Quantitative Proteomic Study Unmasks Fibrinogen Pathway in Polycystic Liver Disease
Abstract
1. Introduction
2. Materials and Methods
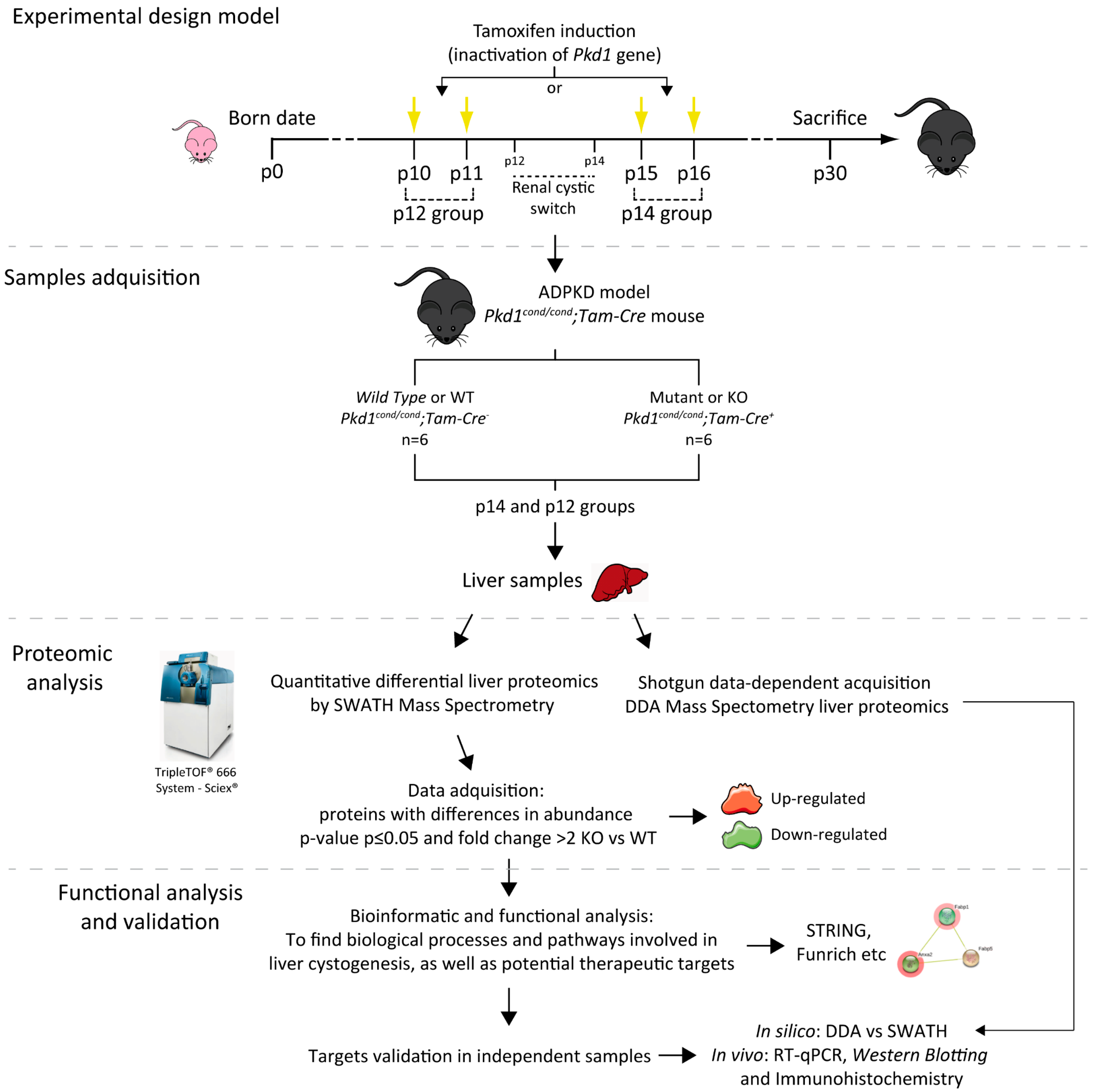
2.1. Murine Model
2.2. Protein Extraction and Digestion
2.3. Proteomics
2.3.1. DDA-Data Dependent Analysis
2.3.2. Generation of the References Spectral Library
2.3.3. Quantification by SWATH and Data Analysis
2.3.4. Data Analysis
2.3.5. Signaling Pathway Analysis
2.4. Histology and Measurement of Cystic Index
2.5. Clinical Chemistry
2.6. RNA Extraction
2.7. Real-Time Quantitative PCR
2.8. Western Blotting
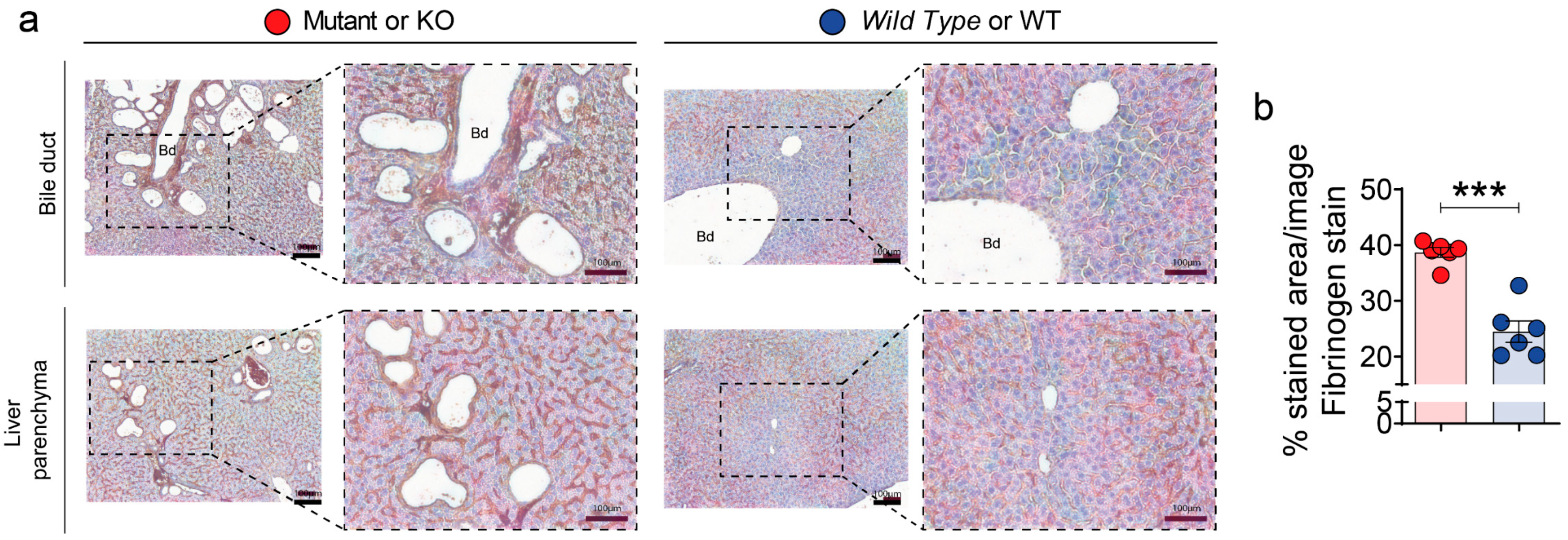
2.9. Immunohistochemistry and Quantification
2.10. Statistics Analysis
3. Results
3.1. Kidney and Liver Does Not Follow the Same Cystic Mechanisms
3.2. Shotgun and SWATH–MS Proteomic Analysis in PLD
Protein Expression Pattern in Hepatic Cystogenesis
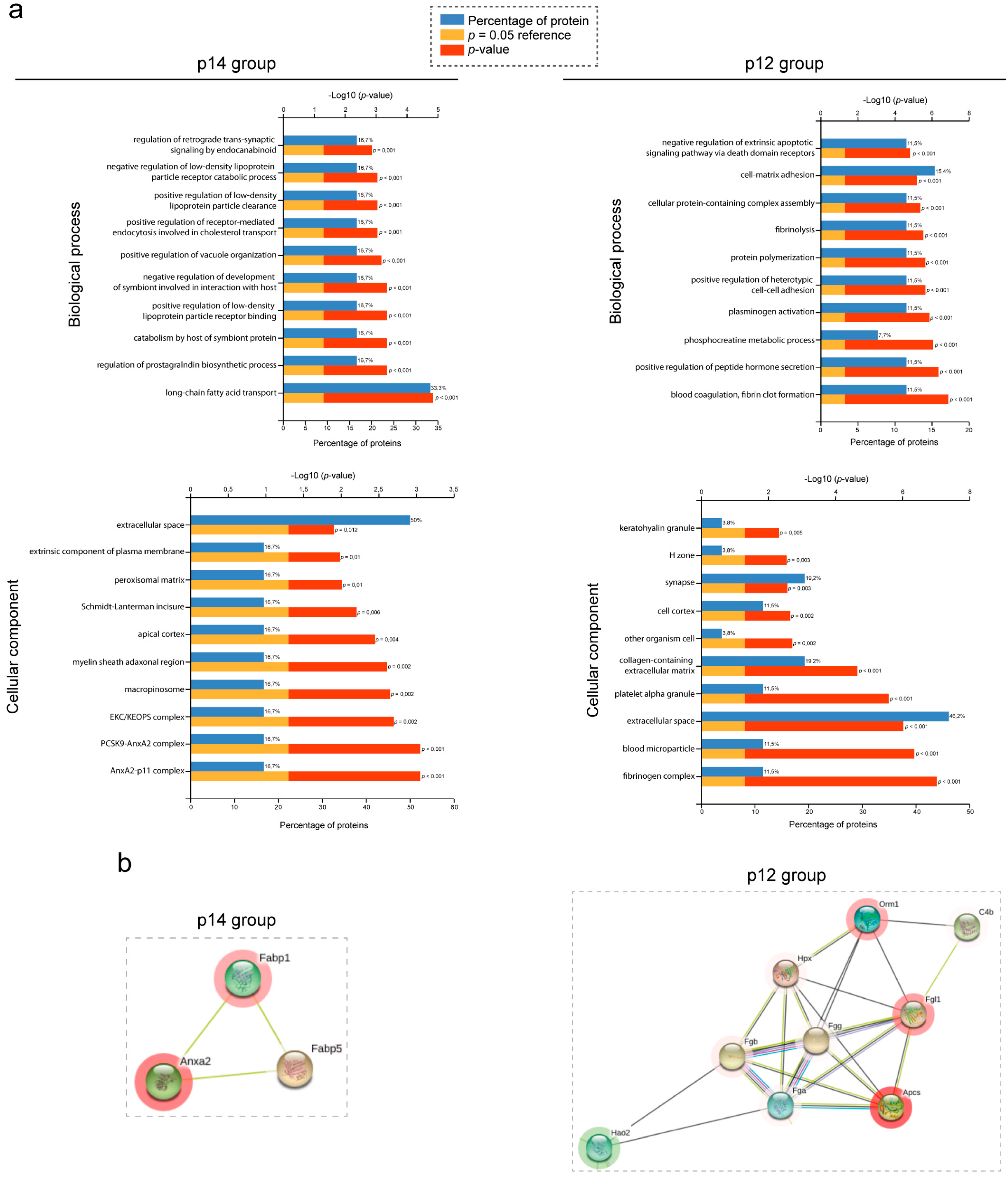
3.3. Clustering, Pathway Enrichment and Protein–Protein Interaction Analysis of Gene Expression in PLD
3.4. Validation of SWATH–MS Analysis Unmasks the Fibrinogen Complex as a New Molecular Mechanism Related to PLD
4. Discussion
5. Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cnossen, W.R.; Drenth, J.P.H. Polycystic liver disease: An overview of pathogenesis, clinical manifestations and management. Orphanet J. Rare Dis. 2014, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Santos-Laso, A.; Izquierdo-Sánchez, L.; Lee-Law, P.Y.; Perugorria, M.J.; Marzioni, M.; Marin, J.J.G.; Bujanda, L.; Banales, J.M. New Advances in Polycystic Liver Diseases. Semin. Liver Dis. 2017, 37, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Suwabe, T.; Shukoor, S.; Chamberlain, A.M.; Killian, J.M.; King, B.F.; Edwards, M.; Senum, S.R.; Madsen, C.D.; Chebib, F.T.; Hogan, M.C.; et al. Epidemiology of autosomal dominant polycystic kidney disease in Olmsted county. Clin. J. Am. Soc. Nephrol. 2020, 15, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Cordido, A.; Besada-Cerecedo, L.; García-González, M.A. The Genetic and Cellular Basis of Autosomal Dominant Polycystic Kidney Disease—A Primer for Clinicians. Front. Pediatr. 2017, 5, 279. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, C.; Guay-Woodford, L.M.; Harris, P.C.; Horie, S.; Peters, D.J.M.; Torres, V.E. Polycystic kidney disease. Nat. Rev. Dis. Prim. 2018, 4, 1–24. [Google Scholar] [CrossRef]
- Cordido, A.; Vizoso-Gonzalez, M.; Garcia-Gonzalez, M.A. Molecular Pathophysiology of Autosomal Recessive Polycystic Kidney Disease. Int. J. Mol. Sci. 2021, 22, 6523. [Google Scholar] [CrossRef]
