CpG Site-Based Signature Predicts Survival of Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of DNA Methylation Data and Characteristics of Study Population
2.2. Bioinformatics and Statistical Analysis of DNA Methylation Data
2.3. Functional Network and Pathway Analysis
3. Results
3.1. Assessing Variability and Differences in Patterns of DNA Methylation Profiles
3.2. Discovery of DNA Methylation Signatures Associated with CRC
3.3. Discovery of A Prognostic Signature and Survival Prediction
3.4. Discovery of Aberrantly Methylated Molecular Networks and Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Ladabaum, U.; Dominitz, J.A.; Kahi, C.; Schoen, R.E. Strategies for Colorectal Cancer Screening. Gastroenterology 2020, 158, 418–432. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef]
- White, A.; Thompson, T.D.; White, M.C.; Sabatino, S.A.; de Moor, J.; Doria-Rose, P.V.; Geiger, A.M.; Richardson, L.C. Cancer screening test use—United States, 2015. MMWR Morb. Mortal Wkly Rep. 2017, 66, 201–206. [Google Scholar] [CrossRef]
- Meester, R.G.; Doubeni, C.; Lansdorp-Vogelaar, I.; Goede, S.L.; Levin, T.R.; Quinn, V.P.; van Ballegooijen, M.; Corley, D.A.; Zauber, A.G. Colorectal cancer deaths attributable to nonuse of screening in the United States. Ann. Epidemiol. 2014, 25, 208–213.e1. [Google Scholar] [CrossRef]
- Mendelaar, P.A.; Smid, M.; van Riet, J.; Angus, L.; Labots, M.; Steeghs, N.; Hendriks, M.P.; Cirkel, G.A.; van Rooijen, J.M.; Ten Tije, A.J.; et al. Whole genome sequencing of metastatic colorectal cancer reveals prior treatment effects and specific metastasis features. Nat Commun. 2021, 12, 574. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Cieslik, M.; Chinnaiyan, A.M. Global genomics project unravels cancer’s complexity at unprecedented scale. Nature 2020, 578, 39–40. [Google Scholar] [CrossRef] [PubMed]
- Komor, M.A.; Bosch, L.J.; Bounova, G.; Bolijn, A.S.; Delis-van Diemen, P.M.; Rausch, C.; Hoogstrate, Y.; Stubbs, A.P.; De Jong, M.; Jenster, G.; et al. Consensus molecular subtype classification of colorectal adenomas. J. Pathol. 2018, 246, 266–276. [Google Scholar] [CrossRef]
- Liu, L.; Liu, J.; Wang, J.; Yuan, W. Machine learning revealed molecular classification of colorectal cancer with negative lymph node metastasis. Biomarkers 2021, 27, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin. Gastroenterol. Hepatol. 2019, 17, 275–289. [Google Scholar] [CrossRef]
- Shiao, S.P.K.; Grayson, J.; Yu, C.H.; Wasek, B.; Bottiglieri, T. Gene Environment Interactions and Predictors of Colorectal Cancer in Family-Based, Multi-Ethnic Groups. J. Pers. Med. 2018, 8, 10. [Google Scholar] [CrossRef]
- You, A.J.; You, C.Y. Biomarkers in Colorectal Cancer. Anticancer Res. 2016, 36, 1093–1102. [Google Scholar]
- Constâncio, V.; Nunes, S.P.; Henrique, R.; Jerónimo, C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells 2020, 9, 624. [Google Scholar] [CrossRef]
- Pidsley, R.; Lawrence, M.G.; Zotenko, E.; Niranjan, B.; Statham, A.; Song, J.; Chabanon, R.M.; Qu, W.; Wang, H.; Richards, M.; et al. Enduring epigenetic landmarks define the cancer microenvironment. Genome Res. 2018, 28, 625–638. [Google Scholar] [CrossRef]
- Ding, D.; Han, S.; Zhang, H.; He, Y.; Li, Y. Predictive biomarkers of colorectal cancer. Comput. Biol. Chem. 2019, 83, 107106. [Google Scholar] [CrossRef]
- Kel, A.; Boyarskikh, U.; Stegmaier, P.; Leskov, L.S.; Sokolov, A.V.; Yevshin, I.; Mandrik, N.; Stelmashenko, D.; Koschmann, J.; Kel-Margoulis, O.; et al. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinform. 2019, 20 (Suppl. 4), 119. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, H.; Zhang, D.; Li, M.; Sun, X.; Wan, L.; Yu, D.; Tian, Y.; Jin, H.; Lin, A.; et al. Integrated analyses of multi-omics reveal global patterns of methylation and hydroxymethylation and screen the tumor suppressive roles of HADHB in colorectal cancer. Clin. Epigenet. 2018, 10, 30. [Google Scholar] [CrossRef]
- Lu, J.; Wilfred, P.; Korbie, D.; Trau, M. Regulation of Canonical Oncogenic Signaling Pathways in Cancer via DNA Methylation. Cancers 2020, 12, 3199. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Program. Available online: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (accessed on 2 June 2022).
- Genomics Data Commons. Available online: https://gdc.cancer.gov/ (accessed on 2 June 2022).
