Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Clinical Examination of Patients
2.3. Biochemical Investigations
2.4. SNP Genotyping
2.5. Statistical Analysis
2.6. Bioinformatics Analysis
3. Results
3.1. Association of Gene Polymorphisms with the Risk of Coronary Artery Disease, Plasma Lipids, and Carotid Intima-Media Thickness
3.2. Smoking as a Triggering Factor Modifying the Genetic Effects on Plasma Lipids, CIMT, and CAD Risk
3.3. Replication for Associations between SNPs and Cardiovacsular Phenotypes in Independent Populations
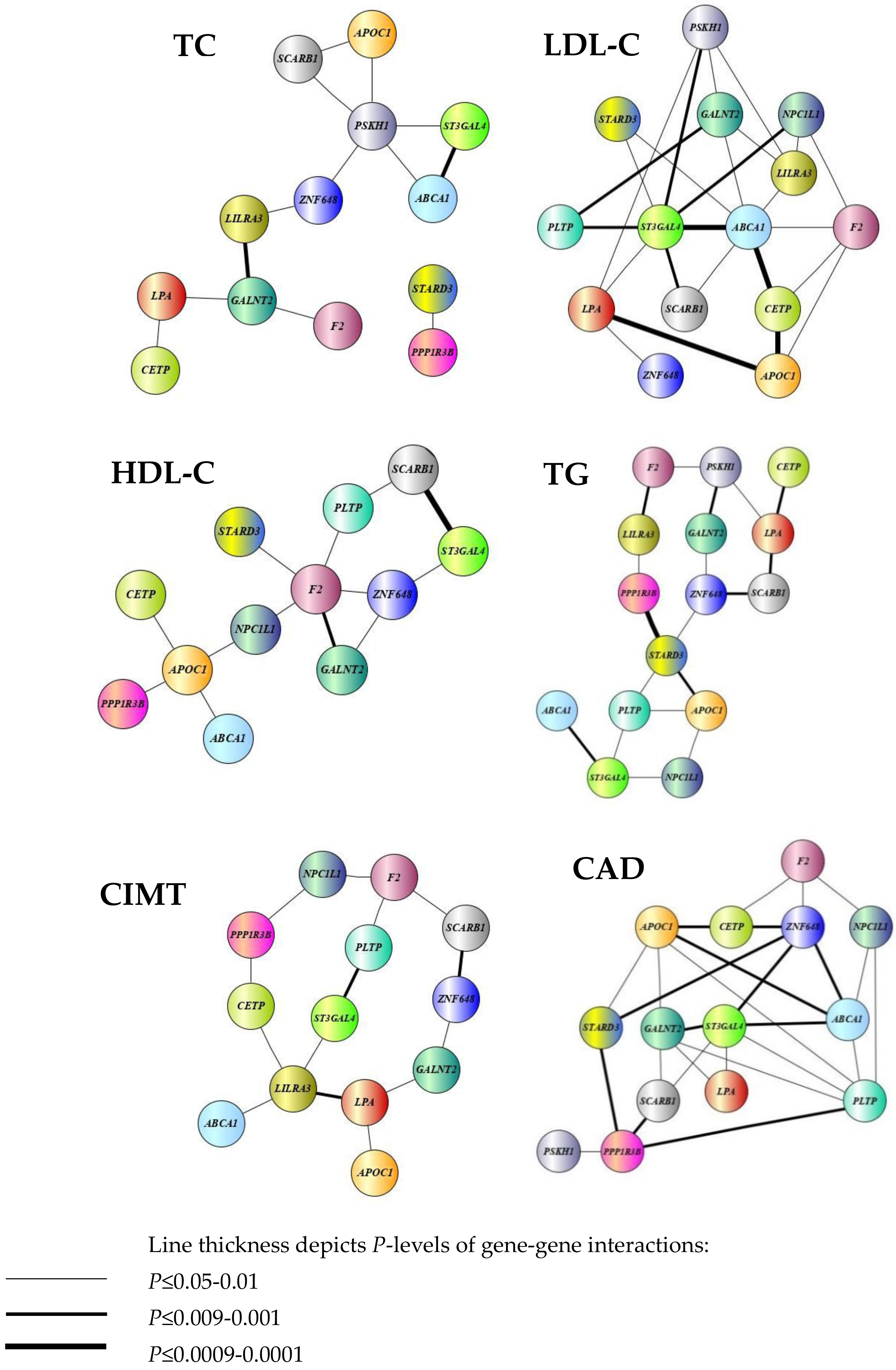
3.4. Analysis of Pairwise SNP-SNP Interactions Contributing to the Studied Cardiovascular Phenotypes
3.5. Modeling for Gene–Gene and Gene–Environment Interactions Determining the Cardiovascular Phenotypes
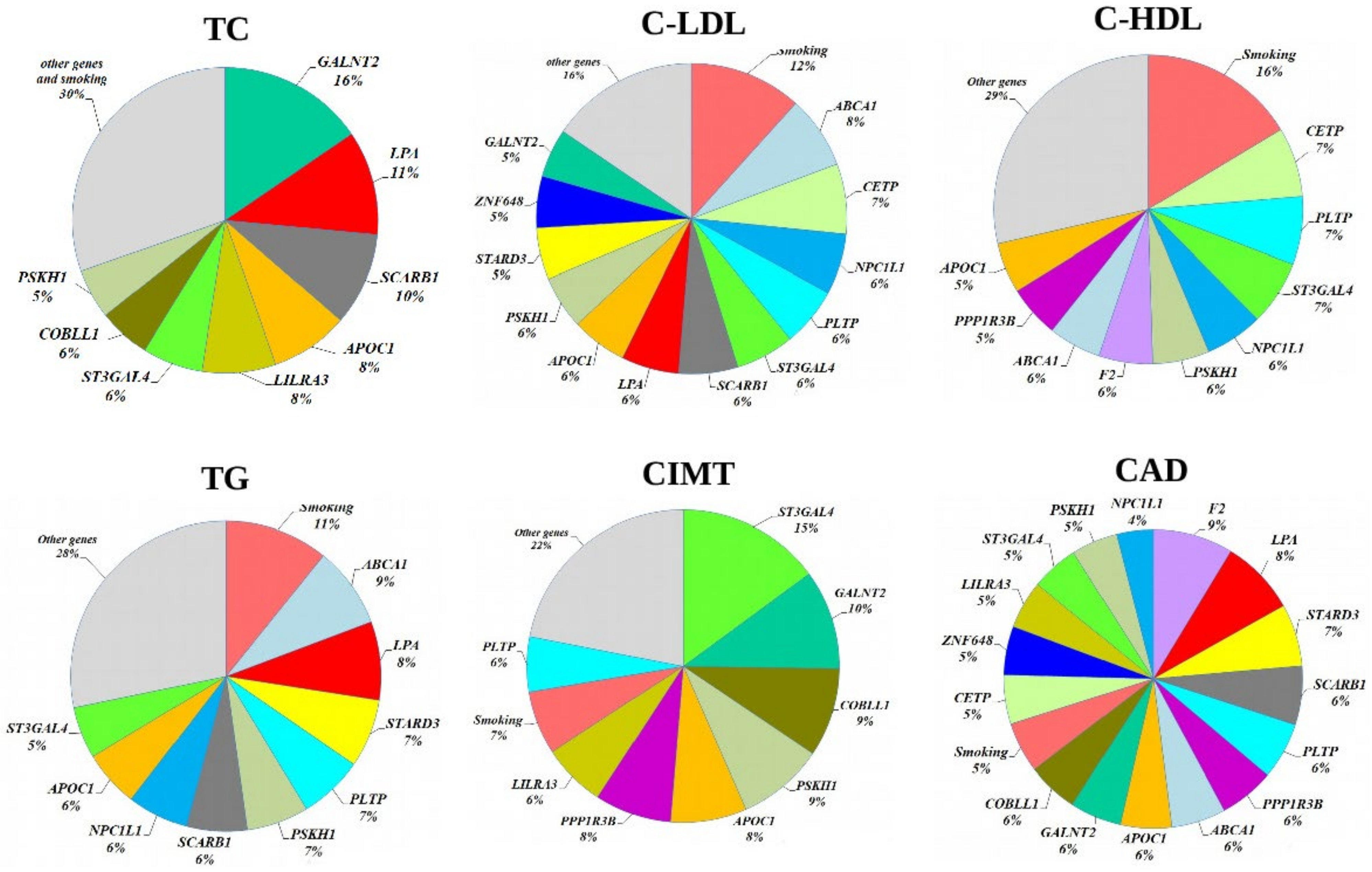
3.6. Functional Annotation of the Studied Gene Polymorphisms
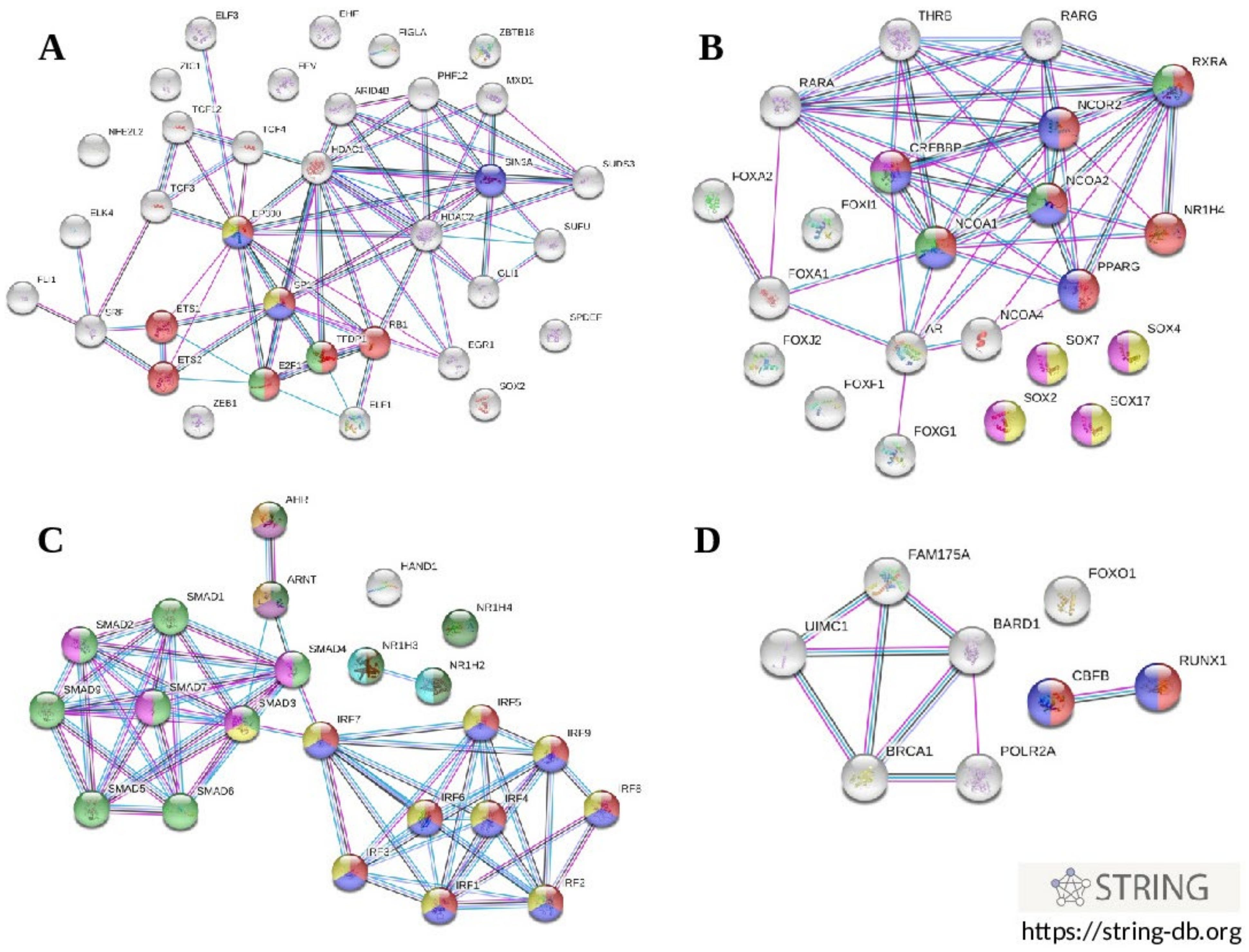
3.7. Enrichment Analysis of Regulatory Gene Networks in Which the Studied Gene Polymorphism Might Be Involved
4. Discussion
4.1. Summary of the Study Findings and Their Comparison with Literature
4.2. The Contribution of Gene–Gene Interactions to the Studied Cardiovascular Phenotypes
4.3. Functional Effects of the Studied SNPs and Their Link to the Pathogenesis of Coronary Artery Disease
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardiovascular Diseases (CVDs) WHO Fact Sheet. 2017. Available online: https://www.who.int/ru/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 21 September 2018).
