Abstract
(1) Background: The main aim was to develop a prototype application that would serve as an open-source repository for a curated subset of predictive and prognostic models regarding oncology, and provide a user-friendly interface for the included models to allow online calculation. The focus of the application is on providing physicians and health professionals with patient-specific information regarding treatment plans, survival rates, and side effects for different expected treatments. (2) Methods: The primarily used models were the ones developed by our research group in the past. This selection was completed by a number of models, addressing the same cancer types but focusing on other outcomes that were selected based on a literature search in PubMed and Medline databases. All selected models were publicly available and had been validated TRIPOD (Transparent Reporting of studies on prediction models for Individual Prognosis Or Diagnosis) type 3 or 2b. (3) Results: The open source repository currently incorporates 18 models from different research groups, evaluated on datasets from different countries. Model types included logistic regression, Cox regression, and recursive partition analysis (decision trees). (4) Conclusions: An application was developed to enable physicians to complement their clinical judgment with user-friendly patient-specific predictions using models that have received internal/external validation. Additionally, this platform enables researchers to display their work, enhancing the use and exposure of their models.
1. Introduction
One of the most challenging areas of modern medicine is oncology [1]. The decision-making process for the best treatment plan is now more difficult than ever because of the inherent heterogeneity of cancer types, patients, and the ever-growing range of available treatments [2]. To choose the “optimal” course of treatment, clinicians must consider evidence from clinical trials, continuing research, their own professional expertise, national guidelines, and the values of the patient [3]. Predictive modeling is becoming a key knowledge-based tool in the healthcare field [4]. The popularity of predictive modeling arises from advancements in a variety of areas, including the availability of health data from electronic health records and databases, a better understanding of causal or statistical predictors of health, disease processes, and multifactorial models of poor health, and advances in nonlinear computer models based on artificial intelligence or neural networks [5]. These new computer-based modeling techniques are gaining credibility in therapeutic settings [6]. However, the current understanding of how so-called machine intelligence will evolve, and thus how current relatively sophisticated predictive models will evolve in response to advances in technology, is difficult to predict [7]. What is known is that despite the fear of the black-box nature of certain AI models, with the introduction of various nomograms dealing with predictive models in oncology, their simplicity and ease of use is helping to cross the bridge of academic/research use to clinical adoption [8,9,10].
There was a need for a public repository of predictive models, and a thorough search was conducted to identify existing applications. However, as the search did not yield useful results, it became our goal to develop a web-based application that would compile and archive publicly available, validated models [11]. The doctors who use such an application would be able to quickly assess the benefits and drawbacks of the chosen models that passed the quality controls. They could securely rely on the application to calculate the output of such models by providing the inputs in a specific wizard rather than designing their own implementation or researching for an appropriate implementation on their own.
The advantages for medical researchers include enhancing the exposure of their model, which should encourage usage and citations, and assisting them in generalizing their models by enabling the models to be evaluated by research groups other than the ones that generated them TRIPOD 4 (Transparent Reporting of studies on prediction models for Individual Prognosis Or Diagnosis) [12]. This paper will describe the foundational work that led to the creation of this application prototype.
2. Materials and Methods
The National Center for Biotechnology Information’s (NCBI) PubMed database and Medline papers from December 2016 to March 2022 were reviewed. The following search terms were utilized to find publications that were pertinent: “validated prognostic models in oncology”, “novel diagnostic tools for cancer with external validation”, “lung, prostate, head and neck predictive models”, and “machine-learning & deep learning models in oncology”. Additionally, all the models included were validated (TRIPOD type 3 or 2b) or open-source repositories or peer-reviewed articles. Since this is not a systematic review, no more than five models for each disease (brain, head and neck, lung, prostate, esophagus, rectum, and endometrium) were selected and translated onto a user interface. Figure 1 illustrates the steps taken from the stage of doing a literature search to that of publishing the results online.
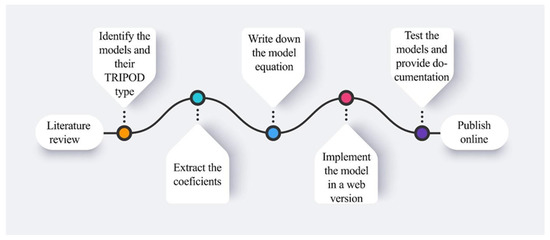
Figure 1.