- Marabita, F.; Almgren, M.; Lindholm, M.E.; Ruhrmann, S.; Fagerström-Billai, F.; Jagodic, M.; Sundberg, C.J.; Ekström, T.J.; Teschendorff, A.E.; Tegnér, J.; et al. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina Human Methylation 450 Bead Chip platform. Epigenetics 2013, 8, 333–346. [Google Scholar] [CrossRef]
- Liu, J.; Siegmund, K.D. An evaluation of processing methods for HumanMethylation450 Bead Chip data. BMC Genom. 2016, 17, 469. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Fortin, J.P.; Triche, T.J., Jr.; Hansen, K.D. Preprocessing, normalization and integration of the Il-lumina Human Methylation EPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [PubMed]
- Maksimovic, J.; Gordon, L.; Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium Human Methylation 450 Bead Chips. Genome Biol. 2012, 13, R44. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yosef, H. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Stalpers, L.J.; Kaplan, E.L.; Edward, L. Kaplan and the Kaplan-Meier Survival Curve. BSHM Bull. J. Br. Soc. Hist. Math. 2018, 33, 109–135. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. The logrank test. BMJ 2004, 328, 1073. [Google Scholar] [CrossRef]
- Ingenuity Pathways Analysis (IPA) System Redwood, CA: Ingenuity Systems, Inc. 2013. Available online: http://www.ingenuity.com (accessed on 6 February 2013).
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kirana, C.; Peng, L.; Miller, R.; Keating, J.P.; Glenn, C.; Shi, H.; Jordan, T.W.; Maddern, G.J.; Stubbs, R.S. Combination of laser microdissection, 2D-DIGE and MALDI-TOF MS to identify protein biomarkers to predict colorectal cancer spread. Clin. Proteom. 2019, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liao, Y.; Huang, J.; Sun, Y.; Chen, H.; Chen, C.; Li, S.; Yang, Z. Epigenetic silencing of ADAMTS5 is associated with increased invasiveness and poor survival in patients with colorectal cancer. J. Cancer Res. Clin. Oncol. 2017, 144, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Y.; Luo, X.; Jin, D.; Zhou, W.; Ju, Z.; Wang, D.; Meng, Q.; Wang, H.; Fu, X.; et al. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics 2021, 11, 7507–7526. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, H.; Song, Q.; Li, G.; Lin, S.; Xiong, S. Long non-coding RNA DPP10-AS1 exerts anti-tumor effects on colon cancer via the upregulation of ADCY1 by regulating microRNA-127-3p. Aging 2021, 13, 9748–9765. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-H.; Ma, S.; Zhang, X.-N.; Zhang, Z.-Y.; Zhu, H.-F.; Ji, Y.-H.; Li, J.; Qian, X.-L.; Wang, Y.-X. Hypermethylation of ADHFE1 Promotes the Proliferation of Colorectal Cancer Cell Via Modulating Cell Cycle Progression. OncoTargets Ther. 2019, 12, 8105–8115. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, S.; Novosad, V.; Engibaryan, N.; Ushkaryov, Y.; Nikulin, S.; Tonevitsky, A. ECM–Receptor Regulatory Network and Its Prognostic Role in Colorectal Cancer. Front. Genet. 2021, 12, 782699. [Google Scholar] [CrossRef]