- Roger, V.L.; Go, A.S.; Lloyd-Jones, D.M.; Benjamin, E.J.; Berry, J.D.; Borden, W.B.; Bravata, D.M.; Dai, S.; Ford, E.S.; Fox, C.S.; et al. Heart Disease and Stroke Statistics—2012 Update: A Report from the American Heart Association. Circulation 2012, 125, e2–e220. [Google Scholar] [CrossRef] [PubMed]
- Gofman, J.W.; Lindgren, F.; Elliott, H.; Mantz, W.; Hewitt, J.; Strisower, B.; Herring, V.; Lyon, T.P. The Role of Lipids and Lipoproteins in Atherosclerosis. 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK343489/ (accessed on 7 December 2020).
- Jin, J.-L.; Zhang, H.-W.; Cao, Y.-X.; Liu, H.-H.; Hua, Q.; Li, Y.-F.; Zhang, Y.; Wu, N.-Q.; Zhu, C.-G.; Xu, R.-X.; et al. Association of small dense low-density lipoprotein with cardiovascular outcome in patients with coronary artery disease and diabetes: A prospective, observational cohort study. Cardiovasc. Diabetol. 2020, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-W.; Jin, J.-L.; Cao, Y.-X.; Liu, H.-H.; Zhang, Y.; Guo, Y.-L.; Wu, N.-Q.; Zhu, C.-G.; Gao, Y.; Xu, R.-X.; et al. Association of small dense LDL-cholesterol with disease severity, hypertension status and clinical outcome in patients with coronary artery disease. J. Hypertens. 2020, 39, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nezu, T.; Hosomi, N.; Aoki, S.; Matsumoto, M. Carotid Intima-Media Thickness for Atherosclerosis. J. Atheroscler. Thromb. 2016, 23, 18–31. [Google Scholar] [CrossRef]
- Khera, A.V.; Kathiresan, A.V.K.S. Genetics of coronary artery disease: Discovery, biology and clinical translation. Nat. Rev. Genet. 2017, 18, 331–344. [Google Scholar] [CrossRef]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [CrossRef]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, A.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2013, 42, D1001–D1006. [Google Scholar] [CrossRef]
- Willer, C.J.; Sanna, S.; Jackson, A.U.; Scuteri, A.; Bonnycastle, L.L.; Clarke, R.; Heath, S.C.; Timpson, N.J.; Najjar, S.S.; Stringham, H.M.; et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008, 40, 161–169. [Google Scholar] [CrossRef]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef]
- Chasman, D.I.; Pare, G.; Zee, R.Y.; Parker, A.N.; Cook, N.R.; Buring, J.E.; Kwiatkowski, D.J.; Rose, L.M.; Smith, J.D.; Williams, P.T.; et al. Genetic Loci Associated with Plasma Concentration of Low-Density Lipoprotein Cholesterol, High-Density Lipoprotein Cholesterol, Triglycerides, Apolipoprotein A1, and Apolipoprotein B Among 6382 White Women in Genome-Wide Analysis with Replication. Circ. Cardiovasc. Genet. 2008, 1, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; Isono, M.; Katsuya, T.; Yokota, M.; Yamamoto, K.; Nabika, T.; Shimokawa, K.; Nakashima, E.; Sugiyama, T.; Rakugi, H.; et al. Association of Genetic Variants Influencing Lipid Levels with Coronary Artery Disease in Japanese Individuals. PLoS ONE 2012, 7, e46385. [Google Scholar] [CrossRef] [PubMed]
- Mirzaev, K.; Abdullaev, S.; Akmalova, K.; Sozaeva, J.; Grishina, E.; Shuev, G.; Bolieva, L.; Sozaeva, M.; Zhuchkova, S.; Gimaldinova, N.; et al. Interethnic differences in the prevalence of main cardiovascular pharmacogenetic biomarkers. Pharmacogenomics 2020, 21, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Heinig, M. Using Gene Expression to Annotate Cardiovascular GWAS Loci. Front. Cardiovasc. Med. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Polonikov, A.; Solodilova, M.; Ivanov, V.P.; Shestakov, A.M.; Ushachev, D.V.; Vialykh, E.K.; Vasil’Eva, O.V.; Poliakova, N.V.; Antsupov, V.V.; Kabanina, V.; et al. A protective effect of GLY272SER polymorphism of GNB3 gene in development of essential hypertension and its relations with environmental hypertension risk factors. Ter. Arkh. 2011, 83, 55–60. [Google Scholar] [PubMed]
- Polonikov, A.V.; Ushachev, D.V.; Ivanov, V.P.; Churnosov, M.I.; Freidin, M.B.; Ataman, A.V.; Harbuzova, V.Y.; Bykanova, M.A.; Bushueva, O.Y.; Solodilova, M.A. Altered erythrocyte membrane protein composition mirrors pleiotropic effects of hypertension susceptibility genes and disease pathogenesis. J. Hypertens. 2015, 33, 2265–2277. [Google Scholar] [CrossRef]
- Polonikov, A.; Kharchenko, A.; Bykanova, M.; Sirotina, S.; Ponomarenko, I.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Bushueva, O.; Churnosov, M.; et al. Polymorphisms of CYP2C8, CYP2C9 and CYP2C19 and risk of coronary heart disease in Russian population. Gene 2017, 627, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Sirotina, S.; Ponomarenko, I.; Kharchenko, A.; Bykanova, M.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Churnosov, M.; Solodilova, M.; Polonikov, A. A Novel Polymorphism in the Promoter of theCYP4A11Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers 2018, 2018, 5812802. [Google Scholar] [CrossRef]
- Azarova, I.E.; Klyosova, E.Y.; Lazarenko, V.A.; Konoplya, A.I.; Polonikov, A.V. rs11927381 Polymorphism and Type 2 Diabetes Mellitus: Contribution of Smoking to the Realization of Susceptibility to the Disease. Bull. Exp. Biol. Med. 2020, 168, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Medvedeva, M.V. Associations of rs2305948 and rs1870377 polymorphic variants of the vascular endothelial growth factor receptor type 2 (KDR) gene with the risk of coronary heart disease. Res. Results Biomed. 2021, 7, 32–43. [Google Scholar] [CrossRef]
- Kononov, S.; Mal, G.; Azarova, I.; Klyosova, E.; Bykanova, M.; Churnosov, M.; Polonikov, A. Pharmacogenetic loci for rosuvastatin are associated with intima-media thickness change and coronary artery disease risk. Pharmacogenomics 2022, 23, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Polonikov, A.V.; Kursk State Medical University; Klyosova, E.Y.; Azarova, I.E. Bioinformatic tools and internet resources for functional annotation of polymorphic loci detected by genome wide association studies of multifactorial diseases (review). Res. Results Biomed. 2021, 7, 15–31. [Google Scholar] [CrossRef]
- Kathiresan, S.; Melander, O.; Guiducci, C.; Surti, A.; Burtt, N.P.; Rieder, M.J.; Cooper, G.M.; Roos, C.; Voight, B.F.; Havulinna, A.S.; et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008, 40, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Mack, S.; Coassin, S.; Rueedi, R.; Yousri, N.A.; Seppälä, I.; Gieger, C.; Schönherr, S.; Forer, L.; Erhart, G.; Marques-Vidal, P.; et al. A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J. Lipid Res. 2017, 58, 1834–1844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gan, W.; Tian, C.; Li, H.; Lin, X.; Chen, Y. Association ofPPP1R3Bpolymorphisms with blood lipid and C-reactive protein levels in a Chinese population. J. Diabetes 2013, 5, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- González, J.R.; Armengol, L.; Solé, X.; Guinó, E.; Mercader, J.M.; Estivill, X.; Moreno, V. SNPassoc: An R package to perform whole genome association studies. Bioinformatics 2007, 23, 654–655. [Google Scholar] [CrossRef]
- Calle, M.L.; Urrea, V.; Vellalta, G.; Malats, N.; Steen, K.V. Improving strategies for detecting genetic patterns of disease susceptibility in association studies. Stat. Med. 2008, 27, 6532–6546. [Google Scholar] [CrossRef]