The pipeline from selecting the convenient models until the publishing phase.
In order to assess the category of the models from the studies, every paper was tested for its compliance with the TRIPOD classification, as shown in Figure 2.

Figure 2.
TRIPOD classifications [13].
The process for extracting the coefficients from a nomogram and implementing them into a web user interface will be described in the following paragraphs.
2.1. Obtaining Model Coefficients from a Nomogram
A large number of predictive models with respect to treatment outcomes (cure rate, cancer recurrence, survival, toxicity) are available in the literature. Regression models are frequently published as nomograms in order to make them easier for medical specialists to read and interpret, often without disclosing the model coefficients. A straightforward technique was utilized to extract the coefficients from nomograms in order to publish the models uniformly on the application. The papers in which nomograms are published will usually report the type of model (e.g., logistic regression); otherwise, this can be determined by examining the part of the nomogram where the total score is mapped to a probability. In the nomogram, numbers of points are assigned to different inputs, for example, female might have more points than male. These points are summed over all the inputs, resulting in a nomogram score. This score is then related to a probability of an outcome. The relationship between the total score and the probability is described by the equation for the model. To obtain an equation describing the nomogram, the total score of the nomogram and the probability are read by digitizing the nomogram, and a linear fit is made using the function describing the model, e.g., Equation (1):
where is the probability, is the intercept, and is the scaling factor. Once the relationship is determined between the model parameters and the total score, the coefficients can be calculated. In order to be as precise as possible when reading the nomograms, any graph digitalization software can be used. An example from one of the utilized models is used to illustrate this approach, and it is represented in Figure 3 below [3].
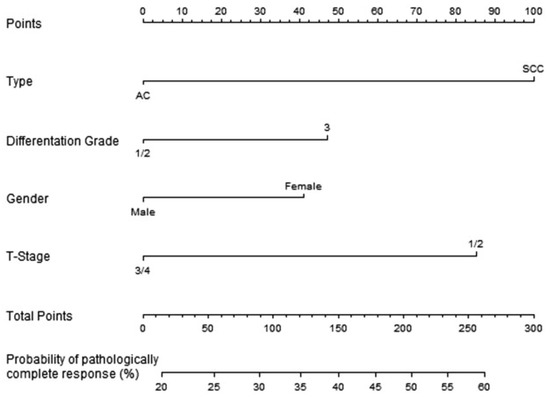
Figure 3.
Nomogram for predicting pathologically complete response after neoadjuvant chemoradiotherapy for oesophageal cancer [14].
In this instance, a logistic regression model is used to determine the link between the coefficients and the likelihood; as a result, the association between the nomogram score (total points) and the standard logistic distribution of the probability is linear, from which the slope and intercept can be determined (Figure 4).
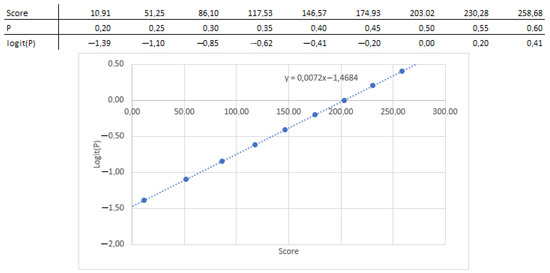
Figure 4.
Coefficients and intercept extracted from the nomogram.
Next, the relationship between the parameters and the nomogram score can be extracted by reading the nomogram, and the coefficient can be obtained by multiplying by the scaling factor (0.307), as shown in Table 1.

Table 1.
Point reading for the nomogram.
The probability can now be obtained through Equations (2) and (3):
where to are 0 or 1 (see Table 1). It is possible to perform a check on how well this equation describes the nomogram by filling in some values into the equation and reading the nomogram to test if the answers are the same. For the example above, you could enter a patient with squamous cell carcinoma, differentiation grade 2, male gender, and a tumor with T-stage 2. Totaling the points (100, 0, 0, and 85) results in a nomogram score of 185, which correlates to a probability of just over 45% when reading the nomogram. Filling this in the equations mentioned above () results in . The exact performance of the equation is often influenced by rounding factors or reading errors.
