hill, S.E.; Nathan, W.J.; Paiano, J.; Callen, E.; Wang, D.; Shinoda, K.; van Wietmarschen, N.; Colón-Mercado, J.M.; Zong, D.; et al. Neuronal enhancers are hotspots for DNA single-strand break repair. Nature 2021, 593, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Demarest, T.G.; Babbar, M.; Kim, E.W.; Okur, M.N.; De, S.; Croteau, D.L.; Bohr, V.A. Cockayne syndrome group B deficiency reduces H3K9me3 chromatin remodeler SETDB1 and exacerbates cellular aging. Nucleic Acids Res. 2019, 47, 8548–8562. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lake, R.J.; Bilkis, R.; Fan, H.-Y. Dynamic Interplay between Cockayne Syndrome Protein B and Poly(ADP-Ribose) Polymerase 1 during Oxidative DNA Damage Repair. Biomedicines 2022, 10, 361. https://doi.org/10.3390/biomedicines10020361
Lake RJ, Bilkis R, Fan H-Y. Dynamic Interplay between Cockayne Syndrome Protein B and Poly(ADP-Ribose) Polymerase 1 during Oxidative DNA Damage Repair. Biomedicines. 2022; 10(2):361. https://doi.org/10.3390/biomedicines10020361
Chicago/Turabian StyleLake, Robert J., Rabeya Bilkis, and Hua-Ying Fan. 2022. "Dynamic Interplay between Cockayne Syndrome Protein B and Poly(ADP-Ribose) Polymerase 1 during Oxidative DNA Damage Repair" Biomedicines 10, no. 2: 361. https://doi.org/10.3390/biomedicines10020361
APA StyleLake, R. J., Bilkis, R., & Fan, H.-Y. (2022). Dynamic Interplay between Cockayne Syndrome Protein B and Poly(ADP-Ribose) Polymerase 1 during Oxidative DNA Damage Repair. Biomedicines, 10(2), 361. https://doi.org/10.3390/biomedicines10020361