- Calle, M.L.; Urrea, V.; Malats, N.; Van Steen, K. mbmdr: An R package for exploring gene–gene interactions associated with binary or quantitative traits. Bioinformatics 2010, 26, 2198–2199. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-Dimensionality Reduction Reveals High-Order Interactions among Estrogen-Metabolism Genes in Sporadic Breast Cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Hudson, R.; Harrison, C.; Craven, M.; Keleş, S. atSNP Search: A web resource for statistically evaluating influence of human genetic variation on transcription factor binding. Bioinformatics 2018, 35, 2657–2659. [Google Scholar] [CrossRef] [PubMed]
- Sing, C.F.; Stengârd, J.H.; Kardia, S.L. Genes, Environment, and Cardiovascular Disease. Arter. Thromb. Vasc. Biol. 2003, 23, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Cervellin, G. The interplay between genetics, epigenetics and environment in modulating the risk of coronary heart disease. Ann. Transl. Med. 2016, 4, 460. [Google Scholar] [CrossRef] [PubMed]
- Teufel, A.; Krupp, M.; Weinmann, A.; Galle, P.R. Current bioinformatics tools in genomic biomedical research (Review). Int. J. Mol. Med. 2006, 17, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Briefings Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Suchindran, S.; Rivedal, D.; Guyton, J.R.; Milledge, T.; Gao, X.; Benjamin, A.; Rowell, J.; Ginsburg, G.S.; McCarthy, J.J. Genome-Wide Association Study of Lp-PLA2 Activity and Mass in the Framingham Heart Study. PLoS Genet. 2010, 6, e1000928. [Google Scholar] [CrossRef][Green Version]
- Saxena, R.; Voight, B.F.; Lyssenko, V.; Burtt, N.P.; de Bakker, P.I.W.; Chen, H.; Roix, J.J.; Kathiresan, S.; Hirschhorn, J.N.; Daly, M.J.; et al. Genome-Wide Association Analysis Identifies Loci for Type 2 Diabetes and Triglyceride Levels. Science 2007, 316, 1331–1336. [Google Scholar] [CrossRef]
- Waterworth, D.M.; Ricketts, S.L.; Song, K.; Chen, L.; Zhao, J.H.; Ripatti, S.; Aulchenko, Y.; Zhang, W.; Yuan, X.; Lim, N.; et al. Genetic Variants Influencing Circulating Lipid Levels and Risk of Coronary Artery Disease. Arter. Thromb. Vasc. Biol. 2010, 30, 2264–2276. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.J.; Theusch, E.; Haldar, T.; Ranatunga, D.K.; Jorgenson, E.; Medina, M.W.; Kvale, M.N.; Kwok, P.-Y.; Schaefer, C.; Krauss, R.M.; et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018, 50, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Rasheed, A.; Tikkanen, E.; Lee, J.-J.; Butterworth, A.S.; Howson, J.M.M.; Assimes, T.L.; Chowdhury, R.; Orho-Melander, M.; Damrauer, S.; et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 2017, 49, 1450–1457. [Google Scholar] [CrossRef]
- Li, H.; Wetten, S.; Li, L.; Jean, P.L.S.; Upmanyu, R.; Surh, L.; Hosford, D.; Barnes, M.R.; Briley, J.D.; Borrie, M.; et al. Candidate Single-Nucleotide Polymorphisms from a Genomewide Association Study of Alzheimer Disease. Arch. Neurol. 2008, 65, 45–53. [Google Scholar] [CrossRef]
- Webster, J.A.; Myers, A.J.; Pearson, J.V.; Craig, D.W.; Hu-Lince, D.; Coon, K.D.; Zismann, V.L.; Beach, T.; Leung, D.; Bryden, L.; et al. Sorl1 as an Alzheimer’s Disease Predisposition Gene? Neurodegener. Dis. 2007, 5, 60–64. [Google Scholar] [CrossRef] [PubMed]
- The AMD Gene Consortium. Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013, 45, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Van der Harst, P.; Verweij, N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018, 122, 433–443. [Google Scholar] [CrossRef]
- Joshi, P.K.; Pirastu, N.; Kentistou, K.A.; Fischer, K.; Hofer, E.; Schraut, K.E.; Clark, D.W.; Nutile, T.; Barnes, C.L.K.; Timmers, P.R.H.J.; et al. Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat. Commun. 2017, 8, 910. [Google Scholar] [CrossRef]
- Pilling, L.C.; Kuo, C.-L.; Sicinski, K.; Tamosauskaite, J.; Kuchel, G.; Harries, L.W.; Herd, P.; Wallace, R.; Ferrucci, L.; Melzer, D. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging 2017, 9, 2504–2520. [Google Scholar] [CrossRef]
- Spracklen, C.N.; Chen, P.; Kim, Y.J.; Wang, X.; Cai, H.; Li, S.; Long, J.; Wu, Y.; Wang, Y.X.; Takeuchi, F.; et al. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 2017, 26, 1770–1784. [Google Scholar] [CrossRef]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef]
- Klarin, D.; Damrauer, S.M.; Cho, K.; Sun, Y.V.; Teslovich, T.M.; Honerlaw, J.; Gagnon, D.R.; Duvall, S.L.; Li, J.; Peloso, G.M.; et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat. Genet. 2018, 50, 1514–1523. [Google Scholar] [CrossRef] [PubMed]
- De Vries, P.S.; Brown, M.R.; Bentley, A.R.; Sung, Y.J.; Winkler, T.W.; Ntalla, I.; Schwander, K.; Kraja, A.T.; Guo, X.; Franceschini, N.; et al. Multiancestry Genome-Wide Association Study of Lipid Levels Incorporating Gene-Alcohol Interactions. Am. J. Epidemiology 2019, 188, 1033–1054. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.H. A global view of epistasis. Nat. Genet. 2005, 37, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.H.; Gilbert, J.C.; Tsai, C.-T.; Chiang, F.-T.; Holden, T.; Barney, N.; White, B.C. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J. Theor. Biol. 2006, 241, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Paththinige, C.; Sirisena, N.; Dissanayake, V. Genetic determinants of inherited susceptibility to hypercholesterolemia—A comprehensive literature review. Lipids Heal. Dis. 2017, 16, 103. [Google Scholar] [CrossRef] [PubMed]
- Jong, M.C.; Hofker, M.H.; Havekes, L.M. Role of ApoCs in Lipoprotein Metabolism. Arter. Thromb. Vasc. Biol. 1999, 19, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Myint, P.K.; Voora, D.; Laskowitz, D.T.; Shi, P.; Ren, F.; Wang, H.; Yang, Y.; Huo, Y.; Gao, W.; et al. Genome-wide association study on progression of carotid artery intima media thickness over 10 years in a Chinese cohort. Atherosclerosis 2015, 243, 30–37. [Google Scholar] [CrossRef]
- De Vries, R.; Dallinga-Thie, G.M.; Smit, A.J.; Wolffenbuttel, B.H.R.; van Tol, A.; Dullaart, R.P.F. Elevated plasma phospholipid transfer protein activity is a determinant of carotid intima-media thickness in type 2 diabetes mellitus. Diabetologia 2005, 49, 398–404. [Google Scholar] [CrossRef]
- Abdushi, S.A.; Nazreku, F.D.; Kryeziu, F.U. Increased carotid intima-media thickness associated with high hs-CRP levels is a predictor of unstable coronary artery disease: Cardiovascular topic. Cardiovasc. J. Afr. 2013, 24, 270–273. [Google Scholar] [CrossRef]
- Paraskevas, K.I. Increased carotid intima-media thickness and coronary artery disease share more links besides inflammation. Int. J. Cardiol. 2015, 204, 54. [Google Scholar] [CrossRef] [PubMed]
- Ligthart, S.; Vaez, A.; Hsu, Y.-H.; Stolk, R.; Uitterlinden, A.G.; Hofman, A.; Alizadeh, B.Z.; Franco, O.; Dehghan, A. Bivariate genome-wide association study identifies novel pleiotropic loci for lipids and inflammation. BMC Genom. 2016, 17, 443. [Google Scholar] [CrossRef]
- Germain, M.; Chasman, D.I.; De Haan, H.; Tang, W.; Lindström, S.; Weng, L.-C.; De Andrade, M.; De Visser, M.C.; Wiggins, K.L.; Suchon, P. Meta-analysis of 65,734 Individuals Identifies TSPAN15 and SLC44A2 as Two Susceptibility Loci for Venous Thromboembolism. Am. J. Hum. Genet. 2015, 96, 532–542. [Google Scholar] [CrossRef]
- Hinds, D.A.; Buil, A.; Ziemek, D.; Martinez-Perez, A.; Malik, R.; Folkersen, L.; Germain, M.; Mälarstig, A.; Brown, A.; Soria, J.M.; et al. Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum. Mol. Genet. 2016, 25, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Nordestgaard, B.G.; Chapman, M.J.; Ray, K.; Borén, J.; Andreotti, F.; Watts, G.; Ginsberg, H.; Amarenco, P.; Catapano, A.L.; Descamps, O.S.; et al. Lipoprotein(a) as a cardiovascular risk factor: Current status. Eur. Hear. J. 2010, 31, 2844–2853. [Google Scholar] [CrossRef]