- Taskoparan, B.; Seza, E.G.; Demirkol, S.; Tunçer, S.; Stefek, M.; Gure, A.O.; Banerjee, S. Opposing roles of the aldo-keto reductases AKR1B1 and AKR1B10 in colorectal cancer. Cell. Oncol. 2017, 40, 563–578. [Google Scholar] [CrossRef]
- Shi, J.; Wang, L.; Yin, X.; Wang, L.; Bo, L.; Liu, K.; Feng, K.; Lin, S.; Xu, Y.; Ning, S.; et al. Comprehensive characterization of clonality of driver genes revealing their clinical relevance in colorectal cancer. J. Transl. Med. 2022, 20, 362. [Google Scholar] [CrossRef]
- Zhao, Z.; Lu, L.; Li, W. TAGLN2 promotes the proliferation, invasion, migration and epithelial-mesenchymal transition of colorectal cancer cells by activating STAT3 signaling through ANXA2. Oncol. Lett. 2021, 22, 737. [Google Scholar] [CrossRef]
- Wang, W.; Li, Q.; Yang, T.; Bai, G.; Li, D.; Sun, H. Expression of AQP5 and AQP8 in human colorectal carcinoma and their clinical significance. World J. Surg. Oncol. 2012, 10, 242. [Google Scholar] [CrossRef]
- Miyoshi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Nagai, K.; Uemura, M.; Sekimoto, M.; Doki, Y.; Mori, M. ATP11A is a novel pre-dictive marker for metachronous metastasis of colorectal cancer. Oncol. Rep. 2010, 23, 505–510. [Google Scholar] [PubMed]
- Jedi, M.; Young, G.P.; Pedersen, S.K.; Symonds, E.L. Methylation and Gene Expression of BCAT1 and IKZF1 in Colorectal Cancer Tissues. Clin. Med. Insights Oncol. 2018, 12, 1179554918775064. [Google Scholar] [CrossRef]
- Lin, R.K.; Hung, W.Y.; Huang, Y.F.; Chang, Y.J.; Lin, C.H.; Chen, W.Y.; Chiu, S.F.; Chang, S.C.; Tsai, S.F. Huang Hypermethylation of BEND5 contributes to cell proliferation and is a prognostic marker of colorectal cancer. Oncotarget 2017, 8, 113431–113443. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.J.; Pyo, J.H.; Ryu, K.J.; Kim, S.J.; Ha, J.M.; Choi, K.; Hong, S.N.; Min, B.-H.; Chang, D.K.; Son, H.J.; et al. Oncogenic Role of BOLL in Colorectal Cancer. Am. J. Dig. Dis. 2015, 60, 1663–1673. [Google Scholar] [CrossRef]
- Skuja, E.; Butane, D.; Nakazawa-Miklasevica, M.; Daneberga, Z.; Purkalne, G.; Miklasevics, E. Deletions in metastatic colorectal cancer with chromothripsis. Exp. Oncol. 2019, 41, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Xu, Q. CASR rs1801725 polymorphism is associated with the risk and prognosis of colorectal cancer: A case-control study. J. Clin. Lab. Anal. 2020, 34, e23463. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, J.; Lu, J.; Wang, P.; Feng, H.; Zong, Y.; Ou, B.; Zheng, M.; Lu, A. Cadherin-12 enhances proliferation in colorectal cancer cells and increases progression by promoting EMT. Tumor Biol. 2016, 37, 9077–9088. [Google Scholar] [CrossRef]
