EMR1/ADGRE1 Expression in Cancer Cells Upregulated by Tumor-Associated Macrophages Is Related to Poor Prognosis in Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. Ethics Approval
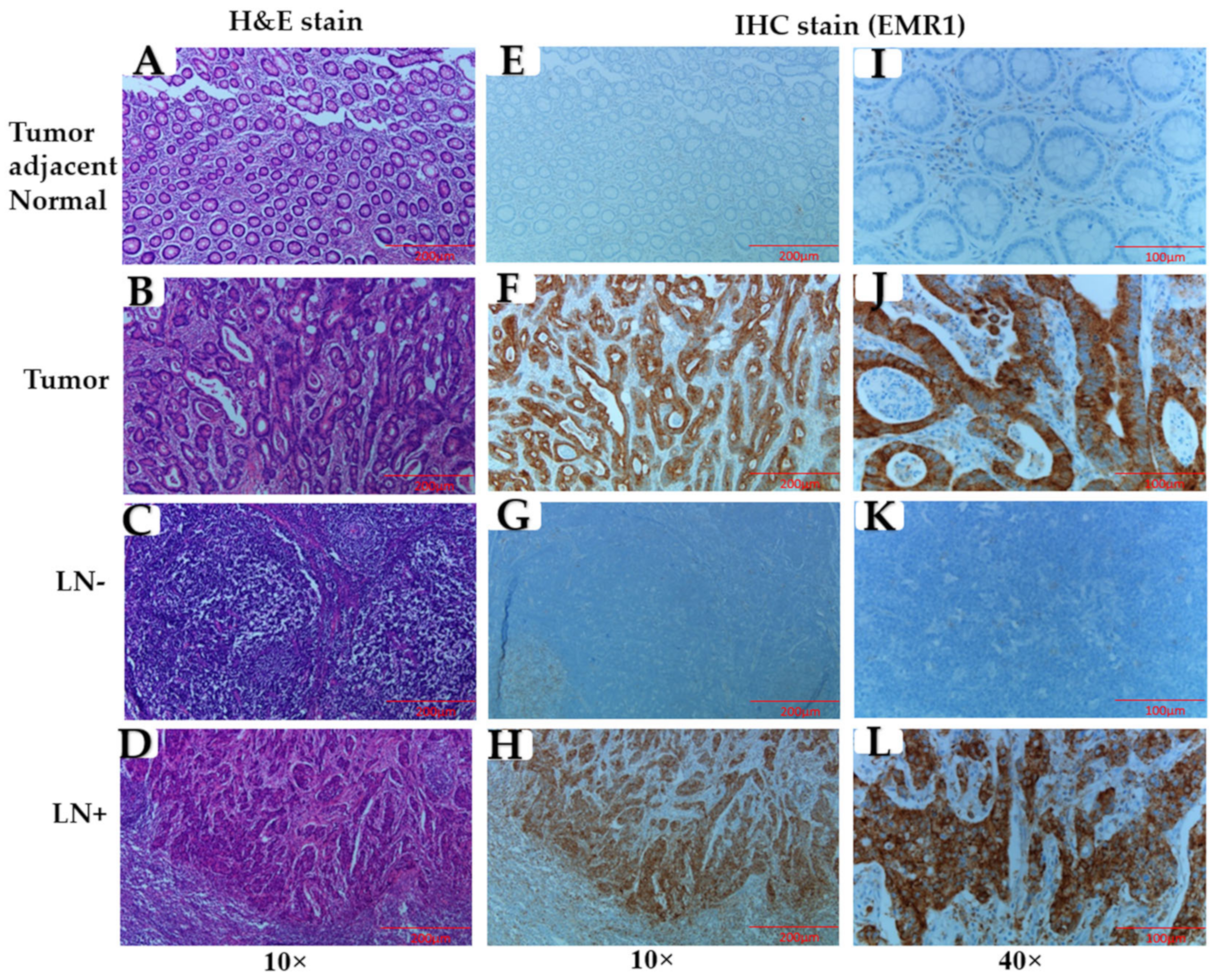
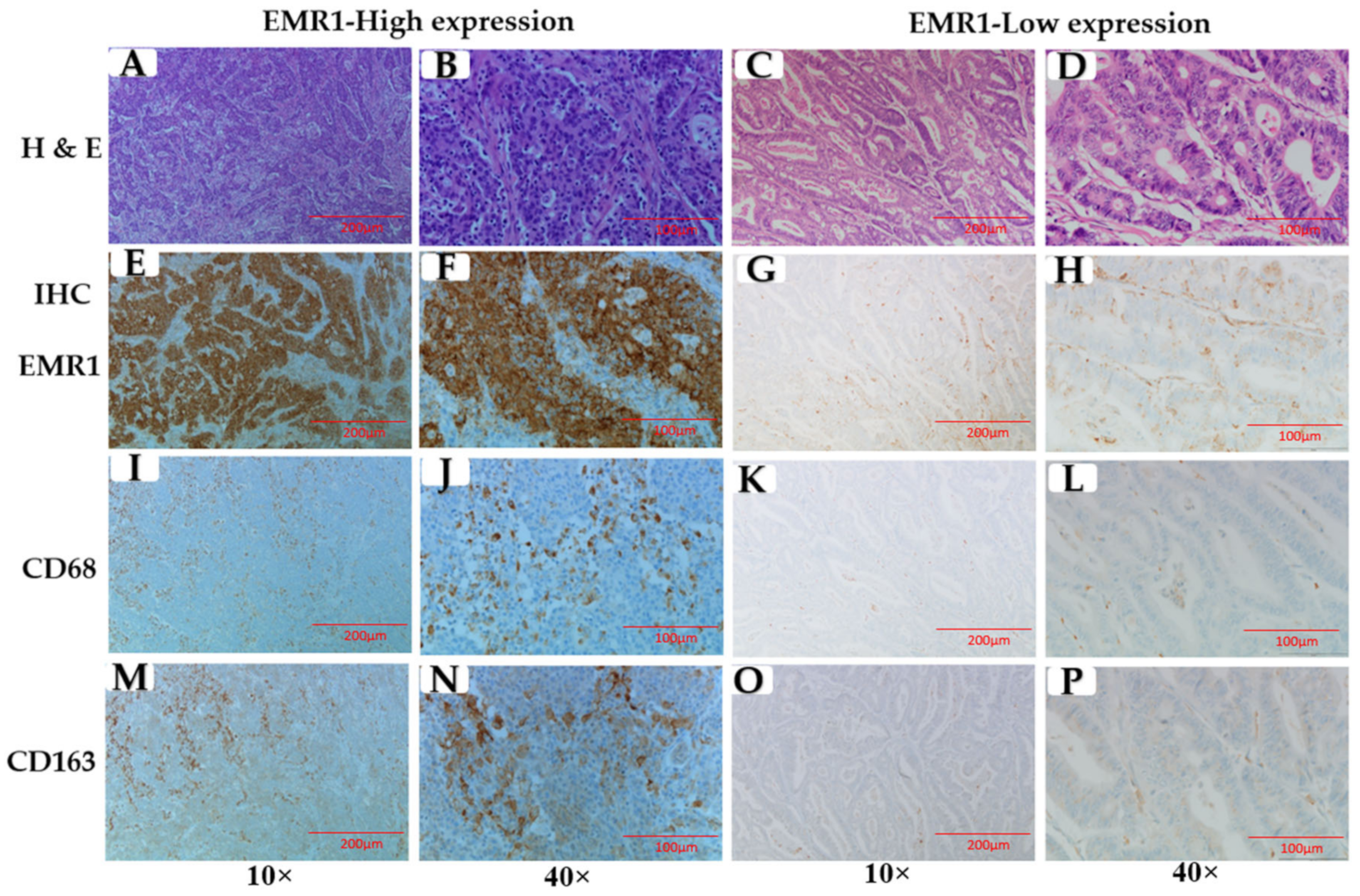
2.3. Immunohistochemical (IHC) Staining
2.4. Cell Culture
2.4.1. Cell Culture and Reagents
2.4.2. Co-Culture Procedures
2.4.3. Flow Cytometry Analysis
2.4.4. RNA Extraction and cDNA Synthesis
2.4.5. Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) mRNA Assay
2.4.6. Western Blot
2.4.7. Statistical Analysis
3. Results
3.1. Association of EMR1-TC with Clinicopathological Variables in Patients with MSI-H and MSS CRC
3.2. Correlation between EMR1-TC and CD68+/CD163+ TAMs in MSI-H and MSS CRC
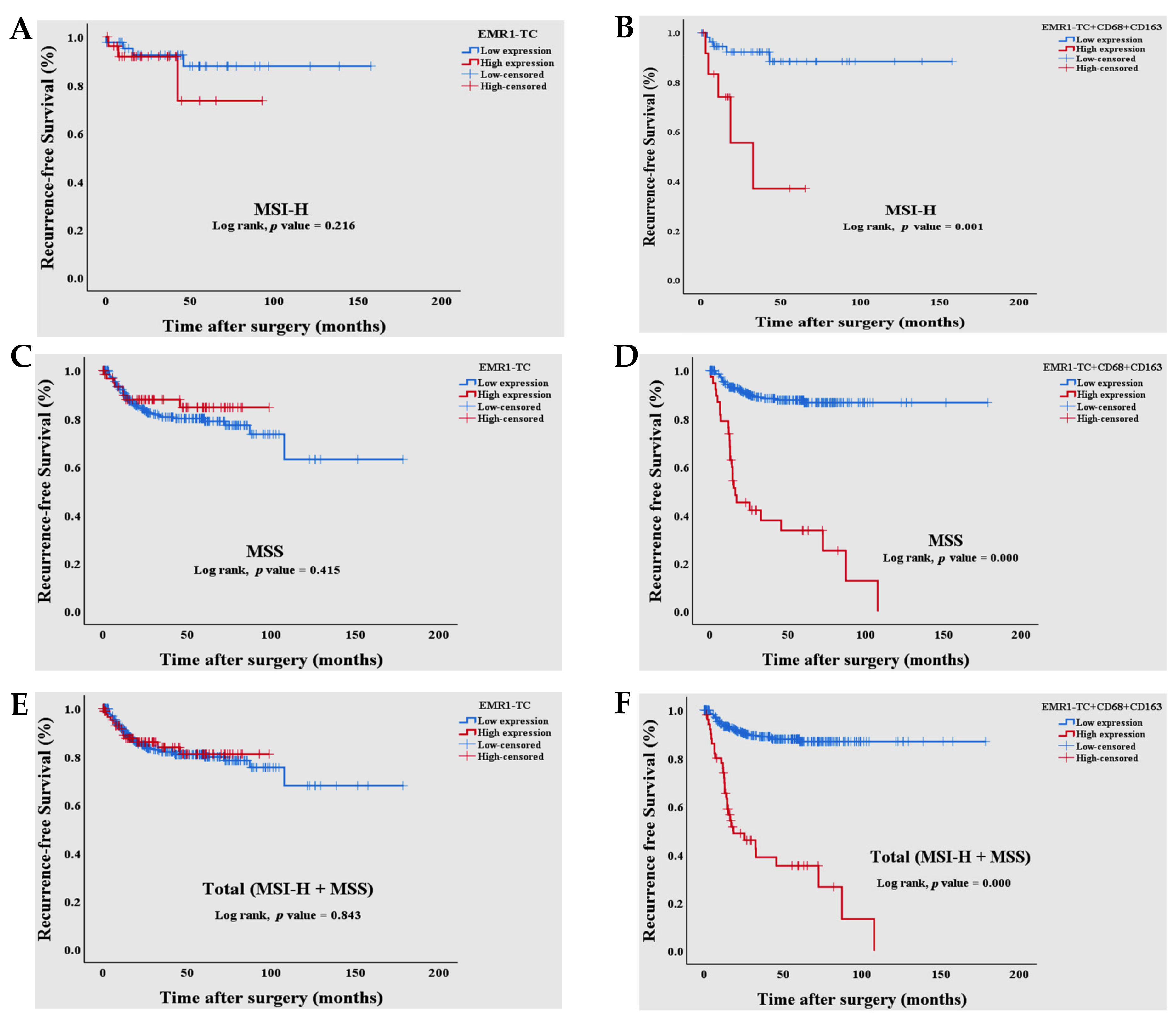
3.3. Survival Analysis of Combined EMR1-TC+CD68+CD163+ Expression in Patients with MSI-H and MSS CRC
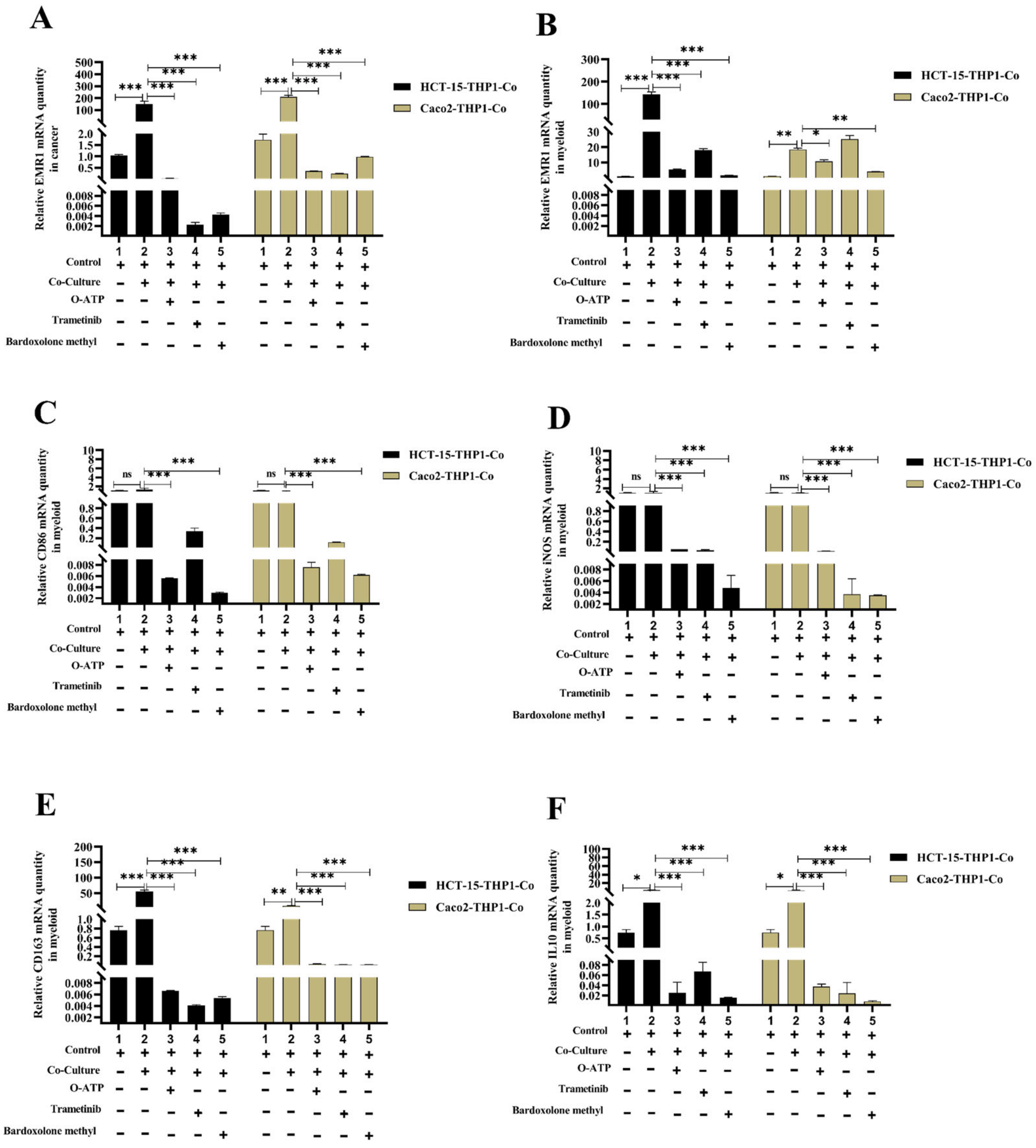
3.4. Macrophage-Induced EMR1 Upregulation in Colon Cancer Cells (CCs) In Vitro
3.5. EMR1 Upregulation in Cancer Cells Correlates with M2 Macrophage Polarization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ranasinghe, R.; Mathai, M.; Zulli, A. A synopsis of modern-day colorectal cancer: Where we stand. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188699. [Google Scholar] [CrossRef] [PubMed]
- Currais, P.; Rosa, I.; Claro, I. Colorectal cancer carcinogenesis: From bench to bedside. World J. Gastrointest. Oncol. 2022, 14, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Keenan, J.I.; Frizelle, F.A. Biomarkers to Detect Early-Stage Colorectal Cancer. Biomedicines 2022, 10, 255. [Google Scholar] [CrossRef] [PubMed]
- Zafari, N.; Khosravi, F.; Rezaee, Z.; Esfandyari, S.; Bahiraei, M.; Bahramy, A.; Ferns, G.A.; Avan, A. The role of the tumor microenvironment in colorectal cancer and the potential therapeutic approaches. J. Clin. Lab. Anal. 2022, 36, e24585. [Google Scholar] [CrossRef]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef]
- Baradaran, A.; Asadzadeh, Z.; Hemmat, N.; Baghbanzadeh, A.; Shadbad, M.A.; Khosravi, N.; Derakhshani, A.; Alemohammad, H.; Nour, M.A.; Safarpour, H. The cross-talk between tumor-associated macrophages and tumor endothelium: Recent advances in macrophage-based cancer immunotherapy. Biomed. Pharmacother. 2022, 146, 112588. [Google Scholar] [CrossRef]
- Jalali, A.; Haji Molla Hoseini, M.; Rezaei, M. Auraptene Promotes THP-1 Polarization to M1 Macrophages and Improves M1 Function. Res. J. Pharmacogn. 2022, 9, 63–75. [Google Scholar]
- Wang, X.; Wang, J.; Zhao, J.; Wang, H.; Chen, J.; Wu, J. HMGA2 facilitates colorectal cancer progression via STAT3-mediated tumor-associated macrophage recruitment. Theranostics 2022, 12, 963. [Google Scholar] [CrossRef]