- Van Aerts, R.M.M.; van de Laarschot, L.F.M.; Banales, J.M.; Drenth, J.P.H. Clinical management of polycystic liver disease. J. Hepatol. 2018, 68, 827–837. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Masyuk, T.V.; Marin, J.J.; Marzioni, M.; Bujanda, L.; Larusso, N.F.; Banales, J.M. Polycystic liver diseases: Advanced insights into the molecular mechanisms. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 750–761. [Google Scholar] [CrossRef]
- Banales, J.M.; Masyuk, T.V.; Gradilone, S.A.; Masyuk, A.I.; Medina, J.F.; LaRusso, N.F. The cAMP effectors Epac and protein kinase a (PKA) are involved in the hepatic cystogenesis of an animal model of autosomal recessive polycystic kidney disease (ARPKD). Hepatology 2009, 49, 160–174. [Google Scholar] [CrossRef]
- Banales, J.; Masyuk, T.V.; Bogert, P.S.; Huang, B.Q.; Gradilone, S.; Lee, S.-O.; Stroope, A.J.; Masyuk, A.I.; Medina, J.F.; LaRusso, N.F. Hepatic cystogenesis is associated with abnormal expression and location of ion transporters and water channels in an animal model of autosomal recessive polycystic kidney disease. Am. J. Pathol. 2008, 173, 1637–1646. [Google Scholar] [CrossRef]
- Spirli, C.; Okolicsanyi, S.; Fiorotto, R.; Fabris, L.; Cadamuro, M.; Lecchi, S.; Tian, X.; Somlo, S.; Strazzabosco, M. Mammalian target of rapamycin regulates vascular endothelial growth factor-dependent liver cyst growth in polycystin-2-defective mice. Hepatology 2010, 51, 1778–1788. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.L.; Grubman, S.A.; Perrone, R.D.; Rojkind, M.; Moy, E.; Lee, D.W.; Jefferson, D.M. Matrix metalloproteinase activity in human intrahepatic biliary epithelial cell lines from patients with autosomal dominant polycystic kidney disease. Connect. Tissue Res. 1996, 33, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Yasoshima, M.; Sato, Y.; Furubo, S.; Kizawa, K.; Sanzen, T.; Ozaki, S.; Harada, K.; Nakanuma, Y. Matrix proteins of basement membrane of intrahepatic bile ducts are degraded in congenital hepatic fibrosis and Caroli’s disease. J. Pathol. 2009, 217, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Masyuk, T.V.; Huang, B.Q.; Ward, C.J.; Masyuk, A.I.; Yuan, D.; Splinter, P.L.; Punyashthiti, R.; Ritman, E.L.; Torres, V.E.; Harris, P.C.; et al. Defects in Cholangiocyte Fibrocystin Expression and Ciliary Structure in the PCK Rat. Gastroenterology 2003, 125, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.Y.K.; Etheridge, N.; Nouwens, A.S.; Dodd, P.R. SWATH analysis of the synaptic proteome in Alzheimer’s disease. Neurochem. Int. 2015, 87, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, 016717. [Google Scholar] [CrossRef]