- Yang, X.; Sethi, A.; Yanek, L.R.; Knapper, C.; Nordestgaard, B.G.; Tybjærg-Hansen, A.; Becker, D.M.; Mathias, R.A.; Remaley, A.T.; Becker, L.C. SCARB1 Gene Variants Are Associated with the Phenotype of Combined High High-Density Lipoprotein Cholesterol and High Lipoprotein (a). Circ. Cardiovasc. Genet. 2016, 9, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Hebbar, P.; Nizam, R.; Melhem, M.; Alkayal, F.; Elkum, N.; John, S.E.; Tuomilehto, J.; Alsmadi, O.; Thanaraj, T.A. Genome-wide association study identifies novel recessive genetic variants for high TGs in an Arab population. J. Lipid Res. 2018, 59, 1951–1966. [Google Scholar] [CrossRef] [PubMed]
- Borthwick, F.; Allen, A.-M.; Taylor, J.M.; Graham, A. Overexpression of STARD3 in human monocyte/macrophages induces an anti-atherogenic lipid phenotype. Clin. Sci. 2010, 119, 265–272. [Google Scholar] [CrossRef]
- Winkler, T.W.; Justice, A.E.; Graff, M.; Barata, L.; Feitosa, M.F.; Chu, S.; Czajkowski, J.; Esko, T.; Fall, T.; Kilpeläinen, T.O.; et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet. 2015, 11, e1005378. [Google Scholar] [CrossRef] [PubMed]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef]
- Rockman, M.V.; Kruglyak, L. Genetics of global gene expression. Nat. Rev. Genet. 2006, 7, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Claudel, T.; Inoue, Y.; Barbier, O.; Duran-Sandoval, D.; Kosykh, V.; Fruchart, J.; Fruchart, J.-C.; Gonzalez, F.J.; Staels, B. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology 2003, 125, 544–555. [Google Scholar] [CrossRef]
- Jakobsson, T.; Venteclef, N.; Toresson, G.; Damdimopoulos, A.E.; Ehrlund, A.; Lou, X.; Sanyal, S.; Steffensen, K.R.; Gustafsson, J.; Treuter, E. GPS2 Is Required for Cholesterol Efflux by Triggering Histone Demethylation, LXR Recruitment, and Coregulator Assembly at the ABCG1 Locus. Mol. Cell 2009, 34, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, R.M.; Van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.L.; Renooij, W.; Murzilli, S.; Klomp, L.W.J.; Siersema, P.D.; Schipper, M.E.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Schulte, K.W.; Green, E.; Wilz, A.; Platten, M.; Daumke, O. Structural Basis for Aryl Hydrocarbon Receptor-Mediated Gene Activation. Structure 2017, 25, 1025–1033.e3. [Google Scholar] [CrossRef] [PubMed]
- Seok, S.-H.; Lee, W.; Jiang, L.; Molugu, K.; Zheng, A.; Li, Y.; Park, S.; Bradfield, C.A.; Xing, Y. Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex. Proc. Natl. Acad. Sci. USA 2017, 114, 5431–5436. [Google Scholar] [CrossRef]
- Alshaarawy, O.; Elbaz, H.; Andrew, M.E. The association of urinary polycyclic aromatic hydrocarbon biomarkers and cardiovascular disease in the US population. Environ. Int. 2016, 89–90, 174–178. [Google Scholar] [CrossRef]
- Amin, M.-M.; Poursafa, P.; Moosazadeh, M.; Abedini, E.; Hajizadeh, Y.; Mansourian, M.; Pourzamani, H. A systematic review on the effects of polycyclic aromatic hydrocarbons on cardiometabolic impairment. Int. J. Prev. Med. 2017, 8, 19. [Google Scholar] [CrossRef]
- Alhamdow, A.; Lindh, C.; Albin, M.; Gustavsson, P.; Tinnerberg, H.; Broberg, K. Early markers of cardiovascular disease are associated with occupational exposure to polycyclic aromatic hydrocarbons. Sci. Rep. 2017, 7, 9426. [Google Scholar] [CrossRef]
- Vogel, C.F.A.; Sciullo, E.; Matsumura, F. Activation of Inflammatory Mediators and Potential Role of Ah-Receptor Ligands in Foam Cell Formation. Cardiovasc. Toxicol. 2004, 4, 363–374. [Google Scholar] [CrossRef]
- Liu, S.; Cai, X.; Wu, J.; Cong, Q.; Chen, X.; Li, T.; Du, F.; Ren, J.; Wu, Y.-T.; Grishin, N.V.; et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 2015, 347, aaa2630. [Google Scholar] [CrossRef] [PubMed]
- Joehanes, R.; Ying, S.; Huan, T.; Johnson, A.D.; Raghavachari, N.; Wang, R.; Liu, P.; Woodhouse, K.A.; Sen, S.K.; Tanriverdi, K.; et al. Gene Expression Signatures of Coronary Heart Disease. Arter. Thromb. Vasc. Biol. 2013, 33, 1418–1426. [Google Scholar] [CrossRef] [PubMed]
- Mishiro, T.; Ishihara, K.; Hino, S.; Tsutsumi, S.; Aburatani, H.; Shirahige, K.; Kinoshita, Y.; Nakao, M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009, 28, 1234–1245. [Google Scholar] [CrossRef] [PubMed]
- Cayrol, C.; Lacroix, C.; Mathe, C.; Ecochard, V.; Ceribelli, M.; Loreau, E.; Lazar, V.; Dessen, P.; Mantovani, R.; Aguilar, L.; et al. The THAP–zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 2006, 109, 584–594. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A.; Larson, S.B. The structure of human apolipoprotein C-1 in four different crystal forms. J. Lipid Res. 2019, 60, 400–411. [Google Scholar] [CrossRef]
- Lowe, J.K.; Maller, J.B.; Pe’Er, I.; Neale, B.M.; Salit, J.; Kenny, E.E.; Shea, J.L.; Burkhardt, R.; Smith, J.G.; Ji, W.; et al. Genome-Wide Association Studies in an Isolated Founder Population from the Pacific Island of Kosrae. PLoS Genet. 2009, 5, e1000365. [Google Scholar] [CrossRef]
- Deelen, J.; Beekman, M.; Uh, H.-W.; Broer, L.; Ayers, K.L.; Tan, Q.; Kamatani, Y.; Bennet, A.M.; Tamm, R.; Trompet, S.; et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum. Mol. Genet. 2014, 23, 4420–4432. [Google Scholar] [CrossRef]
- Cheng, C.; Tempel, D.; Dekker, W.K.D.; Haasdijk, R.; Chrifi, I.; Bos, F.L.; Wagtmans, K.; Van De Kamp, E.H.; Blonden, L.; Biessen, E.A.; et al. Ets2 Determines the Inflammatory State of Endothelial Cells in Advanced Atherosclerotic Lesions. Circ. Res. 2011, 109, 382–395. [Google Scholar] [CrossRef]
- Glenn, K.C.; Frost, G.H.; Bergmann, J.S.; Carney, D.H. Synthetic peptides bind to high-affinity thrombin receptors and modulate thrombin mitogenesis. Pept. Res. 1988, 1, 65–73. [Google Scholar]
- Borissoff, J.I.; Spronk, H.M.; Heeneman, S.; Cate, H.T. Is thrombin a key player in the ’coagulation-atherogenesis’ maze? Cardiovasc. Res. 2009, 82, 392–403. [Google Scholar] [CrossRef]
- Borissoff, J.I.; Spronk, H.M.; Cate, H.T. The Hemostatic System as a Modulator of Atherosclerosis. N. Engl. J. Med. 2011, 364, 1746–1760. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, J.; Deng, S.; Yu, K.; Liu, X.; Deng, Q.; Sun, H.; Zhang, X.; He, M.; Guo, H.; et al. Genome-Wide Analysis of DNA Methylation and Cigarette Smoking in a Chinese Population. Environ. Health Perspect. 2016, 124, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Goodale, B.C.; Tilton, S.C.; Corvi, M.M.; Wilson, G.R.; Janszen, D.B.; Anderson, K.A.; Waters, K.; Tanguay, R.L. Structurally distinct polycyclic aromatic hydrocarbons induce differential transcriptional responses in developing zebrafish. Toxicol. Appl. Pharmacol. 2013, 272, 656–670. [Google Scholar] [CrossRef]
- Jorgensen, E.M.; Alderman, M.H.; Taylor, H.S. Preferential epigenetic programming of estrogen response after in utero xenoestrogen (bisphenol-A) exposure. FASEB J. 2016, 30, 3194–3201. [Google Scholar] [CrossRef] [PubMed]
- Collí-Dulá, R.C.; Friedman, M.A.; Hansen, B.; Denslow, N.D. Transcriptomics analysis and hormonal changes of male and female neonatal rats treated chronically with a low dose of acrylamide in their drinking water. Toxicol. Rep. 2016, 3, 414–426. [Google Scholar] [CrossRef][Green Version]