- Fu, X.-H.; Li, J.-P.; Li, X.-Y.; Tan, Y.; Zhao, M.; Zhang, S.-F.; Wu, X.-D.; Xu, J.-G. M2-Macrophage-Derived Exosomes Promote Meningioma Progression through TGF-β Signaling Pathway. J. Immunol. Res. 2022, 2022, 8326591. [Google Scholar] [CrossRef]
- Fan, C.; Chen, C.; Chen, L.; Chua, K.; Hung, H.; Hsu, J.; Huang, T. Extracellular HSP90α Induces MyD88-IRAK Complex-Associated IKKα/β−NF-κB/IRF3 and JAK2/TYK2−STAT-3 Signaling in Macrophages for Tumor-Promoting M2-Polarization. Cell 2022, 11, 229. [Google Scholar] [CrossRef]
- Kou, Y.; Li, Z.; Sun, Q.; Yang, S.; Wang, Y.; Hu, C.; Gu, H.; Wang, H.; Xu, H.; Li, Y. Prognostic value and predictive biomarkers of phenotypes of tumour-associated macrophages in colorectal cancer. Scand. J. Immunol. 2022, 95, e13137. [Google Scholar] [CrossRef]
- Yi, B.; Dai, K.; Yan, Z.; Yin, Z. Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages. Bioengineered 2022, 13, 6243–6256. [Google Scholar] [CrossRef]
- Lin, A.; Zhang, J.; Luo, P. Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer. Front. Immunol. 2020, 11, 2039. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Y.; Zhang, Y.; Fang, S.; Zhang, M.; Li, H.; Xu, F.; Liu, L.; Liu, J.; Zhao, Q. Subtyping of microsatellite stability colorectal cancer reveals guanylate binding protein 2 (GBP2) as a potential immunotherapeutic target. J. Immunother. Cancer 2022, 10, e004302. [Google Scholar] [CrossRef]
- Ugai, T.; Väyrynen, J.P.; Lau, M.C.; Borowsky, J.; Akimoto, N.; Väyrynen, S.A.; Zhao, M.; Zhong, R.; Haruki, K.; Dias Costa, A. Immune cell profiles in the tumor microenvironment of early-onset, intermediate-onset, and later-onset colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 933–942. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Yang, C.; Wang, Y.; Liu, J.; Shi, Z.; Chen, Y.; Feng, Y.; Ma, X.; Qiao, S. A study of the correlation between M2 macrophages and lymph node metastasis of colorectal carcinoma. World J. Surg. Oncol. 2021, 19, 91. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wei, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Dou, R.; Xiong, B. Elevated CD163+/CD68+ ratio at tumor invasive front is closely associated with aggressive phenotype and poor prognosis in colorectal cancer. Int. J. Biol. Sci. 2019, 15, 984. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, M.; Peng, R.; Liu, J.; Wang, F.; Li, Y.; Zhao, Q.; Liu, J. The prognostic and clinicopathological value of tumor-associated macrophages in patients with colorectal cancer: A systematic review and meta-analysis. Int. J. Colorect. Dis. 2020, 35, 1651–1661. [Google Scholar] [CrossRef]
- Li, J.; Li, L.; Li, Y.; Long, Y.; Zhao, Q.; Ouyang, Y.; Bao, W.; Gong, K. Tumor-associated macrophage infiltration and prognosis in colorectal cancer: Systematic review and meta-analysis. Int. J. Color. Dis. 2020, 35, 1203–1210. [Google Scholar] [CrossRef]