2.2. Application Development Process
In order to automate the self-import process of data models and to create a central model repository, a web-based application has been created that can be accessed by researchers from around the world, as shown in Figure 5.
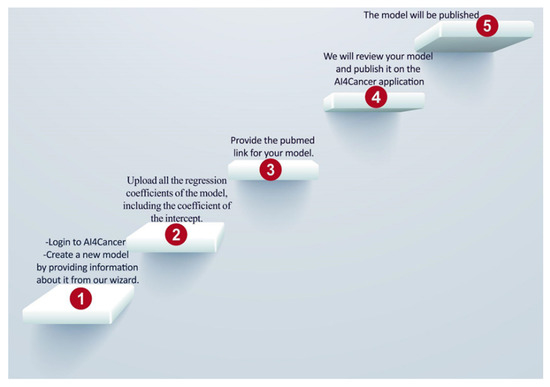
Figure 5.
Shows the steps of user-uploading models in the application.
The development of the web application was done using Python 3.6. and the Django v3.1 web framework. One of the main reasons for choosing Python as the main programming language for this application is because it would allow easier integration of existing research that is based on Python. Furthermore, Django is a highly capable web framework that provides a rich feature set for easy implementation and management of a web app, such as built-in support for a database and an admin console.
For the back-end of the application, a PostgresSQL database was used to store the details related to the model, such as the author of the model, intended end users, and how the model has been developed. In addition, there are stored the parameters (i.e., age, volume, sphere diameter, type of treatment, stage) of each model to dynamically generate the UI in order to facilitate the testing of the model. For the development of the front end of the web application, technologies such as Javascript, Bootstrap 3, HTML5, and CSS3 were used alongside the Django web framework, as shown in Figure 6. The materials and supplements contain comprehensive information about the database (Figure S1).
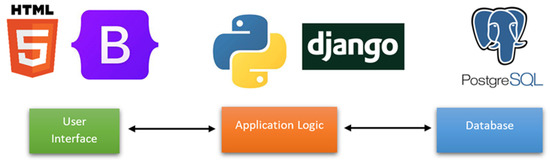
Figure 6.
Illustration of the application architecture.
AI4Cancer is intended for the open-source publishing of prediction models, developed to predict outcomes for cancer patients.
3. Results
The developed open-source application is accessible online at https://ai4cancer-ai.herokuapp.com/models-browser# (accessed on 1 April 2022). As shown in Figure 7, it will serve as a repository for published AI prediction models that address all aspects of various cancer types and stages, including diagnosis, prognosis (patient therapy, risk stratification), and follow-up (treatment result, consequences).
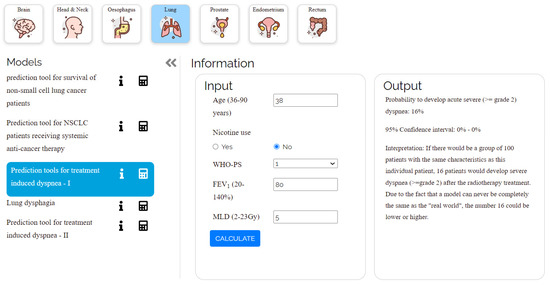
Figure 7.
Shows the user interface of a model (lung cancer) from the website.
Every model that is displayed has an explanation of the technique and clinical datasets that were used to create and validate the model. The limitations of each model are explicitly mentioned. The developed models are lung cancer, rectum, head and neck cancer, and brain metastases, and they are developed to be used only by physicians, not patients, due to the complexity of cancer treatment decision-making options. There are several different types of models that are used, including linear regression, tumor control probability (TCP), linear-quadratic (LQ), Kaplan–Meier, linear-quadratic biologically effective dose (LQ-BED), Cox, and logistic regression [15,16]. As shown in Table 2, there are now 18 models that have been released and deployed.

Table 2.
For every model: cancer type, input features, outcome, cohort type and TRIPOD, and model type.
For each online model given, doctors can find: (a) the intended use (predicted outcome) of the model, (b) to which patients it applies, (c) the information and the parameters that the doctor must enter, and (d) how the model was developed. The doctors can access the website, select a suitable model, and enter the required information to produce the likelihood of survival. The website’s predictive models follow the same calculations as the models outlined in the academic works on which they are based.