- Piontek, K.B.; Huso, D.L.; Grinberg, A.; Liu, L.; Bedja, D.; Zhao, H.; Gabrielson, K.; Qian, F.; Mei, C.; Westphal, H.; et al. A functional floxed allele of Pkd1 that can be conditionally inactivated in vivo. J. Am. Soc. Nephrol. 2004, 15, 3035–3043. [Google Scholar] [CrossRef]
- Piontek, K.; Menezes, L.F.; Garcia-Gonzalez, M.A.; Huso, D.L.; Germino, G.G. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat. Med. 2007, 13, 1490–1495. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Jensen, O.N.; Podtelejnikov, A.V.; Neubauer, G.; Shevchenko, A.; Mortensen, P.; Mann, M. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 1996, 24, 893–896. [Google Scholar] [CrossRef]
- Shilov, I.V.; Seymourt, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The paragon algorithm, a next generation search engine that uses sequence temperature values sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef] [PubMed]
- Del Pilar Chantada-Vázquez, M.; López, A.C.; Vence, M.G.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef] [PubMed]
- Varela-Rodríguez, B.M.; Juiz-Valiña, P.; Varela, L.; Outeiriño-Blanco, E.; Bravo, S.B.; García-Brao, M.J.; Mena, E.; Noguera, J.F.; Valero-Gasalla, J.; Cordido, F.; et al. Beneficial Effects of Bariatric Surgery-Induced by Weight Loss on the Proteome of Abdominal Subcutaneous Adipose Tissue. J. Clin. Med. 2020, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Hermida-Nogueira, L.; Barrachina, M.N.; Izquierdo, I.; García-Vence, M.; Lacerenza, S.; Bravo, S.; Castrillo, A.; García, Á. Proteomic analysis of extracellular vesicles derived from platelet concentrates treated with Mirasol® identifies biomarkers of platelet storage lesion. J. Proteom. 2020, 210, 103529. [Google Scholar] [CrossRef] [PubMed]
- Tamara, C.; Nerea, L.B.; Belén, B.S.; Aurelio, S.; Iván, C.; Fernando, S.; Javier, B.; Felipe, C.F.; María, P. Vesicles shed by pathological murine adipocytes spread pathology: Characterization and functional role of insulin resistant/hypertrophied adiposomes. Int. J. Mol. Sci. 2020, 21, 2252. [Google Scholar] [CrossRef]
- Víctor Álvarez, J.; Bravo, S.B.; García-Vence, M.; De Castro, M.J.; Luzardo, A.; Colón, C.; Tomatsu, S.; Otero-Espinar, F.J.; Couce, M.L. Proteomic analysis in morquio a cells treated with immobilized enzymatic replacement therapy on nanostructured lipid systems. Int. J. Mol. Sci. 2019, 20, 4610. [Google Scholar] [CrossRef]