- Gad, A.A.; Balenga, N. The emerging role of adhesion GPCRs in cancer. ACS Pharmacol. Transl. Sci. 2020, 3, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Waddell, L.A.; Lefevre, L.; Bush, S.J.; Raper, A.; Young, R.; Lisowski, Z.M.; McCulloch, M.E.B.; Muriuki, C.; Sauter, K.A.; Clark, E.L.; et al. ADGRE1 (EMR1, F4/80) Is a Rapidly-Evolving Gene Expressed in Mammalian Monocyte-Macrophages. Front. Immunol. 2018, 9, 2246. [Google Scholar] [CrossRef] [PubMed]
- Legrand, F.; Tomasevic, N.; Simakova, O.; Lee, C.-C.R.; Wang, Z.; Raffeld, M.; Makiya, M.A.; Palath, V.; Leung, J.; Baer, M. The eosinophil surface receptor epidermal growth factor–like module containing mucin-like hormone receptor 1 (EMR1): A novel therapeutic target for eosinophilic disorders. J. Allergy Clin. Immunol. 2014, 133, 1439–1447.e8. [Google Scholar] [CrossRef] [PubMed]
- Hamann, J.; Koning, N.; Pouwels, W.; Ulfman, L.H.; van Eijk, M.; Stacey, M.; Lin, H.H.; Gordon, S.; Kwakkenbos, M.J. EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. Eur. J. Immunol. 2007, 37, 2797–2802. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lin, J.; Wang, G.; Xu, D. Targeting Proliferating Tumor-Infiltrating Macrophages Facilitates Spatial Redistribution of CD8+ T Cells in Pancreatic Cancer. Cancers 2022, 14, 1474. [Google Scholar] [CrossRef]
- Song, C.-H.; Kim, N.; Nam, R.H.; Choi, S.I.; Jang, J.Y.; Kim, J.W.; Na, H.Y.; Lee, H.-N. Combination treatment with 17β-estradiol and anti-PD-L1 suppresses MC38 tumor growth by reducing PD-L1 expression and enhancing M1 macrophage population in MC38 colon tumor model. Cancer Lett. 2022, 543, 215780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Wang, J.; Jiang, Z.; Zhang, L.; Cui, Y.; Zhao, D.; Wang, Y. Atractylenolide II inhibits tumor-associated macrophages (TAMs)-induced lung cancer cell metastasis. Immunopharmacol. Immunotoxicol. 2022, 44, 227–237. [Google Scholar] [CrossRef]
- Soncin, I.; Sheng, J.; Chen, Q.; Foo, S.; Duan, K.; Lum, J.; Poidinger, M.; Zolezzi, F.; Karjalainen, K.; Ruedl, C. The tumour microenvironment creates a niche for the self-renewal of tumour-promoting macrophages in colon adenoma. Nat. Commun. 2018, 9, 582. [Google Scholar] [CrossRef]
- Hall, R.A.; Lala, T. Adhesion G Protein-Coupled Receptors: Structure, Signaling, Physiology and Pathophysiology. Physiol. Rev. 2022, 102, 1587–1624. [Google Scholar]
- Aust, G.; Zhu, D.; Van Meir, E.G.; Xu, L. Adhesion GPCRs in tumorigenesis. Adhes. G Protein Coupled Recept. 2016, 234, 369–396. [Google Scholar]
- Nijmeijer, S.; Vischer, H.F.; Leurs, R. Adhesion GPCRs in immunology. Biochem. Pharmacol. 2016, 114, 88–102. [Google Scholar] [CrossRef]