The primary output of this research is a widely applicable application that comprises verified models for various stages, odds of survival, symptoms, and outcomes. The collection of these predictive models will help doctors make decisions.
4. Discussion
This application can be considered a “model zoo” for academics and medical professionals who are well-versed in the medical ramifications of various cancer forms. None of the models are intended to replace clinical judgment; rather, they are all intended to advance research. The open-source application is not meant for usage by laypeople without assistance (e.g., patients). It is important to emphasize that this manuscript and application are currently only prototypes. The inclusion of all models that satisfy the selection criteria is not claimed. Similarly, any models that are not available on the platform right now should not be viewed as a cause for concern or as a rejection of their validity.
For heightened patient privacy, models that require medical (DICOM) images are not incorporated, as the pipeline does not currently have the ability to check that such uploaded data was properly anonymized. By working together with outside organizations (such as hospitals) that are in charge of preserving the privacy of such data, such machine learning models will be incorporated. In future iterations of the website, it will also be possible to categorize models according to the outcome they are modeling, as well as divide models according to the kind of input data they need. This will result in a more structured semantic-based organization. When the division of models was done based on the result being modeled, there will be dedicated website pages that compare models created to predict a specific outcome.
The models have not been validated for individual use; but rather, only in clinical cohort research. These models should not be used by patients directly and should only be used by doctors who are knowledgeable about the complexity of cancer types. The main intention of these models is to inform doctors and they should not be used for decision support.
Researchers should view this work as an invitation to use this application, with the ability to hide the code from the user while still providing full functionality. Researchers’ models will be successfully integrated with the aid of the application itself. This will generate synergies that will inevitably speed up oncology-related AI research. Additionally, it will prevent models from being unnoticed and underutilized, which frequently occurs when numerous publications on the same general topic are released quickly.
There are certain limitations to the technique employed to extract regression model coefficients from a nomogram. One limitation is that the resolution of the published model significantly impacts accuracy. Another limitation is that even though the model coefficients may be retrieved, a nomogram cannot be used to determine the standard error for the parameter coefficients [48]. However, the technique may be employed with any nomogram, making it a useful tool.
By encouraging more researchers to submit their work via this application, hopefully, the number of curated models will increase and the application can continue to serve the public. This will be accomplished as a result of an increase in staff via the Optima Grant, who will perform monthly checks for the new models. The three prerequisites are (1) open-access papers with a clear, implementable model description; (2) papers with a minimum of 10 citations; (3) models of TRIPOD 3 and 4.
5. Conclusions
The repository accessible at https://ai4cancer-ai.herokuapp.com (accessed on 1 April 2022), in the current prototype level, contains 18 proven machine learning models to assist doctors in making decisions on patient care for a variety of cancer types. Other researchers can make use of this technique for deriving regression coefficients from nomograms. As a result, research teams are being urged to disseminate their models globally.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10112679/s1, Figure S1: Above displayed is the ER Diagram for the Database of the Ai4Cancer web application.
Author Contributions
Conceptualization, I.H. and P.L.; methodology, I.H., Y.v.W. and C.O.; software, P.G. and I.H.; validation, C.O., A.C. and E.L.; formal analysis, Y.v.W., C.O. and A.C.; investigation, I.H. and S.P.; data curation, Y.v.W., C.O.; writing—original draft preparation, I.H., N.M.R., A.W., A.C., C.O. and Y.v.W.; writing—review and editing, Y.W., N.M.R., A.W., A.C. and P.L.; visualization, I.H. and P.G.; supervision, P.L., A.W. and C.O.; project administration, A.W.; funding acquisition, P.L. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge financial support from the European Commission’s Horizon 2020 research and innovation programme under grant agreement MSCA-ITN-PREDICT No. 766276 and IMI-OPTIMA No. 101034347.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge financial support from the European Commission’s Horizon 2020 research.