- Meyer, J.G.; Schilling, B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev. Proteom. 2017, 14, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Mok, T.S.; Lin, X.; Zhang, W.; Cui, Y.; Guo, J.; Chen, X.; Zhang, T.; Wang, T. SWATH-based proteomics identified carbonic anhydrase 2 as a potential diagnosis biomarker for nasopharyngeal carcinoma. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Ortea, I.; Ruiz-Sánchez, I.; Cañete, R.; Caballero-Villarraso, J.; Cañete, M.D. Identification of candidate serum biomarkers of childhood-onset growth hormone deficiency using SWATH-MS and feature selection. J. Proteom. 2018, 175, 105–113. [Google Scholar] [CrossRef]
- Tan, H.T.; Chung, M.C.M. Label-Free Quantitative Phosphoproteomics Reveals Regulation of Vasodilator-Stimulated Phosphoprotein upon Stathmin-1 Silencing in a Pair of Isogenic Colorectal Cancer Cell Lines. Proteomics 2018, 18, 1–13. [Google Scholar] [CrossRef]
- Lambert, J.P.; Ivosev, G.; Couzens, A.L.; Larsen, B.; Taipale, M.; Lin, Z.Y.; Zhong, Q.; Lindquist, S.; Vidal, M.; Aebersold, R.; et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods 2013, 10, 1239–1245. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, M.; Kannan, S.; Chawade, A.; Bhattacharjee, A.; Govekar, R. Clinical biomarker discovery by SWATH-MS based label-free quantitative proteomics: Impact of criteria for identification of differentiators and data normalization method. J. Transl. Med. 2019, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chutipongtanate, S.; Greis, K.D. Multiplex Biomarker Screening Assay for Urinary Extracellular Vesicles Study: A Targeted Label- Free Proteomic Approach. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Warnes, G.R.; Boljer, B.; Bonebakker, L.; Gentleman, R.; Huber Andy Liaw, W.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; Schwartz, M.; et al. gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.1.1. gplots Var. R Program. Tools Plotting Data 2019. Available online: https://www.r-project.org/ (accessed on 12 February 2020).
- Cordido, A.; Cernadas, E.; Fernández-Delgado, M.; García-González, M.A. CystAnalyser: A new software tool for the automatic detection and quantification of cysts in Polycystic Kidney and Liver Disease, and other cystic disorders. PLoS Comput. Biol. 2020, 16, 1–18. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, M.A.; Carette, C.; Bagattin, A.; Chiral, M.; Makinistoglu, M.P.; Garbay, S.; Prévost, G.; Madaras, C.; Hérault, Y.; Leibovici, M.; et al. A suppressor locus for MODY3-diabetes. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Cordido, A.; Nuñez-Gonzalez, L.; Martinez-Moreno, J.M.; Lamas-Gonzalez, O.; Rodriguez-Osorio, L.; Perez-Gomez, M.V.; Martin-Sanchez, D.; Outeda, P.; Chiaravalli, M.; Watnick, T.; et al. TWEAK Signaling Pathway Blockade Slows Cyst Growth and Disease Progression in Autosomal Dominant Polycystic Kidney Disease. J. Am. Soc. Nephrol. 2021, 32, 1913–1932. [Google Scholar] [CrossRef]