- Gai, Z.; Hiller, C.; Chin, S.H.; Hofstetter, L.; Stieger, B.; Konrad, D.; Kullak-Ublick, G.A. Uninephrectomy augments the effects of high fat diet induced obesity on gene expression in mouse kidney. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2014, 1842, 1870–1878. [Google Scholar] [CrossRef]
- Sui, Y.; Park, S.-H.; Wang, F.; Zhou, C. Perinatal Bisphenol A Exposure Increases Atherosclerosis in Adult Male PXR-Humanized Mice. Endocrinology 2018, 159, 1595–1608. [Google Scholar] [CrossRef]
- Yordy, J.S.; Moussa, O.; Pei, H.; Chaussabel, D.; Li, R.; Watson, D.K. SP100 inhibits ETS1 activity in primary endothelial cells. Oncogene 2004, 24, 916–931. [Google Scholar] [CrossRef][Green Version]
- Brasch, J.; Harrison, O.J.; Ahlsen, G.; Carnally, S.M.; Henderson, R.M.; Honig, B.; Shapiro, L. Structure and Binding Mechanism of Vascular Endothelial Cadherin: A Divergent Classical Cadherin. J. Mol. Biol. 2011, 408, 57–73. [Google Scholar] [CrossRef]
- Verschuur, M.; de Jong, M.; Felida, L.; de Maat, M.P.; Vos, H.L. A Hepatocyte Nuclear Factor-3 Site in the Fibrinogen β Promoter Is Important for Interleukin 6-induced Expression, and Its Activity Is Influenced by the Adjacent −148C/T Polymorphism. J. Biol. Chem. 2005, 280, 16763–16771. [Google Scholar] [CrossRef]
- The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature 2010, 467, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Tiret, L.; Poirier, O.; Nicaud, V.; Barbaux, S.; Herrmann, S.M.; Perret, C.; Raoux, S.; Francomme, C.; Lebard, G.; Trégouët, D.; et al. Heterogeneity of linkage disequilibrium in human genes has implications for association studies of common diseases. Hum. Mol. Genet. 2002, 11, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Azarova, I.; Klyosova, E.; Polonikov, A. The Link between Type 2 Diabetes Mellitus and the Polymorphisms of Glutathione-Metabolizing Genes Suggests a New Hypothesis Explaining Disease Initiation and Progression. Life 2021, 11, 886. [Google Scholar] [CrossRef] [PubMed]


| Baseline Characteristics | CAD Patients, n = 991 | Healthy Controls, n = 709 | p-Value | |
|---|---|---|---|---|
| Age, mean ± standard deviation | 59.9 ± 8.8 | 60.4 ± 8.1 | 0.23 | |
| Sex | Males, n (%) | 633 (63.9) | 452 (63.8) | 0.96 |
| Females, n (%) | 358 (36.1) | 257 (36.2) | ||
| Body mass index (kg/m2), mean ± standard deviation | 29.98 ± 6.55 | 27.04 ± 4.48 | <0.001 | |
| Hypertension 1, n (%) | 935 (94.3) | 0 (0.0) | - | |
| Diabetes 2, n (%) | 214 (21.6) | 0 (0.0) | - | |
| Smokers 3 (ever/never), n (%) | 434 (43.9) | 355 (51.5) | 0.002 | |
| TC (mmol/L), Me (Q1; Q3) | 5.76 (4.83; 6.24) | NA | - | |
| LDL-C (mmol/L), Me (Q1; Q3) | 2.20 (1.50; 3.77) | NA | - | |
| HDL-C (mmol/L), Me (Q1; Q3) | 1.30 (1.05; 1.62) | NA | - | |
| TG (mmol/L), Me (Q1; Q3) | 2.40 (1.59; 3.70) | NA | - | |
| CIMT (mm), Me (Q1; Q3) | 0.61 (0.53; 0.80) | NA | - | |
| Gene | Polymorphism (SNP ID) | SNP Location | Association of SNP with Plasma Lipids | References |
|---|---|---|---|---|
| ABCA1 | C > T (rs1883025) | Intron | Decrease in TC and HDL-C | [8,25] |
| APOC1 | A > G (rs4420638) | Intergenic | Increase in LDL-C | [11] |
| CETP | C > A (rs3764261) | Promoter | Increase in TC and HDL-C, decrease in LDL-C and TG | [8] |
| COBLL1 | T > C (rs12328675) | 3′UTR | Increase in HDL-C | [8] |
| F2 | T > C (rs3136441) | Intron | Increase in HDL-C | [8] |
| GALNT2 | A > G (rs4846914) | Intron | Increase in HDL-C, increase in TG | [8,25] |
| LILRA3 | G > C (rs386000) | Intron | Increase in HDL-C | [8] |
| LPA | C > T (rs55730499) | Intron | Increase in Lp(a), Increaed risk of CAD | [9,26] |
| NPC1L1 | C > G (rs217406) | intron | Increase in TC | [8] |
| PLTP | T > C (rs6065906) | Promoter | Decrease in HDL-Cl, increase in TG | [8] |
| PSKH1 | G > A (rs16942887) | Intron | Increase in HDL-C | [8,12] |
| ST3GAL4 | A > T (rs11220463) | Intron | Decrease in TC and LDL-C | [8] |
| STARD3 | G > C (rs881844) | Intron | Decrease in HDL-C | [12] |
| ZNF648 | A > G (rs1689800) | Intron | Decrease in HDL-C | [8,12] |
| SCARB1 | T > C (rs838880) | 3′UTR | Increase in HDL-C | [8] |
| PPP1R3B | G > A (rs9987289) | exon, non-coding region | Decrease in TC and HDL-C, increase in LDL-C | [8,27] |
| Gene (SNP ID) | Minor Allele | Minor Allele Frequencies (MAF) in Populations | p | |
|---|---|---|---|---|
| Central Russia (Sample Size) | European Population | |||
| ABCA1 (rs1883025) | T | 0.222 (1697) | 0.240 | 0.40 |
| APOC1 (rs4420638) | G | 0.153 (1669) | 0.198 | 0.02 |
| CETP (rs3764261) | A | 0.251 (1681) | 0.292 | 0.07 |
| COBLL1 (rs12328675) | C | 0.165 (1340) | 0.157 | 0.68 |
| F2 (rs3136441) | C | 0.205 (1675) | 0.121 | <0.0001 |
| GALNT2 (rs4846914) | A | 0.617 (1686) | 0.601 | 0.52 |
| LILRA3 (rs386000) | C | 0.185 (1699) | 0.191 | 0.76 |
| LPA (rs55730499) | T | 0.077 (1673) | 0.076 | 0.94 |
| NPC1L1 (rs217406) | G | 0.211 (1700) | 0.173 | 0.06 |
| PLTP (rs6065906) | C | 0.168 (1697) | 0.204 | 0.06 |
| PSKH1 (rs16942887) | A | 0.122 (1678) | 0.134 | 0.48 |
| ST3GAL4 (rs11220463) | T | 0.166 (1699) | 0.131 | 0.06 |
| STARD3 (rs881844) | G | 0.663 (1693) | 0.668 | 0.83 |
| ZNF648 (rs1689800) | G | 0.353 (1696) | 0.347 | 0.80 |
| SCARB1 (rs838880) | T | 0.684 (1698) | 0.687 | 0.90 |
| PPP1R3B (rs9987289) | A | 0.076 (1699) | 0.075 | 0.94 |
| Gene (SNP ID) | Genotypes | Healthy Controls N (%) | CAD Patients N (%) | OR (95% CI) 1 | p2 |
|---|---|---|---|---|---|
| ABCA1 (rs1883025) | C/C | 234 (59.2) | 466 (62.4) | 1.00 | 0.19 |
| C/T-T/T | 161 (40.8) | 281 (37.6) | 0.84 (0.65–1.09) | ||
| APOC1 (rs4420638) | A/A-G/G | 295 (78.7) | 535 (71.4) | 1.00 | 0.009 |
| A/G | 80 (21.3) | 214 (28.6) | 1.49 (1.10–2.03) | ||
| CETP (rs3764261) | C/C-A/A | 238 (60.2) | 402 (55.1) | 1.00 | 0.17 |
| C/A | 157 (39.8) | 328 (44.9) | 1.20 (0.92–1.56) | ||
| COBLL1 (rs12328675) | T/T-T/C | 333 (93.8) | 447 (90.5) | 1.00 | 0.08 |
| C/C | 22 (6.2) | 47 (9.5) | 1.61 (0.93–2.81) | ||
| F2 (rs3136441) | T/T | 213 (54.3) | 512 (69) | 1.00 | <0.0001 |
| T/C-C/C | 179 (45.7) | 230 (31) | 0.49 (0.37–0.64) | ||
| GALNT2 (rs4846914) | A/A-A/G | 340 (87.6) | 628 (84.3) | 1.00 | 0.17 |
| G/G | 48 (12.4) | 117 (15.7) | 1.30 (0.89–1.89) | ||
| LILRA3 (rs386000) | G/G-G/C | 382 (96.7) | 728 (97.3) | 1.00 | 0.51 |
| C/C | 13 (3.3) | 20 (2.7) | 0.78 (0.37–1.63) | ||
| LPA (rs55730499) | C/C | 354 (89.6) | 596 (81.8) | 1.00 | 0.0007 |
| C/T-T/T | 41 (10.4) | 133 (18.2) | 1.92 (1.30–2.83) | ||
| NPC1L1 (rs217406) | C/C-G/G | 270 (68.3) | 474 (63.3) | 1.00 | 0.056 |
| C/G | 125 (31.6) | 275 (36.7) | 1.30 (0.99–1.70) | ||
| PLTP (rs6065906) | T/T | 251 (63.5) | 536 (71.8) | 1.00 | 0.002 |
| T/C-C/C | 144 (36.5) | 210 (28.1) | 0.66 (0.50–0.86) | ||
| PSKH1 (rs16942887) | G/G-G/A | 378 (98.4) | 733 (98.8) | 1.00 | 0.47 |
| A/A | 6 (1.6) | 9 (1.2) | 0.66 (0.22–1.98) | ||
| ST3GAL4 (rs11220463) | A/A-A/T | 383 (97) | 735 (98.3) | 1.00 | 0.10 |
| T/T | 12 (3) | 13 (1.7) | 0.50 (0.22–1.14) | ||
| STARD3 (rs881844) | G/G | 155 (39.2) | 333 (44.6) | 1.00 | 0.059 |
| G/C-C/C | 240 (60.8) | 413 (55.4) | 0.78 (0.60–1.01) | ||
| ZNF648 (rs1689800) | A/A-A/G | 349 (88.3) | 669 (89.6) | 1.00 | 0.23 |
| G/G | 46 (11.7) | 78 (10.4) | 0.78 (0.52–1.17) | ||
| SCARB1 (rs838880) | T/T | 191 (48.4) | 327 (43.7) | 1.00 | 0.074 |
| T/C-C/C | 204 (51.6) | 421 (56.3) | 1.26 (0.98–1.63) | ||
| PPP1R3B (rs9987289) | G/G | 348 (88.3) | 632 (84.4) | 1.00 | 0.062 |
| G/A-A/A | 46 (11.7) | 117 (15.6) | 1.43 (0.98–2.08) |