- Naumov, V.A.; Generozov, E.V.; Zaharjevskaya, N.B.; Matushkina, D.S.; Larin, A.K.; Chernyshov, S.V.; Alekseev, M.V.; Shelygin, Y.A.; Govorun, V.M. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics 2013, 8, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Gzil, A.; Szylberg, L.; Jaworski, D.; Dominiak, J.; Zarębska, I.; Grzanka, D. The Essential Role of DCLK1 in Pathogenesis, Diagnostic Procedures and Prognostic Stratification of Colorectal Cancer. Anticancer Res. 2019, 39, 2689–2697. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, X.-Y.; Gong, R.; Peng, K.-W.; Liu, R.-B.; Wang, F. NK homeobox 2.2 functions as tumor suppressor in colorectal cancer due to DNA methylation. J. Cancer 2020, 11, 4791–4800. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, P.; Feng, H.; Wang, P.; Zong, Y.; Ma, J.; Zhang, Z.; Chen, X.; Zheng, M.; Zhu, Z.; et al. Cadherin-12 contributes to tumorigenicity in colorectal cancer by promoting migration, invasion, adhersion and angiogenesis. J. Transl. Med. 2013, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- Long, N.P.; Park, S.; Anh, N.H.; Nghi, T.D.; Yoon, S.J.; Park, J.H.; Lim, J.; Kwon, S.W. High-Throughput Omics and Statistical Learning Integration for the Discovery and Validation of Novel Diagnostic Signatures in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 296. [Google Scholar] [CrossRef]
- Vijai, M.; Baba, M.; Ramalingam, S.; Thiyagaraj, A. DCLK1 and its interaction partners: An effective therapeutic target for colorectal cancer (Review). Oncol. Lett. 2021, 22, 850. [Google Scholar] [CrossRef]
- Kalantari, E.; Ghods, R.; Zanjani, L.S.; Rahimi, M.; Eini, L.; Razmi, M.; Asadi-Lari, M.; Madjd, Z. Cytoplasmic expression of DCLK1-S, a novel DCLK1 isoform, is associated with tumor aggressiveness and worse disease-specific survival in colorectal cancer. Cancer Biomark. 2022, 33, 277–289. [Google Scholar] [CrossRef]
- De Mattia, E.; Polesel, J.; Roncato, R.; Labriet, A.; Bignucolo, A.; Gagno, S.; Buonadonna, A.; D’Andrea, M.; Lévesque, E.; Jonker, D.; et al. IL15RA and SMAD3 Genetic Variants Predict Overall Survival in Metastatic Colorectal Cancer Patients Treated with FOLFIRI Therapy: A New Paradigm. Cancers 2021, 13, 1705. [Google Scholar] [CrossRef]
- Gao, R.; Gao, Z.; Huang, L.; Qin, H. Gut microbiota and colorectal cancer. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 757–769. [Google Scholar] [CrossRef]
- Dariya, B.; Aliya, S.; Merchant, N.; Alam, A.; Nagaraju, G.P. Colorectal Cancer Biology, Diagnosis, and Therapeutic Ap-proaches. Crit Rev Oncog. 2020, 25, 71–94. [Google Scholar] [CrossRef]