Conflicts of Interest
Philippe Lambin reports, within and outside the submitted work, grants/sponsored research agreements from Varian medical, Oncoradiomics, ptTheragnostic/DNAmito, and Health Innovation Ventures. He received an advisor/presenter fee and/or reimbursement of travel costs/external grant writing fee and/or in-kind manpower contribution from Oncoradiomics, BHV, Merck, Varian, Elekta, ptTheragnostic, and Convert pharmaceuticals. Lambin has shares in the company Oncoradiomics SA, Convert pharmaceuticals SA, and The Medical Cloud Company SPRL and is co-inventor of two issued patents with royalties on radiomics (PCT/NL2014/050248, PCT/NL2014/050728) licensed to Oncoradiomics; one issue patent on mtDNA (PCT/EP2014/059089) licensed to ptTheragnostic/DNAmito; three non-patented inventions (software) licensed to ptTheragnostic/DNAmito, Oncoradiomics and Health Innovation Ventures; three non-issued, non-licensed patents on Deep Learning-Radiomics and LSRT (N2024482, N2024889, N2024889). He confirms that none of the above entities or funding were involved in the preparation of this paper.
References
- Craddock, M.; Crockett, C.; McWilliam, A.; Price, G.; Sperrin, M.; van der Veer, S.N.; Faivre-Finn, C. Evaluation of Prognostic and Predictive Models in the Oncology Clinic. Clin. Oncol. 2022, 34, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Chua, I.S.; Gaziel-Yablowitz, M.; Korach, Z.T.; Kehl, K.L.; Levitan, N.A.; Arriaga, Y.E.; Jackson, G.P.; Bates, D.W.; Hassett, M. Artificial intelligence in oncology: Path to implementation. Cancer Med. 2021, 10, 4138–4149. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, R.M.; Karelaia, N. Heuristic and linear models of judgment: Matching rules and environments. Psychol. Rev. 2007, 114, 733–758. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Prosperi, M.; Guo, Y.; Sperrin, M.; Koopman, J.S.; Min, J.S.; He, X.; Rich, S.; Wang, M.; Buchan, I.E.; Bian, J. Causal inference and counterfactual prediction in machine learning for actionable healthcare. Nat. Mach. Intell. 2020, 2, 369–375. [Google Scholar] [CrossRef]
- Wiens, J.; Saria, S.; Sendak, M.; Ghassemi, M.; Liu, V.X.; Doshi-Velez, F.; Jung, K.; Heller, K.; Kale, D.; Saeed, M.; et al. Do no harm: A roadmap for responsible machine learning for health care. Nat. Med. 2019, 25, 1337–1340. [Google Scholar] [CrossRef]
- Dash, S.; Acharya, B.R.; Mittal, M.; Abraham, A.; Kelemen, A. Deep Learning Techniques for Biomedical and Health Informatics; Springer Nature: London, UK, 2019. [Google Scholar]
- Faruqui, N.; Yousuf, M.A.; Whaiduzzaman, M.; Azad, A.K.M.; Barros, A.; Moni, M.A. LungNet: A hybrid deep-CNN model for lung cancer diagnosis using CT and wearable sensor-based medical IoT data. Comput. Biol. Med. 2021, 139, 104961. [Google Scholar] [CrossRef]
- Aurna, N.F.; Yousuf, M.A.; Taher, K.A.; Azad, A.K.M.; Moni, M.A. A classification of MRI brain tumor based on two stage feature level ensemble of deep CNN models. Comput. Biol. Med. 2022, 146, 105539. [Google Scholar] [CrossRef]
- He, B.; Chen, W.; Liu, L.; Hou, Z.; Zhu, H.; Cheng, H.; Zhang, Y.; Zhan, S.; Wang, S. Prediction Models for Prognosis of Cervical Cancer: Systematic Review and Critical Appraisal. Front. Public Health 2021, 9, 654454. [Google Scholar] [CrossRef]
- Dhiman, P.; Ma, J.; Navarro, C.A.; Speich, B.; Bullock, G.; Damen, J.A.; Kirtley, S.; Hooft, L.; Riley, R.D.; Van Calster, B.; et al. Reporting of prognostic clinical prediction models based on machine learning methods in oncology needs to be improved. J. Clin. Epidemiol. 2021, 138, 60–72. [Google Scholar] [CrossRef]