| Gene (SNP ID) | Genotypes | Genotype Frequencies | TC | LDL-C | HDL-C | TG | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | Me | Q1/Q3 1 | p2 | Me | Q1/Q3 1 | p2 | Me | Q1/Q3 1 | p2 | Me | Q1/Q3 1 | p2 | ||
| ABCA1 (rs1883025) | CC | 386 | 60.9 | 5.72 | 4.82/6.27 | 0.67 | 2.13 | 1.50/3.54 | 0.19 | 1.32 | 1.04/1.63 | 0.09 | 2.46 | 1.67/3.72 | 0.26 |
| CT | 212 | 33.4 | 5.78 | 4.76/6.20 | 2.50 | 1.73/3.96 | 1.24 | 1.07/1.53 | 1.94 | 1.50/3.68 | |||||
| TT | 36 | 5.7 | 5.80 | 4.99/6.20 | 2.07 | 1.29/3.89 | 1.37 | 1.07/1.62 | 2.39 | 1.41/3.60 | |||||
| APOC1 (rs4420638) | AA | 446 | 70.2 | 5.80 | 4.90/6.20 | 0.17 | 2.20 | 1.48/3.80 | 0.67 | 1.30 | 1.04/1.62 | 0.30 | 2.45 | 1.63/3.68 | 0.052 |
| AG | 177 | 27.9 | 5.52 | 4.70/6.20 | 2.24 | 1.60/3.55 | 1.30 | 1.07/1.63 | 2.20 | 1.51/3.73 | |||||
| GG | 12 | 1.9 | 5.97 | 5.21/6.50 | 2.09 | 1.89/3.40 | 1.46 | 1.12/1.58 | 3.44 | 1.80/4.01 | |||||
| CETP (rs3764261) | CC | 333 | 54.0 | 5.67 | 4.70/6.30 | 0.25 | 2.46 | 1.72/3.80 | 0.042 D | 1.24 | 1.02/1.58 | 0.005 OD | 2.37 | 1.59/3.70 | 0.17 |
| CA | 275 | 44.6 | 5.73 | 4.90/6.20 | 2.10 | 1.48/3.73 | 1.32 | 1.10/1.67 | 2.40 | 1.59/3.71 | |||||
| AA | 9 | 1.5 | 5.90 | 4.88/6.30 | 4.07 | 3.79/4.74 | 1.03 | 0.87/1.24 | 1.81 | 1.53/1.96 | |||||
| COBLL1 (rs12328675) | TT | 296 | 75.5 | 5.59 | 4.60/6.18 | 0.028 OD | 3.51 | 2.31/4.20 | 0.16 | 1.10 | 0.99/1.34 | 0.17 | 1.80 | 1.472.69 | 0.06 |
| TC | 60 | 15.3 | 5.80 | 4.89/6.29 | 3.76 | 2.50/4.20 | 1.18 | 1.01/1.40 | 1.72 | 1.28/2.14 | |||||
| CC | 36 | 9.2 | 5.25 | 4.43/6.00 | 2.94 | 2.06/3.95 | 1.30 | 1.02/1.48 | 1.47 | 0.90/2.68 | |||||
| F2 (rs3136441) | TT | 436 | 69.1 | 5.69 | 4.80/6.20 | 0.18 | 2.13 | 1.46/3.53 | 0.09 | 1.32 | 1.08/1.66 | 0.12 | 2.55 | 1.59/3.71 | 0.80 |
| TC | 179 | 28.4 | 5.80 | 4.81/6.30 | 2.46 | 1.80/3.94 | 1.29 | 1.01/1.58 | 2.05 | 1.63/3.71 | |||||
| CC | 16 | 2.5 | 5.89 | 5.20/6.20 | 3.27 | 1.90/4.08 | 1.05 | 0.92/1.14 | 1.93 | 1.50/3.50 | |||||
| GALNT2 (rs4846914) | AA | 239 | 37.9 | 5.70 | 4.77/6.20 | 0.02 OD | 2.18 | 1.48/3.52 | 0.31 | 1.30 | 1.04/1.62 | 0.07 | 2.49 | 1.62/3.71 | 0.12 |
| AG | 289 | 45.8 | 5.80 | 4.87/6.30 | 2.30 | 1.50/3.90 | 1.30 | 1.07/1.63 | 2.20 | 1.57/3.71 | |||||
| GG | 103 | 16.3 | 5.70 | 4.73/6.20 | 2.18 | 1.77/3.56 | 1.24 | 1.04/1.53 | 2.40 | 1.60/3.54 | |||||
| LILRA3 (rs386000) | GG | 417 | 65.8 | 5.79 | 4.82/6.27 | 0.18 | 2.18 | 1.48/3.68 | 0.68 | 1.30 | 1.06/1.63 | 0.33 | 2.43 | 1.59/3.73 | 0.14 |
| GC | 198 | 31.2 | 5.70 | 4.87/6.20 | 2.24 | 1.70/3.85 | 1.26 | 1.04/1.60 | 2.38 | 1.62/3.68 | |||||
| CC | 19 | 3.0 | 5.46 | 4.14/5.80 | 2.59 | 1.77/3.90 | 1.31 | 1.06/1.50 | 1.86 | 1.39/2.47 | |||||
| LPA (rs55730499) | CC | 515 | 82.5 | 5.80 | 4.86/6.29 | 0.037 OD | 2.18 | 1.50/3.63 | 0.0007 R | 1.30 | 1.07/1.62 | 0.42 | 2.43 | 1.62/3.73 | 0.06 |
| CT | 108 | 17.3 | 5.43 | 4.59/6.17 | 3.30 | 1.80/4.00 | 1.20 | 1.00/1.44 | 1.88 | 1.34/3.04 | |||||
| TT | 1 | 0.2 | 6.00 | 5.80/7.69 | 4.27 | 4.03/5.80 | 0.95 | 0.93/1.19 | 1.63 | 1.59/6.59 | |||||
| NPC1L1 (rs217406) | CC | 375 | 59.1 | 5.80 | 4.81/6.27 | 0.47 | 2.30 | 1.54/3.88 | 0.63 | 1.30 | 1.05/1.62 | 0.09 | 2.40 | 1.61/3.70 | 0.022 R |
| CG | 233 | 36.7 | 5.71 | 4.84/6.22 | 2.09 | 1.48/3.40 | 1.29 | 1.07/1.62 | 2.40 | 1.60/3.75 | |||||
| GG | 27 | 4.3 | 5.40 | 4.89/6.00 | 2.20 | 1.75/4.02 | 1.28 | 0.91/1.59 | 1.92 | 1.51/3.12 | |||||
| PLTP (rs6065906) | TT | 449 | 71.0 | 5.70 | 4.81/6.24 | 0.50 | 2.20 | 1.51/3.72 | 0.17 | 1.30 | 1.07/1.62 | 0.052 | 2.24 | 1.57/3.53 | 0.035 D |
| TC | 172 | 27.2 | 5.80 | 4.88/6.21 | 2.18 | 1.46/3.82 | 1.24 | 1.02/1.62 | 2.72 | 1.66/3.80 | |||||
| CC | 11 | 1.7 | 5.90 | 4.98/6.20 | 2.82 | 2.06/4.06 | 1.10 | 0.96/1.40 | 1.92 | 1.57/3.75 | |||||
| PSKH1 (rs16942887) | GG | 495 | 78.7 | 5.70 | 4.80/6.20 | 0.13 | 2.20 | 1.48/3.69 | 0.06 | 1.28 | 1.03/1.60 | 0.18 | 2.38 | 1.57/3.70 | 0.23 |
| GA | 127 | 20.2 | 5.80 | 4.89/6.30 | 2.26 | 1.61/3.88 | 1.31 | 1.10/1.63 | 2.40 | 1.63/3.53 | |||||
| AA | 7 | 1.1 | 6.27 | 5.40/6.50 | 3.90 | 2.20/4.33 | 1.32 | 0.92/1.53 | 2.00 | 1.62/3.80 | |||||
| ST3GAL4 (rs11220463) | AA | 431 | 68.0 | 5.80 | 4.88/6.30 | 0.25 | 2.25 | 1.50/3.90 | 0.62 | 1.29 | 1.05/1.62 | 0.20 | 2.37 | 1.62/3.71 | 0.52 |
| AT | 192 | 30.3 | 5.56 | 4.75/6.20 | 2.10 | 1.49/3.15 | 1.32 | 1.07/1.62 | 2.45 | 1.48/3.54 | |||||
| TT | 11 | 1.7 | 5.60 | 4.88/6.02 | 2.49 | 1.48/4.54 | 0.94 | 0.83/1.63 | 3.12 | 2.31/4.22 | |||||
| STARD3 (rs881844) | GG | 283 | 44.8 | 5.78 | 4.79/6.27 | 0.59 | 2.18 | 1.51/3.73 | 0.82 | 1.30 | 1.04/1.66 | 0.27 | 2.40 | 1.62/3.73 | 0.57 |
| GC | 285 | 45.1 | 5.70 | 4.87/6.23 | 2.18 | 1.48/3.66 | 1.30 | 1.09/1.60 | 2.55 | 1.59/3.70 | |||||
| CC | 64 | 10.1 | 5.81 | 4.84/6.20 | 2.80 | 1.61/4.08 | 1.25 | 1.05/1.62 | 1.90 | 1.50/3.25 | |||||
| ZNF648 (rs1689800) | AA | 254 | 254 | 5.69 | 4.80/6.30 | 0.53 | 2.20 | 1.50/3.80 | 0.026 R | 1.31 | 1.03/1.58 | 0.18 | 2.44 | 1.62/3.74 | 0.62 |
| AG | 318 | 318 | 5.80 | 4.86/6.20 | 2.10 | 1.45/3.40 | 1.30 | 1.07/1.66 | 2.41 | 1.59/3.70 | |||||
| GG | 61 | 61 | 5.70 | 4.71/6.18 | 3.00 | 2.00/3.93 | 1.20 | 1.05/1.41 | 2.00 | 1.59/3.70 | |||||
| SCARB1 (rs838880) | TT | 275 | 43.4 | 5.80 | 4.80/6.20 | 0.035 R | 2.31 | 1.69/3.93 | 0.043 OD | 1.27 | 1.04/1.62 | 0.65 | 2.20 | 1.59/3.50 | 0.30 |
| TC | 301 | 47.5 | 5.70 | 4.80/6.29 | 2.06 | 1.45/3.67 | 1.30 | 1.07/1.62 | 2.59 | 1.62/3.75 | |||||
| CC | 58 | 9.1 | 5.81 | 5.00/6.32 | 2.70 | 2.10/3.88 | 1.30 | 1.10/1.58 | 2.24 | 1.50/3.70 | |||||
| PPP1R3B (rs9987289) | GG | 538 | 84.7 | 5.70 | 4.81/6.20 | 0.10 | 2.18 | 1.49/3.74 | - | 1.30 | 1.06/1.61 | - | 2.45 | 1.60/3.71 | 0.51 |
| GA | 96 | 15.1 | 5.90 | 5.00/6.30 | 2.40 | 1.69/3.84 | 1.30 | 1.03/1.70 | 2.03 | 1.59/3.68 | |||||
| AA | 1 | 0.2 | 4.06 | 3.97/4.14 | - | - | - | - | - | - | |||||
| Gene (SNP ID) | Genotypes | Genotype Frequencies | CIMT, mm | ||||
|---|---|---|---|---|---|---|---|
| N | % | Me | Q1/Q3 1 | p2 | padj3 | ||
| ABCA1 (rs1883025) | CC | 386 | 60.9 | 0.62 | 0.53/0.80 | 0.48 | 0.30 |
| CT | 212 | 33.4 | 0.60 | 0.52/0.79 | |||
| TT | 36 | 5.7 | 0.68 | 0.58/0.80 | |||
| APOC1 (rs4420638) | AA | 446 | 70.2 | 0.63 | 0.53/0.80 | 0.68 | 0.65 |
| AG | 177 | 27.9 | 0.60 | 0.52/0.80 | |||
| GG | 12 | 1.9 | 0.61 | 0.55/0.63 | |||
| CETP (rs3764261) | CC | 333 | 54.0 | 0.60 | 0.52/0.80 | 0.62 | 0.40 |
| CA | 275 | 44.6 | 0.63 | 0.55/0.80 | |||
| AA | 9 | 1.5 | 0.57 | 0.57/0.57 | |||
| COBLL1 (rs12328675) | TT | 296 | 75.5 | 0.70 | 0.55/0.85 | 0.05 | 0.009 R |
| TC | 60 | 15.3 | 0.65 | 0.50/0.78 | |||
| CC | 36 | 9.2 | 0.56 | 0.48/0.69 | |||
| F2 (rs3136441) | TT | 436 | 69.1 | 0.63 | 0.55/0.80 | 0.21 | 0.13 |
| TC | 179 | 28.4 | 0.60 | 0.50/0.75 | |||
| CC | 16 | 2.5 | 0.60 | 0.50/0.69 | |||
| GALNT2 (rs4846914) | AA | 239 | 37.9 | 0.60 | 0.54/0.75 | 0.39 | 0.18 |
| AG | 289 | 45.8 | 0.64 | 0.55/0.80 | |||
| GG | 103 | 16.3 | 0.61 | 0.50/0.85 | |||
| LILRA3 (rs386000) | GG | 417 | 65.8 | 0.60 | 0.53/0.80 | 0.49 | 0.13 |
| GC | 198 | 31.2 | 0.63 | 0.53/0.80 | |||
| CC | 19 | 3.0 | 0.55 | 0.44/0.73 | |||
| LPA (rs55730499) | CC | 515 | 82.5 | 0.62 | 0.53/0.80 | 0.31 | 0.31 |
| CT | 108 | 17.3 | 0.60 | 0.55/0.78 | |||
| TT | 1 | 0.2 | 0.40 | 0.40/0.40 | |||
| NPC1L1 (rs217406) | CC | 375 | 59.1 | 0.61 | 0.53/0.80 | 0.19 | 0.10 |
| CG | 233 | 36.7 | 0.62 | 0.54/0.80 | |||
| GG | 27 | 4.3 | 0.55 | 0.45/0.72 | |||
| PLTP (rs6065906) | TT | 449 | 71.0 | 0.61 | 0.53/0.80 | 0.34 | 0.48 |