- Consortium, T.U.; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Fonseka, P.; Pathan, M.; Chitti, S.V.; Kang, T.; Mathivanan, S. FunRich enables enrichment analysis of OMICs datasets. J. Mol. Biol. 2021, 433, 166747. [Google Scholar] [CrossRef]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Poirier, S.; Seidah, N.G. Annexin A2 is a C-terminal PCSK9-binding protein that regulates endogenous low density lipoprotein receptor levels. J. Biol. Chem. 2008, 283, 31791–31801. [Google Scholar] [CrossRef] [PubMed]
- Kopec, A.K.; Luyendyk, J.P. Role of Fibrin(ogen) in Progression of Liver Disease: Guilt by Association? Semin. Thromb. Hemost. 2016, 42, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Abu-Wasel, B.; Walsh, C.; Keough, V.; Molinari, M. Pathophysiology, epidemiology, classification and treatment options for polycystic liver diseases. World J. Gastroenterol. 2013, 19, 5775–5786. [Google Scholar] [CrossRef] [PubMed]
- Happé, H.; Peters, D.J.M. Translational research in ADPKD: Lessons from animal models. Nat. Rev. Nephrol. 2014, 10, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Hajjar, K.A. Annexin A2 system in human biology: Cell surface and beyond. Semin. Thromb. Hemost. 2013, 39, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Flood, E.C.; Almeida, D.; Yan, L.B.; Berlin, D.A.; Heerdt, P.M.; Hajjar, K.A. Annexin A2 supports pulmonary microvascular integrity by linking vascular endothelial cadherin and protein tyrosine phosphatases. J. Exp. Med. 2017, 214, 2535–2545. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, X.; Zhang, Z.; Feng, Y.; Jia, X.; Shi, Y.; Yang, H.; Zhang, Z.; Zhang, X.; Liu, L.; et al. Subcellular proteome analysis unraveled annexin A2 related to immune liver fibrosis. J. Cell. Biochem. 2010, 110, 219–228. [Google Scholar] [CrossRef]
- Yang, M.; Wang, C.; Li, S.; Xv, X.; She, S.; Ran, X.; Li, S.; Hu, H.; Hu, P.; Zhang, D.; et al. Annexin A2 promotes liver fibrosis by mediating von Willebrand factor secretion. Dig. Liver Dis. 2017, 49, 780–788. [Google Scholar] [CrossRef]
- Fernández, G.C.; Ilarregui, J.M.; Rubel, C.J.; Toscano, M.A.; Gómez, S.A.; Bompadre, M.B.; Isturiz, M.A.; Rabinovich, G.A.; Palermo, M.S. Galectin-3 and soluble fibrinogen act in concert to modulate neutrophil activation and survival: Involvement of alternative MAPK pathways. Glycobiology 2005, 15, 519–527. [Google Scholar] [CrossRef]