- van Wijk, Y.; Ramaekers, B.; Vanneste, B.G.L.; Halilaj, I.; Oberije, C.; Chatterjee, A.; Marcelissen, T.; Jochems, A.; Woodruff, H.C.; Lambin, P. Modeling-Based Decision Support System for Radical Prostatectomy Versus External Beam Radiotherapy for Prostate Cancer Incorporating an Clinical Trial and a Cost-Utility Study. Cancers 2021, 13, 2687. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M.; members of the TRIPOD group. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. Eur. Urol. 2015, 67, 1142–1151. [Google Scholar] [CrossRef]
- Toxopeus, E.L.A.; Nieboer, D.; Shapiro, J.; Biermann, K.; van der Gaast, A.; van Rij, C.M.; Steyerberg, E.W.; van Lanschot, J.J.B.; Wijnhoven, B.P.L. Nomogram for predicting pathologically complete response after neoadjuvant chemoradiotherapy for oesophageal cancer. Radiother. Oncol. 2015, 115, 392–398. [Google Scholar] [CrossRef]
- Cox, D.R.; Cox, D. Selected Statistical Papers of Sir David Cox: Volume 1, Design of Investigations, Statistical Methods and Applications; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Bonnin, R. Machine Learning for Developers: Uplift Your Regular Applications with the Power of Statistics, Analytics, and Machine Learning; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- Hendriks, L.; Keek, S.A.; Chatterjee, A.; Belderbos, J.; Bootsma, G.; van den Borne, B.; Dingemans, A.-M.C.; Gietema, H.; Groen, H.J.M.; Herder, G.; et al. 127P Does radiomics have added value in predicting the development of brain metastases in patients with radically treated stage III non-small cell lung cancer (NSCLC)? Ann. Oncol. 2022, 33, S91. [Google Scholar] [CrossRef]
- Rodrigues, G.; Warner, A.; Zindler, J.; Slotman, B.; Lagerwaard, F. A clinical nomogram and recursive partitioning analysis to determine the risk of regional failure after radiosurgery alone for brain metastases. Radiother. Oncol. 2014, 111, 52–58. [Google Scholar] [CrossRef]
- Molenaar, R.; Wiggenraad, R.; Verbeek-de Kanter, A.; Walchenbach, R.; Vecht, C. Relationship between volume, dose and local control in stereotactic radiosurgery of brain metastasis. Br. J. Neurosurg. 2009, 23, 170–178. [Google Scholar] [CrossRef]
- Chang, E.L.; Hassenbusch, S.J., 3rd; Shiu, A.S.; Lang, F.F.; Allen, P.K.; Sawaya, R.; Maor, M.H. The role of tumor size in the radiosurgical management of patients with ambiguous brain metastases. Neurosurgery 2003, 53, 272–280, discussion 280–281. [Google Scholar] [CrossRef]
- Chang, E.L.; Selek, U.; Hassenbusch SJ 3rd Maor, M.H.; Allen, P.K.; Mahajan, A.; Sawaya, R.; Woo, S.Y. Outcome variation among “radioresistant” brain metastases treated with stereotactic radiosurgery. Neurosurgery 2005, 56, 936–945, discussion 936–945. [Google Scholar]
- Chao, S.T.; Barnett, G.H.; Vogelbaum, M.A.; Angelov, L.; Weil, R.J.; Neyman, G.; Reuther, A.M.; Suh, J.H. Salvage stereotactic radiosurgery effectively treats recurrences from whole-brain radiation therapy. Cancer 2008, 113, 2198–2204. [Google Scholar] [CrossRef]
- Ernst-Stecken, A.; Ganslandt, O.; Lambrecht, U.; Sauer, R.; Grabenbauer, G. Phase II trial of hypofractionated stereotactic radiotherapy for brain metastases: Results and toxicity. Radiother. Oncol. 2006, 81, 18–24. [Google Scholar] [CrossRef]
- Higuchi, Y.; Serizawa, T.; Nagano, O.; Matsuda, S.; Ono, J.; Sato, M.; Iwadate, Y.; Saeki, N. Three-staged stereotactic radiotherapy without whole brain irradiation for large metastatic brain tumors. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1543–1548. [Google Scholar] [CrossRef]