| TC | 172 | 27.2 | 0.60 | 0.50/0.75 | |||
| CC | 11 | 1.7 | 0.79 | 0.63/0.85 | |||
| PSKH1 (rs16942887) | GG | 495 | 78.7 | 0.62 | 0.53/0.80 | 0.93 | 0.88 |
| GA | 127 | 20.2 | 0.61 | 0.54/0.80 | |||
| AA | 7 | 1.1 | 0.60 | 0.55/0.78 | |||
| ST3GAL4 (rs11220463) | AA | 431 | 68.0 | 0.64 | 0.55/0.80 | 0.046 | 0.01 OD |
| AT | 192 | 30.3 | 0.60 | 0.50/0.73 | |||
| TT | 11 | 1.7 | 0.55 | 0.55/0.80 | |||
| STARD3 (rs881844) | GG | 283 | 44.8 | 0.63 | 0.55/0.78 | 0.48 | 0.20 |
| GC | 285 | 45.1 | 0.60 | 0.52/0.80 | |||
| CC | 64 | 10.1 | 0.57 | 0.50/0.78 | |||
| ZNF648 (rs1689800) | AA | 254 | 254 | 0.62 | 0.55/0.80 | 0.20 | 0.02 D |
| AG | 318 | 318 | 0.60 | 0.52/0.78 | |||
| GG | 61 | 61 | 0.65 | 0.52/0.85 | |||
| SCARB1 (rs838880) | TT | 275 | 43.4 | 0.62 | 0.53/0.78 | 0.98 | 0.51 |
| TC | 301 | 47.5 | 0.61 | 0.51/0.80 | |||
| CC | 58 | 9.1 | 0.60 | 0.53/0.78 | |||
| PPP1R3B (rs9987289) | GG | 538 | 84.7 | 0.62 | 0.55/0.80 | - | - |
| GA | 96 | 15.1 | 0.60 | 0.48/0.75 | |||
| AA | 1 | 0.2 | - | - | |||
| Gene (SNP ID) | Smoking Habit | Cardiovascular Phenotypes | |||||
|---|---|---|---|---|---|---|---|
| TC | LDL-C | HDL-C | TG | CIMT | CAD | ||
| ABCA1 (rs1883025) | Smokers | - | 0.003 D | - | - | - | - |
| Non-smokers | - | - | - | - | - | - | |
| APOC1 (rs4420638) | Smokers | - | - | - | - | - | - |
| Non-smokers | - | - | - | - | - | 0.009 AD | |
| CETP (rs3764261) | Smokers | - | - | - | - | - | - |
| Non-smokers | - | - | 0.006 OD | - | - | - | |
| COBLL1 (rs12328675) | Smokers | 0.03 OD | - | - | - | - | - |
| Non-smokers | - | - | - | - | - | 0.02 R | |
| F2 (rs3136441) | Smokers | 0.02 AD | - | - | - | - | 0.004 D |
| Non-smokers | - | - | - | - | - | 0.02 R | |
| GALNT2 (rs4846914) | Smokers | - | - | - | - | 0.05 OD | - |
| Non-smokers | - | - | - | 0.008 R | - | - | |
| LILRA3 (rs386000) | Smokers | - | - | - | - | - | - |
| Non-smokers | - | - | - | 0.03 R | - | - | |
| LPA (rs55730499) | Smokers | - | 0.0001 R | - | - | - | - |
| Non-smokers | - | - | - | - | - | 0.01 OD | |
| NPC1L1 (rs217406) | Smokers | - | - | - | - | - | 0.01 OD |
| Non-smokers | - | - | - | - | 0.02 R | - | |
| PSKH1 (rs16942887) | Smokers | 0.049 D | - | - | - | - | - |
| Non-smokers | - | - | - | - | - | - | |
| STARD3 (rs881844) | Smokers | - | - | - | - | - | - |
| Non-smokers | - | - | - | - | 0.03 R | - | |
| ZNF648 (rs1689800) | Smokers | - | 0.02 R | - | - | - | - |
| Non-smokers | - | - | - | - | - | - | |
| Gene, Effective Allele | Phenotype | p-Value * | Beta/Odds Ratio | Sample Size |
|---|---|---|---|---|
| ABCA1 rs1883025-T | Coronary artery disease | 0.0000218 | ▼0.9790 | 1,524,980 |
| Total cholesterol | 3.40 × 10−91 | ▼−0.0583 | 431,334 | |
| LDL cholesterol | 4.80 × 10−43 | ▼−0.0296 | 682,058 | |
| HDL cholesterol | 2.19 × 10−139 | ▼−0.0679 | 385,758 | |
| Triglycerides | 1.92 × 10−18 | ▼−0.0190 | 711,468 | |
| APOC1 rs4420638-G | Coronary artery disease | 5.80 × 10−32 | ▲1.0814 | 1,477,190 |
| Total cholesterol | 1.15 × 10−203 | ▲0.1357 | 314,177 | |
| LDL cholesterol | 1.77 × 10−37 | ▲0.1941 | 113,518 | |
| HDL cholesterol | 1.21 × 10−54 | ▼−0.0624 | 252,659 | |
| Triglycerides | 1.06 × 10−259 | ▲0.0605 | 598,528 | |
| CETP rs3764261-A | Coronary artery disease | 3.57 × 10−10 | ▼0.9671 | 1,591,550 |
| Total cholesterol | 1.15 × 10−61 | ▲0.0469 | 410,790 | |
| LDL cholesterol | 1.49 × 10−192 | ▼−0.0356 | 662,996 | |
| HDL cholesterol | 7.08 × 10−36 | ▲0.2124 | 14,126 | |
| Triglycerides | 5.97 × 10−68 | ▼−0.0362 | 692,195 | |
| COBLL1 (GRB14) rs12328675-C | Coronary artery disease | 0.004766 | ▼0.9835 | 1,481,940 |
| Total cholesterol | 0.1169 | ▼−0.0097 | 303,083 | |
| LDL cholesterol | 0.00017 | ▼−0.0142 | 609,213 | |
| HDL cholesterol | 3.99 × 10−13 | ▲0.0381 | 315,152 | |
| Triglycerides | 4.45 × 10−31 | ▼−0.0406 | 605,928 | |
| F2 rs3136441-C | Coronary artery disease | 0.6976 | ▲1.0021 | 1,513,820 |
| Total cholesterol | 0.0002989 | ▲0.0096 | 418,936 | |
| LDL cholesterol | 0.2641 | ▼−0.0031 | 668,268 | |
| HDL cholesterol | 4.24 × 10−23 | ▲0.0289 | 371,896 | |
| Triglycerides | 5.12 × 10−23 | ▼−0.0234 | 699,079 | |
| GALNT2 rs4846914-G | Coronary artery disease | 1.14 × 10−8 | ▼0.9757 | 1,593,010 |
| Total cholesterol | 0.04971 | ▲0.0054 | 433,614 | |
| LDL cholesterol | 0.000917 | ▼−0.0071 | 684,307 | |
| HDL cholesterol | 7.61 × 10−47 | ▲0.0401 | 388,040 | |
| Triglycerides | 4.74 × 10−227 | ▼−0.0404 | 713,750 | |
| LILRA3 rs386000-C | Coronary artery disease | 0.5958 | ▲1.0029 | 1,448,870 |
| Total cholesterol | 0.0000164 | ▲0.0143 | 379,606 | |
| LDL cholesterol | 0.08641 | ▲0.0047 | 624,565 | |
| HDL cholesterol | 6.26 × 10−23 | ▲0.0314 | 324,003 | |
| Triglycerides | 0.02057 | ▼−0.0056 | 663,279 | |
| LPA rs55730499-T | Coronary artery disease | 1.45 × 10−174 | ▲1.3562 | 1,010,500 |
| Total cholesterol | 7.87 × 10−11 | ▲0.0776 | 45,549 | |
| LDL cholesterol | 4.16 × 10−298 | ▲0.1235 | 356,869 | |
| HDL cholesterol | 0.8817 | ▲0.0067 | 45,509 | |
| Triglycerides | 6.52 × 10−14 | ▼−0.0357 | 360,181 | |
| NPC1L1 rs217406-G | Coronary artery disease | 0.0008186 | ▲1.0234 | 1,543,540 |
| Total cholesterol | 4.77 × 10−13 | ▲0.0321 | 243,481 | |
| LDL cholesterol | 3.23 × 10−36 | ▲0.0364 | 542,160 | |
| HDL cholesterol | 0.03886 | ▼−0.0128 | 245,165 | |
| Triglycerides | 0.01255 | ▲0.0073 | 548,639 | |
| PLTP (PCIF1) rs6065906-C | Coronary artery disease | 0.009999 | ▼0.9865 | 1,593,110 |
| Total cholesterol | 0.627 | ▼−0.0021 | 380,779 | |
| LDL cholesterol | 6.84 × 10−8 | ▲0.0156 | 625,258 | |
| HDL cholesterol | 3.62 × 10−36 | ▼−0.0484 | 326,104 | |
| Triglycerides | 1.59 × 10−201 | ▲0.0515 | 663,221 | |
| PSKH1 rs16942887-A | Coronary artery disease | 0.03214 | ▲1.0153 | 1,524,990 |
| Total cholesterol | 2.80 × 10−6 | ▲0.0220 | 385,717 | |
| LDL cholesterol | 0.7133 | ▲0.0015 | 637,521 | |
| HDL cholesterol | 5.23 × 10−41 | ▲0.0641 | 338,751 | |
| Triglycerides | 0.0000296 | ▼−0.0132 | 668,145 | |
| ST3GAL4 rs11220463-T | Coronary artery disease | 0.0000982 | ▲1.0234 | 1,591,520 |
| Total cholesterol | 3.52 × 10−14 | ▲0.0220 | 382,747 | |
| LDL cholesterol | 4.62 × 10−47 | ▲0.0378 | 625,938 | |
| HDL cholesterol | 0.06697 | ▼−0.0055 | 326,790 | |
| Triglycerides | 0.03181 | ▲0.0065 | 665,187 | |
| STARD3 rs881844-G | Coronary artery disease | 0.0001801 | ▼0.9825 | 1,592,970 |
| Total cholesterol | 5.93 × 10−6 | ▲0.0122 | 374,624 | |
| LDL cholesterol | 0.05305 | ▲0.0043 | 617,962 | |
| HDL cholesterol | 4.85 × 10−20 | ▲0.0262 | 318,709 | |
| Triglycerides | 0.1204 | ▼−0.0036 | 657,086 | |
| ZNF648 rs1689800-G | Coronary artery disease | 0.2033 | ▲1.0063 | 1,524,990 |
| Total cholesterol | 0.8575 | ▲0.0006 | 433,743 | |
| LDL cholesterol | 2.72 × 10−9 | ▲0.0120 | 684,428 | |
| HDL cholesterol | 3.72 × 10−23 | ▼−0.0237 | 388,167 | |
| Triglycerides | 0.00359 | ▲0.0064 | 713,877 | |
| SCARB1 rs838880-T | Coronary artery disease | 0.0000238 | ▲1.0190 | 1,524,990 |
| Total cholesterol | 0.0000147 | ▼−0.0139 | 292,669 | |
| LDL cholesterol | 0.6628 | ▲0.0009 | 597,781 | |
| HDL cholesterol | 1.12 × 10−31 | ▼−0.0316 | 303,498 | |
| Triglycerides | 0.03555 | ▲0.0053 | 595,585 | |
| PPP1R3B (RP11-10A14.4) rs9987289-G | Coronary artery disease | 0.284 | ▲1.0118 | 1,516,240 |
| Total cholesterol | 3.89 × 10−25 | ▲0.0675 | 373,464 | |
| LDL cholesterol | 1.35 × 10−47 | ▲0.0520 | 618,405 | |