- Lei, P.; Wang, H.; Yu, L.; Xu, C.; Sun, H.; Lyu, Y.; Li, L.; Zhang, D.-L. A correlation study of adhesion G protein-coupled receptors as potential therapeutic targets in Uterine Corpus Endometrial cancer. Int. Immunopharmacol. 2022, 108, 108743. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Olsson, L.; Lindmark, G.; Hammarström, M.-L.; Hammarström, S.; Sitohy, B. The myeloid cell biomarker EMR1 is ectopically expressed in colon cancer. Tumor Biol. 2021, 43, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lee, M.-R.; Choi, E.; Cho, M.-Y. Clinicopathologic significance of survivin expression in relation to CD133 expression in surgically resected stage II or III colorectal cancer. J. Pathol. Transl. Med. 2017, 51, 17. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jung, P.Y.; Lee, J.S.; Hwang, S.; Sohn, J.H.; Yoon, Y.; Bae, K.S.; Eom, Y.W. Cultured human skeletal muscle satellite cells exhibit characteristics of mesenchymal stem cells and play anti-inflammatory roles through prostaglandin E2 and hepatocyte growth factors. Cell Biol. Int. 2021, 45, 2443–2451. [Google Scholar] [CrossRef]
- Kim, D.-H.; Park, J.C.; Lee, J.-S. G protein-coupled receptors (GPCRs) in rotifers and cladocerans: Potential applications in ecotoxicology, ecophysiology, comparative endocrinology, and pharmacology. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2022, 256, 109297. [Google Scholar] [CrossRef] [PubMed]
- Usman, S.; Khawer, M.; Rafique, S.; Naz, Z.; Saleem, K. The current status of anti-GPCR drugs against different cancers. J. Pharm. Anal. 2020, 10, 517–521. [Google Scholar] [CrossRef]
- Huang, Y.-S.; Hu, C.-H.; Tseng, W.-Y.; Cheng, C.-H.; Stacey, M.; Gordon, S.; Chang, G.-W.; Lin, H.-H. Activation of Adhesion GPCR EMR2/ADGRE2 Induces Macrophage Differentiation and Inflammatory Responses via Gα16/Akt/MAPK/NF-κB Signaling Pathways. Front. Immunol. 2017, 8, 0373. [Google Scholar] [CrossRef]
- Davies, J.Q.; Lin, H.-H.; Stacey, M.; Yona, S.; Chang, G.-W.; Gordon, S.; Hamann, J.; Campo, L.; Han, C.; Chan, P. Leukocyte adhesion-GPCR EMR2 is aberrantly expressed in human breast carcinomas and is associated with patient survival. Oncol. Rep. 2011, 25, 619–627. [Google Scholar]
- Rutkowski, M.J.; Sughrue, M.E.; Kane, A.J.; Kim, J.M.; Bloch, O.; Parsa, A.T. Epidermal growth factor module-containing mucin-like receptor 2 is a newly identified adhesion G protein-coupled receptor associated with poor overall survival and an invasive phenotype in glioblastoma. J. Neuro-Oncol. 2011, 105, 165–171. [Google Scholar] [CrossRef]
- Aust, G.; Hamann, J.; Schilling, N.; Wobus, M. Detection of alternatively spliced EMR2 mRNAs in colorectal tumor cell lines but rare expression of the molecule in colorectal adenocarcinomas. Virchows Arch. 2003, 443, 32–37. [Google Scholar] [CrossRef]
- Peng, P.; Hu, C.; Han, Y.; Xu, Y.; Shen, H.; Yu, Y.; Zhou, H. Expressions of CD97 isoforms in colon cancer tissues and their clinical significances. Cancer Res. Clin. 2019, 6, 662–665. [Google Scholar]