- Tang, M.; Cao, X.; Li, P.; Zhang, K.; Li, Y.; Zheng, Q.Y.; Li, G.Q.; Chen, J.; Xu, G.L.; Zhang, K.Q. Increased expression of Fibrinogen-Like Protein 2 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hantgan, R.R.; Stahle, M.C.; Lord, S.T. Dynamic regulation of fibrinogen: Integrin αiIbβ3 binding. Biochemistry 2010, 49, 9217–9225. [Google Scholar] [CrossRef] [PubMed]
- Shillingford, J.M.; Murcia, N.S.; Larson, C.H.; Low, S.H.; Hedgepeth, R.; Brown, N.; Flask, C.A.; Novick, A.C.; Goldfarb, D.A.; Kramer-Zucker, A.; et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc. Natl. Acad. Sci. USA 2006, 103, 5466–5471. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Boctor, S.; Barisoni, L.M.C.; Gusella, G.L. Inactivation of Integrin- 1 Prevents the Development of Polycystic Kidney Disease after the Loss of Polycystin-1. J. Am. Soc. Nephrol. 2015, 26, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Bugge, T.H.; Kombrinck, K.W.; Flick, M.J.; Daugherty, C.C.; Danton, M.J.S.; Degen, J.L. Loss of fibrinogen rescues mice from the pleiotropic effects of plasminogen deficiency. Cell 1996, 87, 709–719. [Google Scholar] [CrossRef]
- Masyuk, T.; Masyuk, A.; Larusso, N. Polycystic Liver Diseases: Genetics, Mechanisms, and Therapies and Therapies. In The Liver: Biology and Pathobiology; Arias, I.M., Alter, H.J., Boyer, J.L., Cohen, D.E., Shafritz, D.A., Thorgeirsson, S.S., Wolkoff, A.W., Eds.; Wiley: Hoboken, NJ, USA, 2020. [Google Scholar]
- Zhang, Y.; Kim, D.K.; Lu, Y.; Jung, Y.S.; Lee, J.-M.; Kim, Y.H.; Lee, Y.S.; Kim, J.; Dewidar, B.; Jeong, W.-I.; et al. Orphan nuclear receptor ERRγ is a key regulator of human fibrinogen gene expression. PLoS ONE 2017, 12, e0182141. [Google Scholar] [CrossRef] [PubMed]
- Barutta, F.; Bruno, G.; Mastrocola, R.; Bellini, S.; Gruden, G. The role of cannabinoid signaling in acute and chronic kidney diseases. Kidney Int. 2018, 94, 252–258. [Google Scholar] [CrossRef]
- Zhang, Y.; Kim, D.K.; Jung, Y.S.; Kim, Y.H.; Lee, Y.S.; Kim, J.; Jeong, W.I.L.; Lee, I.K.; Cho, S.J.; Dooley, S.; et al. Inverse agonist of ERRγ reduces cannabinoid receptor type 1-mediated induction of fibrinogen synthesis in mice with a high-fat diet-intoxicated liver. Arch. Toxicol. 2018, 92, 2885–2896. [Google Scholar] [CrossRef]
- Kim, D.K.; Gang, G.T.; Ryu, D.; Koh, M.; Kim, Y.N.; Kim, S.S.; Park, J.; Kim, Y.H.; Sim, T.; Lee, I.K.; et al. Inverse agonist of nuclear receptor ERRγ mediates antidiabetic effect through inhibition of hepatic gluconeogenesis. Diabetes 2013, 62, 3093–3102. [Google Scholar] [CrossRef][Green Version]