- Han, F.T.; Wu, G.; Zhang, S.; Zhang, J.; Zhao, Y.; Xu, J. he association of Metabolic Syndrome and its Components with the Incidence and Survival of Colorectal Cancer: A Systematic Review and Meta-analysis. Int. J. Biol. Sci. 2021, 17, 487–497. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kawada, K.; Obama, K. Inflammation-Related Biomarkers for the Prediction of Prognosis in Colorectal Cancer Patients. Int. J. Mol. Sci. 2021, 22, 8002. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.G.; Ahnen, D.J. Colorectal Cancer in the Young. Curr. Gastroenterol. Rep. 2018, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Connell, L.C.; Mota, J.M.; Braghiroli, M.I.; Hoff, P.M. The Rising Incidence of Younger Patients with Colorectal Cancer: Questions About Screening, Biology, and Treatment. Curr. Treat. Options Oncol. 2017, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.G.; Boland, C.R. Colorectal Cancer in Persons Under Age 50: Seeking Causes and Solutions. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 441–455. [Google Scholar] [CrossRef]
- Soofiyani, S.R.; Ahangari, H.; Soleimanian, A.; Babaei, G.; Ghasemnejad, T.; Safavi, S.E.; Eyvazi, S.; Tarhriz, V. The role of circadian genes in the pathogenesis of colorectal cancer. Gene 2021, 804, 145894. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Pazienza, V.; Vinciguerra, M. Clock Genes and Clock-Controlled Genes in the Regulation of Metabolic Rhythms. J. Biol. Med. Rhythm Res. 2012, 29, 227–251. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Panza, A.; Valvano, M.R.; Palumbo, O.; Carella, M.; Pazienza, V.; Biscaglia, G.; Tavano, F.; Di Sebastiano, P.; Andriulli, A.; et al. Clock Gene Expression Levels and Relationship with Clinical and Pathological Features in Colorectal Cancer Patients. J. Biol. Med. Rhythm Res. 2011, 28, 841–851. [Google Scholar] [CrossRef]
- Lee, C.C. Tumor Suppression by the Mammalian Period Genes. Cancer Causes Control 2006, 17, 525–530. [Google Scholar] [CrossRef]
- Schernhammer, E.S.; Laden, F.; Speizer, F.E.; Willett, W.C.; Hunter, D.J.; Kawachi, I.; Fuchs, C.S.; Colditz, G.A. Night-shift work and risk of colorectal cancer in the nurses’ health study. J. Natl. Cancer Inst. 2003, 95, 825–828. [Google Scholar] [CrossRef]
- Deng, W.-W.; Hu, Q.; Liu, Z.-R.; Chen, Q.-H.; Wang, W.-X.; Zhang, H.-G.; Zhang, Q.; Huang, Y.-L.; Zhang, X.-K. KDM4B promotes DNA damage response via STAT3 signaling and is a target of CREB in colorectal cancer cells. Mol. Cell. Biochem. 2018, 449, 81–90. [Google Scholar] [CrossRef]
- Nishihara, H.; Kizaka-Kondoh, S.; Insel, P.A.; Eckmann, L. Inhibition of apoptosis in normal and transformed intestinal epithelial cells by cAMP through induction of inhibitor of apoptosis protein (IAP)-2. Proc. Natl. Acad. Sci. USA 2003, 100, 8921–8926. [Google Scholar] [CrossRef] [PubMed]
- Proto, M.C.; Gazzerro, P.; Di Croce, L.; Santoro, A.; Malfitano, A.M.; Pisanti, S.; Laezza, C.; Bifulco, M. Interaction of endocannabinoid system and steroid Hormones in the control of colon cancer cell growth. J. Cell. Physiol. 2011, 227, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Nimri, L.; Saadi, J.; Peri, I.; Yehuda-Shnaidman, E.; Schwartz, B. Mechanisms linking obesity to altered metabolism in mice colon carcinogenesis. Oncotarget 2015, 6, 38195–38209. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Shim, S.; Kong, J.S.; Kim, M.; Park, S.; Lee, S.; Kim, A. Overexpression of dopamine receptor D2 promotes colorectal cancer progression by activating the β-catenin/ZEB1 axis. Cancer Sci. 2021, 112, 3732–3743. [Google Scholar] [CrossRef]