- Hilbig, D.; Dietrich, N.; Wandel, E.; Gonsior, S.; Sittig, D.; Hamann, J.; Aust, G. The interaction of CD97/ADGRE5 With β-Catenin in adherens junctions Is lost during colorectal carcinogenesis. Front. Oncol. 2018, 8, 182. [Google Scholar] [CrossRef]
- Ji, B.; Feng, Y.; Sun, Y.; Ji, D.; Qian, W.; Zhang, Z.; Wang, Q.; Zhang, Y.; Zhang, C.; Sun, Y. GPR56 promotes proliferation of colorectal cancer cells and enhances metastasis via epithelial-mesenchymal transition through PI3K/AKT signaling activation. Oncol. Rep. 2018, 40, 1885–1896. [Google Scholar] [CrossRef]
- Tang, X.; Jin, R.; Qu, G.; Wang, X.; Li, Z.; Yuan, Z.; Zhao, C.; Siwko, S.; Shi, T.; Wang, P. GPR116, an adhesion G-protein–coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 2013, 73, 6206–6218. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Z.; Chen, J.; Xue, M.; Pan, X. ELTD1 promotes invasion and metastasis by activating MMP2 in colorectal cancer. Int. J. Biol. Sci. 2021, 17, 3048. [Google Scholar] [CrossRef]
- Fu, L.-Q.; Du, W.-L.; Cai, M.-H.; Yao, J.-Y.; Zhao, Y.-Y.; Mou, X.-Z. The roles of tumor-associated macrophages in tumor angiogenesis and metastasis. Cell. Immunol. 2020, 353, 104119. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, K.; Kunisho, S.; Takigawa, H.; Yuge, R.; Oka, S.; Tanaka, S.; Shimamoto, F.; Chayama, K.; Kitadai, Y. Role of tumor-associated macrophages at the invasive front in human colorectal cancer progression. Cancer Sci. 2021, 112, 2692–2704. [Google Scholar] [CrossRef] [PubMed]
- Herrera, M.; Herrera, A.; Domínguez, G.; Silva, J.; García, V.; García, J.M.; Gómez, I.; Soldevilla, B.; Muñoz, C.; Provencio, M. Cancer-associated fibroblast and M 2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013, 104, 437–444. [Google Scholar] [CrossRef]
- Koelzer, V.H.; Canonica, K.; Dawson, H.; Sokol, L.; Karamitopoulou-Diamantis, E.; Lugli, A.; Zlobec, I. Phenotyping of tumor-associated macrophages in colorectal cancer: Impact on single cell invasion (tumor budding) and clinicopathological outcome. Oncoimmunology 2016, 5, e1106677. [Google Scholar] [CrossRef]
- Feng, Q.; Chang, W.; Mao, Y.; He, G.; Zheng, P.; Tang, W.; Wei, Y.; Ren, L.; Zhu, D.; Ji, M.; et al. Tumor-associated Macrophages as Prognostic and Predictive Biomarkers for Postoperative Adjuvant Chemotherapy in Patients with Stage II Colon Cancer. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 2019, 25, 3896–3907. [Google Scholar] [CrossRef] [PubMed]
- Aasebø, K.; Bruun, J.; Bergsland, C.H.; Nunes, L.; Eide, G.E.; Pfeiffer, P.; Dahl, O.; Glimelius, B.; Lothe, R.A.; Sorbye, H. Prognostic role of tumour-infiltrating lymphocytes and macrophages in relation to MSI, CDX2 and BRAF status: A population-based study of metastatic colorectal cancer patients. Br. J. Cancer 2022, 126, 48–56. [Google Scholar] [CrossRef]