- Kim, D.K.; Jeong, J.H.; Lee, J.M.; Kim, K.S.; Park, S.H.; Kim, Y.D.; Koh, M.; Shin, M.; Jung, Y.S.; Kim, H.S.; et al. Inverse agonist of estrogen-related receptor γ controls Salmonella typhimurium infection by modulating host iron homeostasis. Nat. Med. 2014, 20, 419–424. [Google Scholar] [CrossRef]
- Kim, J.; Im, C.Y.; Yoo, E.K.; Ma, M.J.; Kim, S.-B.; Hong, E.; Chin, J.; Hwang, H.; Lee, S.; Kim, N.D.; et al. Identification of Selective ERRγ Inverse Agonists. Molecules 2016, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.D.; Jeong, S.Y.; Lee, S.W.; Ha, J.H.; Lee, I.K.; Kim, S.H.; Kim, J.; Cho, S.J.; Ahn, B.C.; Lee, J.; et al. Inverse agonist of estrogen-related receptor G enhances sodium iodide symporter function through mitogen-activated protein kinase signaling in anaplastic thyroid cancer cells. J. Nucl. Med. 2015, 56, 1690–1696. [Google Scholar] [CrossRef] [PubMed]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.G.; Negrini, M.; et al. E2F1-Regulated MicroRNAs Impair TGFβ-Dependent Cell-Cycle Arrest and Apoptosis in Gastric Cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.H.; Choi, H.; Oshima, M.; Cheong, J.H.; Kim, S.; Lee, J.H.; Park, Y.S.; Choi, H.S.; Kweon, M.N.; Pack, C.G.; et al. Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Kim, J.; Hwang, H.; Yoon, H.; Lee, J.E.; Oh, J.M.; An, H.; Ji, H.D.; Lee, S.; Cha, E.; Ma, M.J.; et al. An orally available inverse agonist of estrogen-related receptor gamma showed expanded efficacy for the radioiodine therapy of poorly differentiated thyroid cancer. Eur. J. Med. Chem. 2020, 205, 112501. [Google Scholar] [CrossRef]
- Singh, T.D.; Song, J.; Kim, J.; Chin, J.; Ji, H.D.; Lee, J.E.; Lee, S.B.; Yoon, H.; Yu, J.H.; Kim, S.K.; et al. A Novel Orally Active Inverse Agonist of Estrogen-related Receptor Gamma (ERRg), DN200434, A Booster of NIS in Anaplastic Thyroid Cancer. Clin. Cancer Res. 2019, 25, 5069–5081. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]




| Protein Accession 1 | Protein Code | Gene Name | Fold-Change WT to Mutant | p-Value (p ≤ 0.05 Student’s t-Test) |
|---|---|---|---|---|
| Pkd1 deletion at postnatal day 14 (p14 group) | ||||
| Q00898 | A1AT5 | Serpina1e | 2.33 | 1.32 × 10−3 |
| P07356 | ANXA2 | Anxa2 | 2.27 | 3.72 × 10−6 |
| P12710 | FABPL | Fabp1 | 2.20 | 6.58 × 10−7 |
| Q8BWU5 | OSGEP | Osgep | 2.19 | 4.45 × 10−3 |
| Q05816 | FABP5 | Fabp5 | 2.04 | 3.76 × 10−4 |
| Q8VED5 | K2C79 | Krt79 | 0.43 | 2.82 × 10−3 |