- Gemignani, F.; Landi, S.; Moreno, V.; Gioia-Patricola, L.; Chabrier, A.; Guino, E.; Navarro, M.; Cambray, M.; Capellà, G.; Canzian, F.; et al. Polymorphisms of the Dopamine Receptor Gene DRD2 and Colorectal Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1633–1638. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Di Prima, M.; Wang, D.; Tröster, A.; Maric, D.; Terrades-Garcia, N.; Ha, T.; Kwak, H.; Sanchez-Martin, D.; Kudlinzki, D.; Schwalbe, H.; et al. Identification of Eph receptor signaling as a regulator of autophagy and a therapeutic target in colorectal carcinoma. Mol. Oncol. 2019, 13, 2441–2459. [Google Scholar] [CrossRef] [PubMed]
- Strimpakos, A.; Pentheroudakis, G.; Kotoula, V.; De Roock, W.; Kouvatseas, G.; Papakostas, P.; Makatsoris, T.; Papamichael, D.; Andreadou, A.; Sgouros, J.; et al. The Prognostic Role of Ephrin A2 and Endothelial Growth Factor Receptor Pathway Mediators in Patients with Advanced Colorectal Cancer Treated with Cetuximab. Clin. Color. Cancer 2013, 12, 267–274.e2. [Google Scholar] [CrossRef]
- Eddy, K.; Eddin, M.N.; Fateeva, A.; Pompili, S.V.B.; Shah, R.; Doshi, S.; Chen, S. Implications of a Neuronal Receptor Family, Metabotropic Glutamate Receptors, in Cancer Development and Progression. Cells 2022, 11, 2857. [Google Scholar] [CrossRef]
- Chang, H.J.; Yoo, B.C.; Lim, S.-B.; Jeong, S.-Y.; Kim, W.H.; Park, J.-G. Metabotropic Glutamate Receptor 4 Expression in Colorectal Carcinoma and Its Prognostic Significance. Clin. Cancer Res. 2005, 11, 3288–3295. [Google Scholar] [CrossRef]
- Yoo, B.C.; Jeon, E.; Hong, S.H.; Shin, Y.K.; Chang, H.J.; Park, J.G. Metabotropic glutamate receptor 4-mediated 5-Fluorouracil resistance in a human colon cancer cell line. Clin. Cancer Res. 2004, 10, 4176–4184. [Google Scholar] [CrossRef]
- Finci, L.; Zhang, Y.; Meijers, R.; Wang, J.-H. Signaling mechanism of the netrin-1 receptor DCC in axon guidance. Prog. Biophys. Mol. Biol. 2015, 118, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Solimando, A.; Pezzella, F. The Anti-VEGF(R) Drug Discovery Legacy: Improving Attrition Rates by Breaking the Vicious Cycle of Angiogenesis in Cancer. Cancers 2021, 13, 3433. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.R.S.; Douglas, L.R.; Duriez, P.J.; Balourdas, D.-I.; Joerger, A.C.; Khadiullina, R.; Bulatov, E.; Baud, M.G.J. Discovery of Nanomolar-Affinity Pharmacological Chaperones Stabilizing the Oncogenic p53 Mutant Y220C. ACS Pharmacol. Transl. Sci. 2022, 5, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Laugsand, E.A.; Brenne, S.S.; Skorpen, F. DNA methylation markers detected in blood, stool, urine, and tissue in colorectal cancer: A systematic review of paired samples. Int. J. Color. Dis. 2020, 36, 239–251. [Google Scholar] [CrossRef]