- Nagorsen, D.; Voigt, S.; Berg, E.; Stein, H.; Thiel, E.; Loddenkemper, C. Tumor-infiltrating macrophages and dendritic cells in human colorectal cancer: Relation to local regulatory T cells, systemic T-cell response against tumor-associated antigens and survival. J. Transl. Med. 2007, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- De Smedt, L.; Lemahieu, J.; Palmans, S.; Govaere, O.; Tousseyn, T.; Van Cutsem, E.; Prenen, H.; Tejpar, S.; Spaepen, M.; Matthijs, G. Microsatellite instable vs stable colon carcinomas: Analysis of tumour heterogeneity, inflammation and angiogenesis. Br. J. Cancer 2015, 113, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Kaidi, D.; Szeponik, L.; Yrlid, U.; Wettergren, Y.; Bexe Lindskog, E. Impact of thymidine phosphorylase and CD163 expression on prognosis in stage II colorectal cancer. Clin. Transl. Oncol. 2022, 24, 1818–1827. [Google Scholar] [CrossRef]
- Lin, H.-H.; Faunce, D.E.; Stacey, M.; Terajewicz, A.; Nakamura, T.; Zhang-Hoover, J.; Kerley, M.; Mucenski, M.L.; Gordon, S.; Stein-Streilein, J. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J. Exp. Med. 2005, 201, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Nakao, T.; Shimada, M.; Yoshikawa, K.; Tokunaga, T.; Nishi, M.; Kashihara, H.; Takasu, C.; Wada, Y.; Yoshimoto, T.; Yamashita, S. The correlation of immunoscore and frailty in colorectal cancer. Int. J. Clin. Oncol. 2022, 27, 528–537. [Google Scholar] [CrossRef]
- Bell, P.D.; Pai, R.K. Immune response in colorectal carcinoma: A review of its significance as a predictive and prognostic biomarker. Histopathology 2022, 81, 696–714. [Google Scholar] [CrossRef]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Liu, Q.; Dou, R.; Xiong, B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol. Cancer 2019, 18, 64. [Google Scholar] [CrossRef]
- Zheng, B.-W.; Yang, M.-L.; Huang, W.; Zheng, B.-Y.; Zhang, T.-L.; Li, J.; Lv, G.-H.; Yan, Y.-G.; Zou, M.-X. Prognostic Significance of Tumor-Associated Macrophages in Chondroblastoma and Their Association with Response to Adjuvant Radiotherapy. J. Inflamm. Res. 2021, 14, 1991. [Google Scholar] [CrossRef]
- Mao, H.; Pan, F.; Guo, H.; Bu, F.; Xin, T.; Chen, S.; Guo, Y. Feedback mechanisms between M2 macrophages and Th17 cells in colorectal cancer patients. Tumor Biol. 2016, 37, 12223–12230. [Google Scholar] [CrossRef]
- He, L.; Jhong, J.-H.; Chen, Q.; Huang, K.-Y.; Strittmatter, K.; Kreuzer, J.; DeRan, M.; Wu, X.; Lee, T.-Y.; Slavov, N. Global characterization of macrophage polarization mechanisms and identification of M2-type polarization inhibitors. Cell Rep. 2021, 37, 109955. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhang, X.; Tan, B.; Zhang, S.; Yin, C.; Xue, Q.; Zhang, Z.; Ren, H.; Chen, J.; Liu, M. Blocking P2X7-Mediated Macrophage Polarization Overcomes Treatment Resistance in Lung CancerP2X7 Inhibition Facilitates Lung Cancer Treatment. Cancer Immunol. Res. 2020, 8, 1426–1439. [Google Scholar] [CrossRef] [PubMed]