| Pkd1 deletion at postnatal day 12 (p12 group) | ||||
| P12246 | SAMP | Apcs | 4.37 | 4.79 × 10−7 |
| Q71KU9 | FGL1 | Fgl1 | 3.52 | 5.02 × 10−16 |
| Q60590 | A1AG1 | Orm1 | 3.33 | 2.99 × 10−9 |
| Q9JHU9 | INO1 | Isyna1 | 3.32 | 8.19 × 10−9 |
| Q6P8J7 | KCRS | Ckmt2 | 2.71 | 3.01 × 10−2 |
| O55222 | ILK | Ilk | 2.67 | 7.46 × 10−4 |
| P07310 | KCRM | Ckm | 2.67 | 1.13 × 10−2 |
| Q61024 | ASNS | Asns | 2.58 | 1.21 × 10−9 |
| Q05816 | FABP5 | Fabp5 | 2.51 | 2.28 × 10−6 |
| P31725 | S10A9 | S100a9 | 2.39 | 1.45 × 10−6 |
| Q8K0E8 | FIBB | Fgb | 2.33 | 1.21 × 10−14 |
| Q64464 | CP3AD | Cyp3a13 | 2.18 | 3.77 × 10−16 |
| Q91X72 | HEMO | Hpx | 2.16 | 1.08 × 10−13 |
| Q8R429 | AT2A1 | Atp2a1 | 2.15 | 2.20 × 10−2 |
| P70697 | DCUP | Urod | 2.15 | 3.57 × 10−6 |
| E9PV24 | FIBA | Fga | 2.14 | 4.78 × 10−11 |
| Q8VCM7 | FIBG | Fgg | 2.09 | 2.84 × 10−12 |
| P01029 | CO4B | C4b | 2.08 | 1.51 × 10−14 |
| O09012 | PEX5 | Pex5 | 2.01 | 8.43 × 10−3 |
| P00405 | COX2 | Mtco2 | 2.01 | 1.35 × 10−4 |
| P05208 | CEL2A | Cela2a | 0.35 | 2.37 × 10−2 |
| Q63836 | SBP2 | Selenbp2 | 0.39 | 3.27 × 10−5 |
| Q9NYQ2 | HAOX2 | Hao2 | 0.42 | 2.40 × 10−11 |
| Q9D1L0 | CHCH2 | Chchd2 | 0.45 | 1.79 × 10−7 |
| P16015 | CAH3 | Ca3 | 0.47 | 1.19 × 10−4 |
| Q8R1F5 | HYI | Hyi | 0.49 | 5.79 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordido, A.; Vizoso-Gonzalez, M.; Nuñez-Gonzalez, L.; Molares-Vila, A.; Chantada-Vazquez, M.d.P.; Bravo, S.B.; Garcia-Gonzalez, M.A. Quantitative Proteomic Study Unmasks Fibrinogen Pathway in Polycystic Liver Disease. Biomedicines 2022, 10, 290. https://doi.org/10.3390/biomedicines10020290
Cordido A, Vizoso-Gonzalez M, Nuñez-Gonzalez L, Molares-Vila A, Chantada-Vazquez MdP, Bravo SB, Garcia-Gonzalez MA. Quantitative Proteomic Study Unmasks Fibrinogen Pathway in Polycystic Liver Disease. Biomedicines. 2022; 10(2):290. https://doi.org/10.3390/biomedicines10020290
Chicago/Turabian StyleCordido, Adrian, Marta Vizoso-Gonzalez, Laura Nuñez-Gonzalez, Alberto Molares-Vila, Maria del Pilar Chantada-Vazquez, Susana B. Bravo, and Miguel A. Garcia-Gonzalez. 2022. "Quantitative Proteomic Study Unmasks Fibrinogen Pathway in Polycystic Liver Disease" Biomedicines 10, no. 2: 290. https://doi.org/10.3390/biomedicines10020290
APA StyleCordido, A., Vizoso-Gonzalez, M., Nuñez-Gonzalez, L., Molares-Vila, A., Chantada-Vazquez, M. d. P., Bravo, S. B., & Garcia-Gonzalez, M. A. (2022). Quantitative Proteomic Study Unmasks Fibrinogen Pathway in Polycystic Liver Disease. Biomedicines, 10(2), 290. https://doi.org/10.3390/biomedicines10020290