| HDL cholesterol | 1.72 × 10−32 | ▲0.0640 | 314,374 | |
| Triglycerides | 0.01654 | ▼−0.0066 | 656,355 |
| G × G/G × E Interaction Models | NH | β-H | WH | NL | β-L | WL | Pperm | |
|---|---|---|---|---|---|---|---|---|
| Two-order models | ||||||||
| 1 | LILRA3 rs386000 × GALNT2 rs4846914 | 3 | 0.352 | 17.72 | 0 | - | - | 0.001 |
| 2 | LPA rs55730499 × GALNT2 rs4846914 | 1 | 0.883 | 12.48 | 2 | −0.164 | 5.09 | 0.004 |
| 3 | SCARB1 rs838880 × COBLL1 rs12328675 | 0 | - | - | 2 | −0.372 | 9.46 | 0.04 |
| 4 | SCARB1 rs838880 × LPA rs55730499 | 1 | 0.377 | 8.78 | 1 | −2.089 | 4.42 | 0.044 |
| Three-order models | ||||||||
| 1 | SCARB1 rs838880 × LPA rs55730499 × APOC1 rs4420638 | 3 | 0.436 | 23.04 | 5 | −0.248 | 9.45 | <0.002 |
| 2 | LPA rs55730499 × LILRA3 rs386000 × GALNT2 rs4846914 | 4 | 0.549 | 22.04 | 3 | −0.287 | 9.45 | <0.002 |
| 3 | LILRA3 rs386000 × GALNT2 rs4846914 × SMOKING | 4 | 0.426 | 21.87 | 1 | −0.443 | 3.13 | <0.002 |
| 4 | LPA rs55730499 × GALNT2 rs4846914× ABCA1 rs1883025 | 4 | 0.947 | 20.61 | 3 | −0.199 | 6.14 | <0.002 |
| Four-order models | ||||||||
| 1 | LILRA3 rs386000 × GALNT2 rs4846914 × COBLL1 rs12328675 × SMOKING | 6 | 0.688 | 32.02 | 2 | −0.859 | 12.06 | <0.002 |
| 2 | STARD3 rs881844 × ST3GAL4 rs11220463× LPA rs55730499 × APOC1 rs4420638 | 5 | 0.544 | 25.21 | 2 | −0.206 | 4.43 | <0.002 |
| 3 | SCARB1 rs838880 × LPA rs55730499 × APOC1 rs4420638 × SMOKING | 4 | 0.562 | 23.56 | 4 | −0.870 | 12.15 | <0.002 |
| 4 | SCARB1 rs838880 × LILRA3 rs386000 × GALNT2 rs4846914 × COBLL1 rs12328675 | 6 | 0.856 | 35.96 | 3 | −0.759 | 13.91 | 0.002 |
| G × G/G × E Interaction Models | NH | β-H | WH | NL | β-L | WL | Pperm | |
|---|---|---|---|---|---|---|---|---|
| Two-order models | ||||||||
| 1 | PSKH1 rs16942887 × SMOKING | 2 | 0.596 | 47.25 | 2 | −0.561 | 40.94 | <0.001 |
| 2 | ABCA1 rs1883025 × SMOKING | 3 | 0.569 | 43.18 | 3 | −0.569 | 43.18 | <0.001 |
| 3 | ST3GAL4 rs11220463 × SMOKING | 2 | 0.563 | 42.46 | 3 | −0.566 | 42.68 | <0.001 |
| 4 | SCARB1 rs838880 × SMOKING | 2 | 0.547 | 40.48 | 3 | −0.566 | 42.68 | <0.001 |
| Three-order models | ||||||||
| 1 | PSKH1 rs16942887 × F2 rs3136441 × SMOKING | 3 | 0.626 | 53.01 | 3 | −0.498 | 28.45 | <0.002 |
| 2 | ABCA1 rs1883025 × PLTP rs6065906 × SMOKING | 3 | 0.609 | 51.58 | 3 | −0.619 | 39.64 | <0.002 |
| 3 | PSKH1 rs16942887 × LILRA3 rs386000 × SMOKING | 4 | 0.615 | 51.45 | 3 | −0.521 | 33.49 | <0.002 |
| 4 | SCARB1 rs838880 × ABCA1 rs1883025 × SMOKING | 4 | 0.532 | 37.84 | 7 | −0.663 | 50.48 | <0.002 |
| Four-order models | ||||||||
| 1 | SCARB1 rs838880 × PSKH1 rs16942887 × NPC1L1 rs217406 × SMOKING | 7 | 0.662 | 57.46 | 7 | −0.685 | 47.82 | <0.002 |
| 2 | STARD3 rs881844 × NPC1L1 rs217406 × LILRA3 rs386000 × SMOKING | 6 | 0.681 | 56.28 | 4 | −0.547 | 19.32 | <0.002 |
| 3 | SCARB1 rs838880 × ST3GAL4 rs11220463 × GALNT2 rs4846914 × SMOKING | 3 | 0.473 | 17.22 | 10 | −0.696 | 55.97 | <0.002 |
| 4 | PSKH1 rs16942887 × PLTP rs6065906 × F2 rs3136441 × SMOKING | 5 | 0.648 | 55.90 | 7 | −0.579 | 36.11 | <0.002 |
| G × G/G × E Interaction Models | NH | β-H | WH | NL | β-L | WL | Pperm | |
|---|---|---|---|---|---|---|---|---|
| Two-order models | ||||||||
| 1 | LPA rs55730499 × COBLL1 rs12328675 | 2 | 0.220 | 41.65 | 2 | −0.209 | 43.84 | <0.001 |
| 2 | STARD3 rs881844 × F2 rs3136441 | 1 | 0.119 | 20.13 | 4 | −0.161 | 40.10 | <0.001 |
| 3 | LILRA3 rs386000 × F2 rs3136441 | 2 | 0.142 | 33.29 | 3 | −0.128 | 25.97 | <0.001 |
| 4 | LPA rs55730499 × F2 rs3136441 | 3 | 0.133 | 28.12 | 2 | −0.150 | 33.18 | <0.001 |
| Three-order models | ||||||||
| 1 | PLTP rs6065906 × LPA rs55730499 × COBLL1 rs12328675 | 3 | 0.280 | 52.83 | 3 | −0.099 | 13.26 | <0.002 |
| 2 | LPA rs55730499 × COBLL1 rs12328675 × SMOKING | 3 | 0.265 | 51.27 | 2 | −0.104 | 13.95 | <0.002 |
| 3 | LPA rs55730499 × LILRA3 rs386000 × COBLL1 rs12328675 | 4 | 0.253 | 50.63 | 3 | −0.132 | 23.58 | <0.002 |
| 4 | SCARB1 rs838880 × LPA rs55730499 × COBLL1 rs12328675 | 4 | 0.277 | 48.58 | 2 | −0.127 | 19.67 | <0.002 |
| Four-order models | ||||||||
| 1 | SCARB1 rs838880 × STARD3 rs881844 × LPA rs55730499 × COBLL1 rs12328675 | 9 | 0.345 | 68.48 | 4 | −0.157 | 29.20 | <0.002 |
| 2 | SCARB1 rs838880 × LPA rs55730499 × COBLL1 rs12328675 × SMOKING | 6 | 0.331 | 67.94 | 2 | −0.147 | 16.68 | <0.002 |
| 3 | STARD3 rs881844 × PLTP rs6065906 × NPC1L1 rs217406 NPC1L1 × F2 rs3136441 | 3 | 0.139 | 16.98 | 12 | −0.221 | 64.49 | <0.002 |
| 4 | LPA rs55730499 × F2 rs3136441 COBLL1 rs12328675 × SMOKING | 6 | 0.312 | 63.97 | 5 | −0.189 | 38.10 | <0.002 |
| SNP ID | Gene | FuncPred 1 | Number eQTL (GTEx 2) | Binding Sites for TF 3 | Regulatory Potential (rSNPbase 4) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Regulatory Potential | Conservatism | cis | trans | Loss | Gain | rSNP | rSNP in LD | Post-Transcriptional Regulation | Circular RNA Binding Regions (circRNA) | ||
| rs1883025 | ABCA1 | 0.149 | 0.001 | −/− | - | 24 | 9 | + | 18 | + | 5 |
| rs4420638 | APOC1 | - | 0 | −/− | - | 2 | 25 | - | 7 | - | 4 |
| rs3764261 | CETP | 0 | 0.001 | −/+ | 1 | 11 | 13 | + | 1 | - | - |
| rs12328675 | COBLL1 | 0 | 0 | −/+ | 1 | 8 | 10 | + | 3 | - | 1 |
| rs3136441 | F2 | - | 0.017 | −/+ | 33 | 13 | 8 | + | 49 | + | 1 |
| rs4846914 | GALNT2 | 0.199 | 0 | −/− | - | 5 | 7 | + | 21 | + | 3 |
| rs386000 | LILRA3 | 0 | 0.001 | 26/+ | - | - | - | - | 19 | - | 10 |
| rs55730499 | LPA | - | - | −/− | 1 | 11 | 5 | - | 2 | - | 3 |
| rs217406 | NPC1L1 | 0.339 | 0 | 1/+ | 17 | 9 | 7 | + | 18 | + | 4 |
| rs6065906 | PLTP | - | 0 | 13/+ | 7 | 41 | 3 | + | 20 | - | - |
| rs16942887 | PSKH1 | - | 0.006 | −/+ | 45 | 12 | 4 | + | 81 | + | - |
| rs11220463 | ST3GAL4 | 0 | 0.001 | 8/+ | - | 19 | 9 | + | 24 | + | - |
| rs881844 | STARD3 | 0.273 | 0.002 | 1/+ | 38 | 26 | 11 | + | 73 | + | 1 |
| rs1689800 | ZNF648 | 0 | 0 | −/− | - | 8 | 21 | - | 5 | - | - |
| rs838880 | SCARB1 | - | 0 | −/− | - | 3 | 22 | + | 1 | - | - |
| rs9987289 | PPP1R3B | 0.045 | 0.001 | −/− | - | 9 | 26 | + | 51 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazarenko, V.; Churilin, M.; Azarova, I.; Klyosova, E.; Bykanova, M.; Ob'edkova, N.; Churnosov, M.; Bushueva, O.; Mal, G.; Povetkin, S.; et al. Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease. Biomedicines 2022, 10, 259. https://doi.org/10.3390/biomedicines10020259
Lazarenko V, Churilin M, Azarova I, Klyosova E, Bykanova M, Ob'edkova N, Churnosov M, Bushueva O, Mal G, Povetkin S, et al. Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease. Biomedicines. 2022; 10(2):259. https://doi.org/10.3390/biomedicines10020259
Chicago/Turabian StyleLazarenko, Victor, Mikhail Churilin, Iuliia Azarova, Elena Klyosova, Marina Bykanova, Natalia Ob'edkova, Mikhail Churnosov, Olga Bushueva, Galina Mal, Sergey Povetkin, and et al. 2022. "Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease" Biomedicines 10, no. 2: 259. https://doi.org/10.3390/biomedicines10020259
APA StyleLazarenko, V., Churilin, M., Azarova, I., Klyosova, E., Bykanova, M., Ob'edkova, N., Churnosov, M., Bushueva, O., Mal, G., Povetkin, S., Kononov, S., Luneva, Y., Zhabin, S., Polonikova, A., Gavrilenko, A., Saraev, I., Solodilova, M., & Polonikov, A. (2022). Comprehensive Statistical and Bioinformatics Analysis in the Deciphering of Putative Mechanisms by Which Lipid-Associated GWAS Loci Contribute to Coronary Artery Disease. Biomedicines, 10(2), 259. https://doi.org/10.3390/biomedicines10020259