| Variables | EMR1-TC (Total) | EMR1-TC (MSI-H) | EMR1-TC (MSS) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | p Value | N (%) | p Value | N (%) | p Value | |||||
| Low | High | Low | High | Low | High | |||||
| Gender | Male | 189 (47.5) | 54 (13.6) | 0.715 | 21 (29.6) | 11 (15.5) | 0.458 | 168 (51.4) | 43 (13.1) | 0.775 |
| Female | 118 (29.6) | 37 (9.3) | 24 (33.8) | 15 (21.1) | 94 (28.7) | 22 (6.7) | ||||
| Location | Right | 120 (30.2) | 52 (13.1) | 0.003 | 33 (46.5) | 23 (32.4) | 0.113 | 88 (26.9) | 29 (8.9) | 0.112 |
| Left | 187 (47.0) | 39 (9.8) | 12 (16.9) | 3 (4.2) | 174 (53.2) | 36 (11.0) | ||||
| T_stage | Tis, T1, T2 | 43 (10.8) | 21 (5.3) | 0.039 | 2 (2.8) | 5 (7.0) | 0.044 | 41 (12.5) | 16 (4.9) | 0.088 |
| T3, T4 | 264 (66.3) | 70 (17.6) | 43 (60.6) | 21 (29.6) | 221 (67.6) | 49 (15.0) | ||||
| LNM | LN− | 187 (47.0) | 39 (9.8) | 0.003 | 38 (53.5) | 12 (16.9) | 0.001 | 149 (45.6) | 27 (8.3) | 0.037 |
| LN+ | 120 (30.2) | 52 (13.1) | 7 (9.9) | 14 (19.7) | 113 (34.6) | 38 (11.6) | ||||
| Lym_inv | Not | 169 (42.5) | 40 (10.1) | 0.073 | 29 (40.8) | 10 (14.1) | 0.030 | 140 (42.8) | 30 (9.2) | 0.332 |
| Yes | 138 (34.7) | 51 (12.8) | 16 (22.5) | 16 (22.5) | 122 (37.3) | 35 (10.7) | ||||
| EMR1-SC | Low | 278 (69.8) | 46 (11.6) | 0.000 | 34 (47.9) | 12 (16.9) | 0.020 | 245 (74.9) | 34 (10.4) | 0.000 |
| High | 29 (7.3) | 45 (11.3) | 11 (15.5) | 14 (19.7) | 17 (5.2) | 31 (9.5) | ||||
| CD163-SC | Low | 257 (64.7) | 57 (14.4) | 0.000 | 39 (54.9) | 12 (16.9) | 0.001 | 218 (66.9) | 45 (13.8) | 0.013 |
| High | 49 (12.3) | 34 (8.6) | 6 (8.5) | 14 (19.7) | 43 (13.2) | 20 (6.1) | ||||
| CD68 | Low | 204 (51.3) | 31 (7.8) | 0.000 | 19 (26.8) | 3 (4.2) | 0.008 | 185 (56.6) | 28 (8.6) | 0.000 |
| High | 103 (25.9) | 60 (15.1) | 26 (36.6) | 23 (32.4) | 77 (23.5) | 37 (11.3) | ||||
| Parameter | Total (MSI-H+MSS) | MSI-H | MSS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p Value | HR | 95% C.I. | p Value | HR | 95% C.I. | p Value | HR | 95% C.I. | ||||
| Lower | Upper | Lower | Upper | Lower | Upper | |||||||
| LNM | 0.001 | 2.295 | 1.377 | 3.825 | 0.002 | 10.183 | 2.344 | 44.244 | 0.021 | 1.983 | 1.111 | 3.540 |
| CD68 | 0.000 | 3.359 | 2.014 | 5.603 | 0.007 | 7.279 | 1.723 | 30.756 | 0.001 | 2.771 | 1.560 | 4.924 |
| CD163 | 0.001 | 2.575 | 1.456 | 4.556 | 0.045 | 4.163 | 1.029 | 16.838 | 0.034 | 2.045 | 1.054 | 3.969 |
| Variables | (EMR1-TC+CD68+CD163+) Total (MSI-H+MSS) | (EMR1-TC+CD68+CD163+) (MSI-H) | (EMR1-TC+CD68+CD163+) (MSS) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | p Value | N (%) | p Value | N (%) | p Value | |||||
| Low | High | Low | High | Low | High | |||||