- Müller, D.; Győrffy, B. DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochim. Biophys. Acta 2022, 1877, 188722. [Google Scholar] [CrossRef]
- Jensen, S.Ø.; Øgaard, N.; Ørntoft, M.B.W.; Rasmussen, M.H.; Bramsen, J.B.; Kristensen, H.; Mouritzen, P.; Madsen, M.R.; Madsen, A.H.; Sunesen, K.G.; et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer-a clinical biomarker discovery and validation study. Clin. Epigenetics 2019, 11, 158. [Google Scholar] [CrossRef]
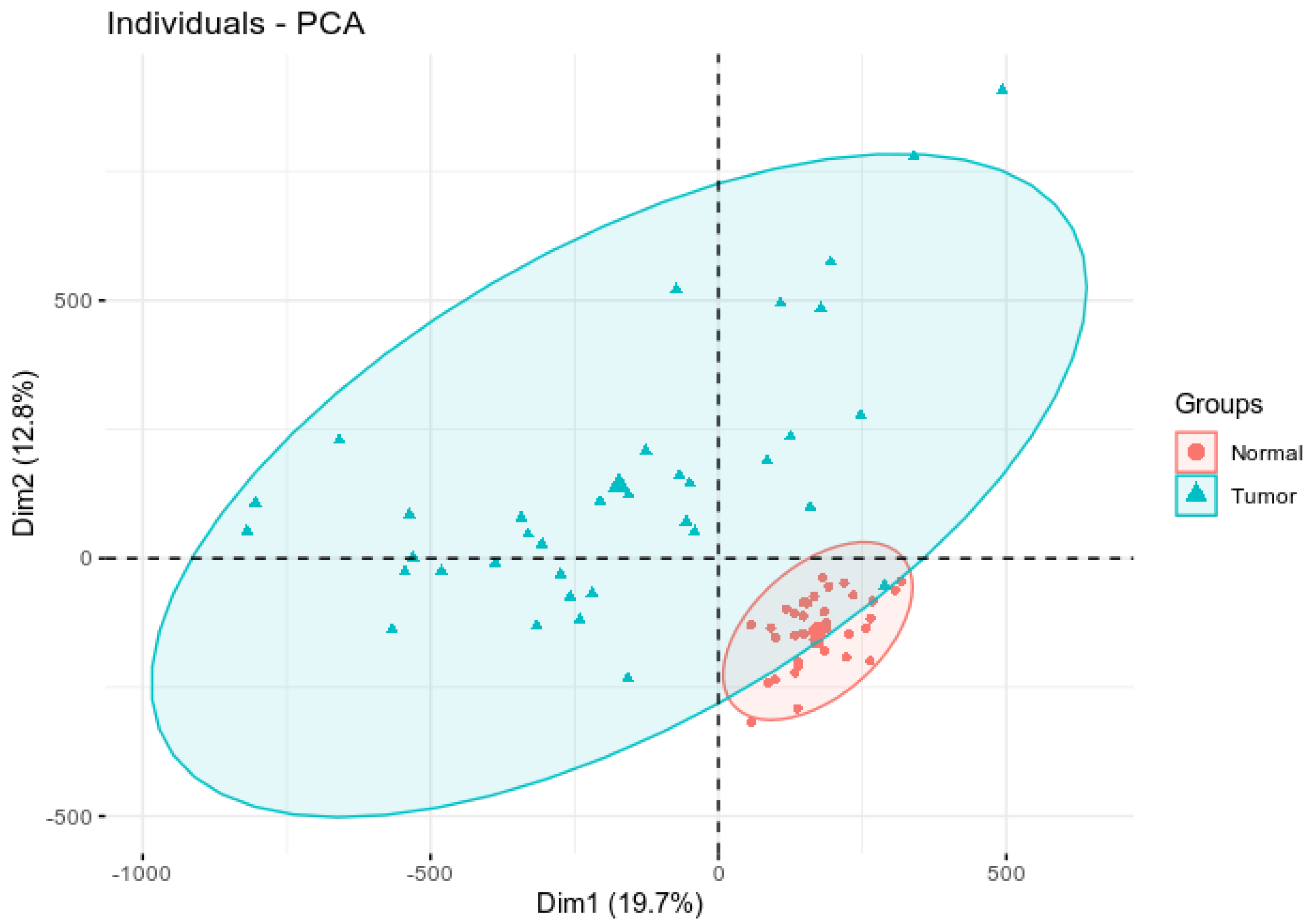
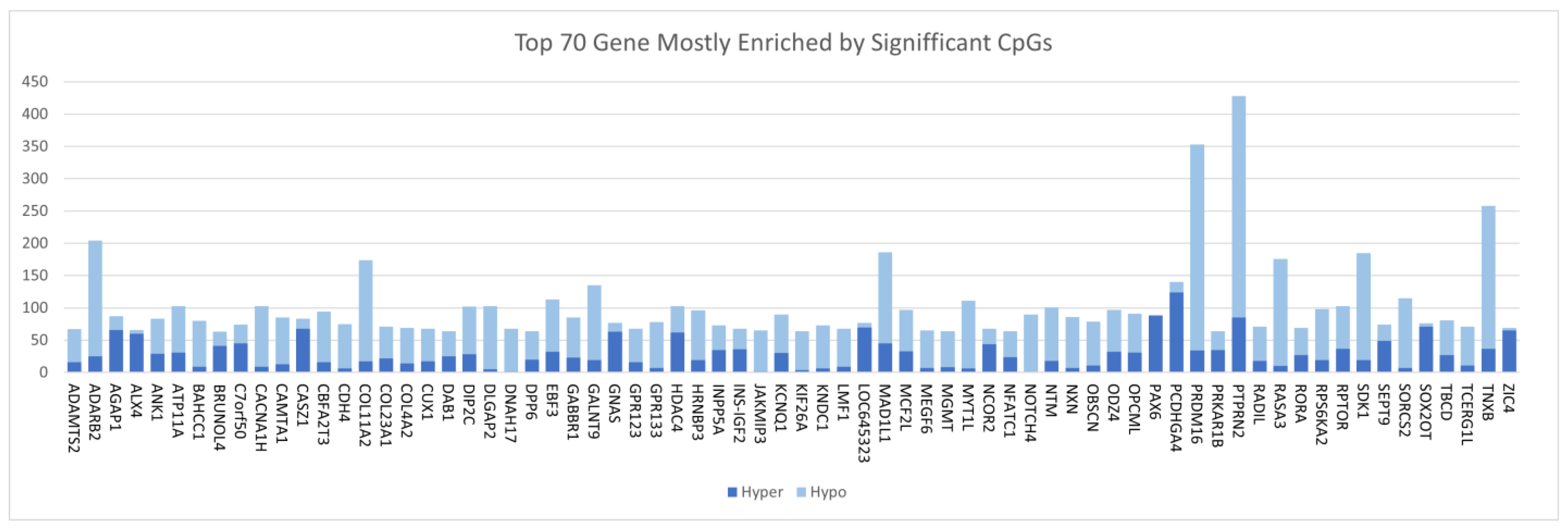
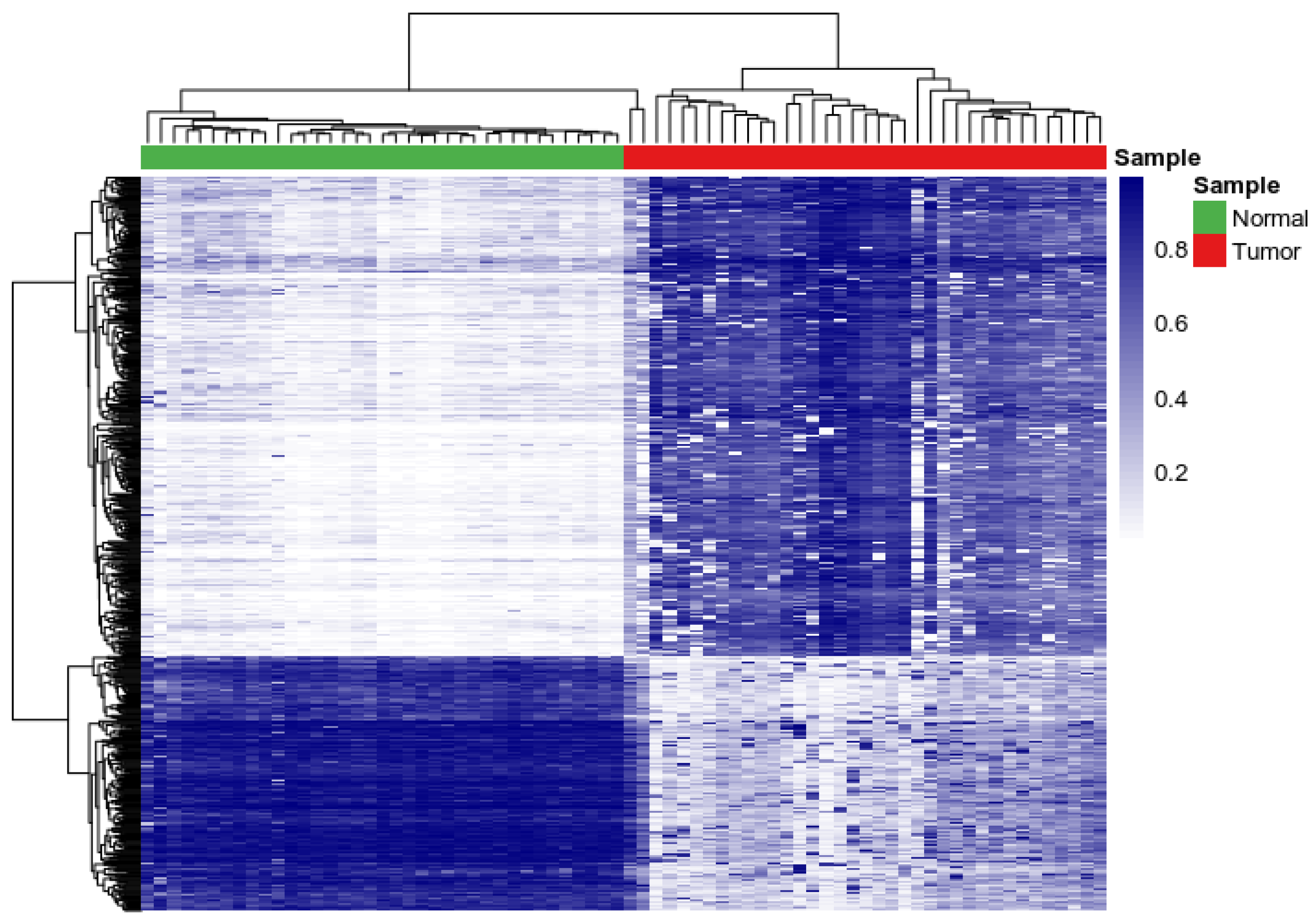
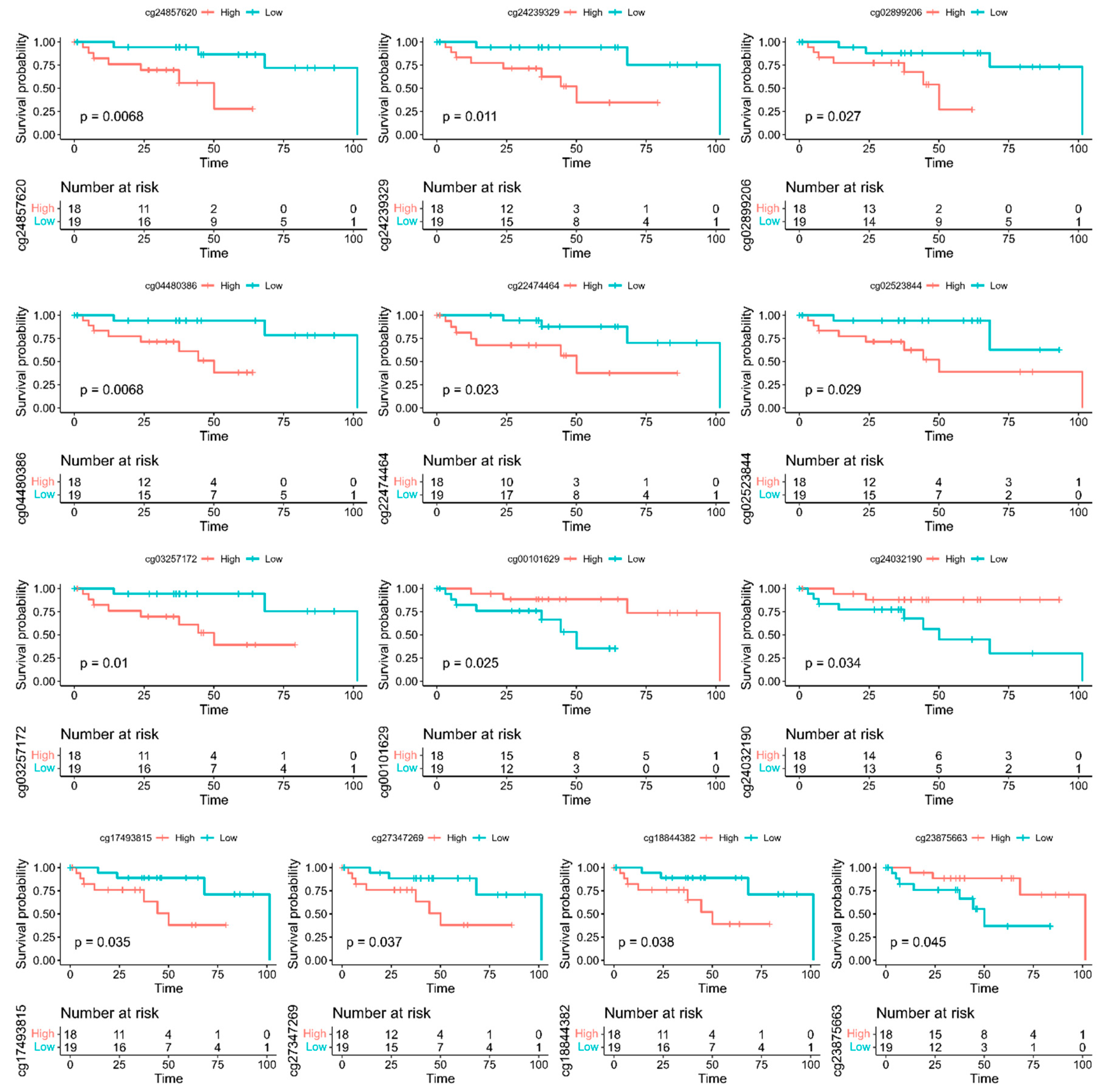
| Gene | Chromosome | CpG Site Probe | Feat.cgi | p-Value |
|---|---|---|---|---|
| ADAMTS2 | 5q35.3 | cg14409941 | Body-island | 1.46 × 10−33 |
| ADAMTS5 | 21q21.3 | cg08190291 | TSS200-island | 2.21 × 10−18 |
| ADAMTS5 | 21q21.3 | cg21646598 | TSS1500-island | 1.46 × 10−24 |
| ADARB2 | 10p15.3 | cg02899206 | TSS200-island | 4.62 × 10−30 |
| ADARB2 | 10p15.3 | cg23684973 | TSS1500-island | 1.55 × 10−29 |
| ADCY1 | 7p12.3 | cg25322847 | Body-shelf | 1.13 × 10−28 |
| ADHFE1 | 8q13.1 | cg01588438 | TSS200-island | 1.07 × 10−36 |
| ADHFE1 | 8q13.1 | cg01988129 | Body-island | 6.84 × 10−32 |
| ADHFE1 | 8q13.1 | cg08090772 | TSS200-island | 8.19 × 10−32 |
| ADHFE1 | 8q13.1 | cg09383816 | TSS200-island | 7.76 × 10−34 |
| ADHFE1 | 8q13.1 | cg18065361 | TSS200-island | 1.68 × 10−29 |
| ADHFE1 | 8q13.1 | cg19283840 | TSS200-island | 2.53 × 10−31 |
| ADHFE1 | 8q13.1 | cg20295442 | TSS200-island | 3.59 × 10−35 |
| ADHFE1 | 8q13.1 | cg20912169 | 5′UTR-island | 7.50 × 10−35 |
| ADRB3 | 8p11.23 | cg09258813 | 1stExon-island | 3.29 × 10−20 |
| AGRN | 1p36.33 | cg09248054 | Body-island | 7.98 × 10−30 |
| AGRN | 1p36.33 | cg16318112 | Body-island | 2.59 × 10−30 |
| AHRR | 5p15.33 | cg14453201 | Body-island | 1.07 × 10−17 |
| AKR1B1 | 7q33 | cg08167706 | TSS1500-shore | 1.86 × 10−16 |
| AMPH | 7p14.1 | cg02383130 | 5′UTR-island | 4.73 × 10−20 |
| AMPH | 7p14.1 | cg07034660 | Body-shore | 1.10 × 10−22 |
| AMPH | 7p14.1 | cg07926691 | 5′UTR-island | 2.18 × 10−27 |
| AMPH | 7p14.1 | cg10293925 | 5′UTR-island | 2.30 × 10−27 |
| AMPH | 7p14.1 | cg19875547 | Body-island | 8.40 × 10−31 |
| AMPH | 7p14.1 | cg26122980 | 5′UTR-island | 7.20 × 10−30 |
| ANK1 | 8p11.21 | cg17331296 | 1stExon-island | 7.24 × 10−28 |
| ANKRD13B | 17q11.2 | cg21101720 | Body-island | 2.94 × 10−23 |
| ANXA2 | 15q22.2 | cg22365276 | 5′UTR-shore | 8.76 × 10−26 |
| AQP5 | 12q13.12 | cg26328335 | TSS1500-island | 4.63 × 10−19 |
| ATP11A | 13q34 | cg08162124 | Body-shore | 1.02 × 10−22 |
| ATP8B2 | 1q21.3 | cg00581482 | 5′UTR-island | 1.24 × 10−22 |