| T_stage | Tis, T1, T2 | 58 (14.6) | 6 (1.5) | 0.422 | 4 (5.6) | 3 (4.2) | 0.110 | 54 (16.5) | 3 (0.9) | 0.114 |
| T3, T4 | 289 (72.6) | 45 (11.3) | 54 (76.1) | 10 (14.1) | 235 (71.9) | 35 (10.7) | ||||
| LNM | LN- | 207 (52.0) | 19 (4.8) | 0.004 | 45 (63.4) | 5 (7.0) | 0.015 | 162 (49.5) | 14 (4.3) | 0.037 |
| LN+ | 140 (35.2) | 32 (8.0) | 13 (18.3) | 8 (11.3) | 127 (38.8) | 24 (7.3) | ||||
| Lym_inv | Not | 192 (48.2) | 17 (4.3) | 0.004 | 37 (52.1) | 2 (2.8) | 0.002 | 155 (47.4) | 15 (4.6) | 0.121 |
| Yes | 155 (38.9) | 34 (8.5) | 21 (29.6) | 11 (15.5) | 134 (41.0) | 23 (7.0) | ||||
| Recurrence or not | Not | 311 (78.3) | 20 (5.0) | 0.000 | 53 (74.6) | 8 (11.3) | 0.014 | 258 (79.1) | 12 (3.7) | 0.000 |
| Yes | 35 (8.8) | 31 (7.8) | 5 (7.0) | 5 (7.0) | 30 (9.2) | 26 (8.0) | ||||
| Parameters | Recurrence Free Survival (RFS) | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||||||
| p-Value | HR | 95.0% CI | p-Value | HR | 95.0% CI | |||
| Lower | Upper | Lower | Upper | |||||
| T_Stage (l-ll vs lll-lV) | 0.008 | 14.776 | 2.050 | 106.515 | 0.023 | 9.981 | 1.365 | 73.015 |
| LNM (LN+ vs LN−) | 0.000 | 2.659 | 1.609 | 4.394 | 0.229 | 1.405 | 0.807 | 2.447 |
| Lymph_inv (No vs Yes) | 0.000 | 2.662 | 1.597 | 4.436 | 0.077 | 1.657 | 0.947 | 2.900 |
| EMR1-TC (Low vs High) | 0.843 | 0.940 | 0.512 | 1.728 | ||||
| EMR1-SC (Low vs High) | 0.446 | 0.769 | 0.392 | 1.509 | ||||
| CD68 (Low vs High) | 0.070 | 0.610 | 0.357 | 1.041 | ||||
| CD163-SC (Low vs High) | 0.241 | 0.678 | 0.354 | 1.298 | ||||
| Combined EMR1-TC+CD68+CD163+ (Low vs High) | 0.000 | 8.169 | 5.011 | 13.318 | 0.000 | 6.565 | 3.988 | 10.806 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akter, R.; Kim, K.; Kwon, H.Y.; Kim, Y.; Eom, Y.W.; Cho, H.-m.; Cho, M.-Y. EMR1/ADGRE1 Expression in Cancer Cells Upregulated by Tumor-Associated Macrophages Is Related to Poor Prognosis in Colorectal Cancer. Biomedicines 2022, 10, 3121. https://doi.org/10.3390/biomedicines10123121
Akter R, Kim K, Kwon HY, Kim Y, Eom YW, Cho H-m, Cho M-Y. EMR1/ADGRE1 Expression in Cancer Cells Upregulated by Tumor-Associated Macrophages Is Related to Poor Prognosis in Colorectal Cancer. Biomedicines. 2022; 10(12):3121. https://doi.org/10.3390/biomedicines10123121
Chicago/Turabian StyleAkter, Rokeya, Kwangmin Kim, Hye Youn Kwon, Youngwan Kim, Young Woo Eom, Hye-mi Cho, and Mee-Yon Cho. 2022. "EMR1/ADGRE1 Expression in Cancer Cells Upregulated by Tumor-Associated Macrophages Is Related to Poor Prognosis in Colorectal Cancer" Biomedicines 10, no. 12: 3121. https://doi.org/10.3390/biomedicines10123121
APA StyleAkter, R., Kim, K., Kwon, H. Y., Kim, Y., Eom, Y. W., Cho, H.-m., & Cho, M.-Y. (2022). EMR1/ADGRE1 Expression in Cancer Cells Upregulated by Tumor-Associated Macrophages Is Related to Poor Prognosis in Colorectal Cancer. Biomedicines, 10(12), 3121. https://doi.org/10.3390/biomedicines10123121