| ATP8B2 | 1q21.3 | cg08190044 | 5′UTR-island | 1.72 × 10−22 |
| AUTS2 | 7q11.22 | cg21393713 | 1stExon-island | 9.58 × 10−32 |
| AVP | 20p13 | cg23035419 | TSS1500-shore | 4.05 × 10−20 |
| AVPR1A | 12q14.2 | cg12516059 | 1stExon-shore | 3.35 × 10−23 |
| B3GNTL1 | 17q25.3 | cg10344477 | Body-shelf | 1.50 × 10−27 |
| BARHL2 | 1p22.2 | cg26332310 | 1stExon-island | 2.48 × 10−19 |
| BCAT1 | 12p12.1 | cg13980808 | Body-shore | 1.10 × 10−18 |
| BEND5 | 1p33 | cg11666087 | 1stExon-island | 3.86 × 10−19 |
| BEND5 | 1p33 | cg16573178 | 1stExon-island | 7.30 × 10−17 |
| BOLL | 2q33.1 | cg03774803 | 1stExon-island | 1.99 × 10−25 |
| BOLL | 2q33.1 | cg13356896 | TSS200-island | 2.03 × 10−28 |
| BOLL | 2q33.1 | cg24589459 | TSS1500-island | 4.46 × 10−19 |
| C9orf50 | 9q34.11 | cg09731694 | 1stExon-island | 1.81 × 10−32 |
| C9orf50 | 9q34.11 | cg13405887 | 1stExon-island | 4.58 × 10−47 |
| C9orf50 | 9q34.11 | cg14015706 | 1stExon-island | 8.68 × 10−39 |
| CADM2 | 3p12.1 | cg05152589 | 5′UTR-island | 1.17 × 10−18 |
| CALCR | 7q21.3 | cg20276156 | TSS1500-shore | 2.43 × 10−20 |
| CASR | 3q13.33-q21.1 | cg05937969 | 5′UTR-island | 1.37 × 10−22 |
| CASR | 3q13.33-q21.1 | cg25729826 | 5′UTR-island | 9.43 × 10−24 |
| Gene | Probe | Feat.cgi | p-Value | Methylated |
|---|---|---|---|---|
| ADARB2 | cg02899206 | TSS200-island | 0.027 | Low |
| CDH12 | cg04480386 | 5′UTR-opensea | 0.0068 | Low |
| DCLK1 | cg24239329 | TSS200-island | 0.011 | Low |
| DOK6 | cg27347269 | TSS1500-island | 0.037 | Low |
| EFS | cg18844382 | TSS200-island | 0.038 | Low |
| GUCY1B3 | cg17493815 | Body-island | 0.035 | Low |
| KIAA1026 | cg00101629 | Body-opensea | 0.025 | High |
| MAGI2 | cg02523844 | TSS1500-island | 0.029 | Low |
| NKX2-2 | cg22474464 | Body-island | 0.023 | Low |
| NPBWR1 | cg24857620 | TSS1500-island | 0.0068 | Low |
| NR5A2 | cg03257172 | Body-shore | 0.01 | Low |
| PCSK2 | cg23875663 | Body-shelf | 0.045 | High |
| SMAD3 | cg24032190 | Body-opensea | 0.034 | High |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Zhang, L.; Kuchi, A.; Otohinoyi, D.; Hicks, C. CpG Site-Based Signature Predicts Survival of Colorectal Cancer. Biomedicines 2022, 10, 3163. https://doi.org/10.3390/biomedicines10123163
Wu J, Zhang L, Kuchi A, Otohinoyi D, Hicks C. CpG Site-Based Signature Predicts Survival of Colorectal Cancer. Biomedicines. 2022; 10(12):3163. https://doi.org/10.3390/biomedicines10123163
Chicago/Turabian StyleWu, Jiande, Lu Zhang, Aditi Kuchi, David Otohinoyi, and Chindo Hicks. 2022. "CpG Site-Based Signature Predicts Survival of Colorectal Cancer" Biomedicines 10, no. 12: 3163. https://doi.org/10.3390/biomedicines10123163
APA StyleWu, J., Zhang, L., Kuchi, A., Otohinoyi, D., & Hicks, C. (2022). CpG Site-Based Signature Predicts Survival of Colorectal Cancer. Biomedicines, 10(12), 3163. https://doi.org/10.3390/biomedicines10123163