- Lutterbach, J.; Cyron, D.; Henne, K.; Ostertag, C.B. Radiosurgery followed by planned observation in patients with one to three brain metastases. Neurosurgery 2008, 62 (Suppl. 2), 776–784. [Google Scholar] [CrossRef]
- Matsuo, T.; Shibata, S.; Yasunaga, A.; Iwanaga, M.; Mori, K.; Shimizu, T.; Hayashi, N.; Ochi, M.; Hayashi, K. Dose optimization and indication of Linac radiosurgery for brain metastases. Int. J. Radiat. Oncol. Biol. Phys. 1999, 45, 931–939. [Google Scholar] [CrossRef]
- Narayana, A.; Chang, J.; Yenice, K.; Chan, K.; Lymberis, S.; Brennan, C.; Gutin, P.H. Hypofractionated stereotactic radiotherapy using intensity-modulated radiotherapy in patients with one or two brain metastases. Stereotact. Funct. Neurosurg. 2007, 85, 82–87. [Google Scholar] [CrossRef]
- Saitoh, J.-I.; Saito, Y.; Kazumoto, T.; Kudo, S.; Ichikawa, A.; Hayase, N.; Kazumoto, K.; Sakai, H.; Shibuya, K. Therapeutic effect of linac-based stereotactic radiotherapy with a micro-multileaf collimator for the treatment of patients with brain metastases from lung cancer. Jpn. J. Clin. Oncol. 2010, 40, 119–124. [Google Scholar] [CrossRef][Green Version]
- Vogelbaum, M.A.; Angelov, L.; Lee, S.-Y.; Li, L.; Barnett, G.H.; Suh, J.H. Local control of brain metastases by stereotactic radiosurgery in relation to dose to the tumor margin. J. Neurosurg. 2006, 104, 907–912. [Google Scholar] [CrossRef]
- Wiggenraad, R.; Verbeek-de Kanter, A.; Kal, H.B.; Taphoorn, M.; Vissers, T.; Struikmans, H. Dose-effect relation in stereotactic radiotherapy for brain metastases. A systematic review. Radiother. Oncol. 2011, 98, 292–297. [Google Scholar] [CrossRef]
- Zindler, J.D.; Jochems, A.; Lagerwaard, F.J.; Beumer, R.; Troost, E.G.C.; Eekers, D.B.P.; Compter, I.; van der Toorn, P.-P.; Essers, M.; Oei, B.; et al. Individualized early death and long-term survival prediction after stereotactic radiosurgery for brain metastases of non-small cell lung cancer: Two externally validated nomograms. Radiother. Oncol. 2017, 123, 189–194. [Google Scholar] [CrossRef]
- Thor, M.; Owosho, A.A.; Clark, H.D.; Oh, J.H.; Riaz, N.; Hovan, A.; Tsai, J.; Thomas, S.D.; Yom, S.H.K.; Wu, J.S.; et al. Internal and external generalizability of temporal dose-response relationships for xerostomia following IMRT for head and neck cancer. Radiother. Oncol. 2017, 122, 200–206. [Google Scholar] [CrossRef]
- Ramaekers, B.L.T.; Grutters, J.P.C.; Pijls-Johannesma, M.; Lambin, P.; Joore, M.A.; Langendijk, J.A. Protons in head-and-neck cancer: Bridging the gap of evidence. Int. J. Radiat. Oncol. Biol. Phys. 2013, 85, 1282–1288. [Google Scholar] [CrossRef] [PubMed]
- Ramaekers, B.L.T.; Joore, M.A.; Grutters, J.P.C.; van den Ende, P.; de Jong, J.; Houben, R.; Lambin, P.; Christianen, M.; Beetz, I.; Pijls-Johannesma, M.; et al. The impact of late treatment-toxicity on generic health-related quality of life in head and neck cancer patients after radiotherapy. Oral Oncol. 2011, 47, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Ramaekers, B.L.T.; Pijls-Johannesma, M.; Joore, M.A.; van den Ende, P.; Langendijk, J.A.; Lambin, P.; Kessels, A.G.H.; Grutters, J.P.C. Systematic review and meta-analysis of radiotherapy in various head and neck cancers: Comparing photons, carbon-ions and protons. Cancer Treat. Rev. 2011, 37, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Beetz, I.; Schilstra, C.; van der Schaaf, A.; van den Heuvel, E.R.; Doornaert, P.; van Luijk, P.; Vissink, A.; van der Laan, B.F.A.M.; Leemans, C.R.; Bijl, H.P.; et al. NTCP models for patient-rated xerostomia and sticky saliva after treatment with intensity modulated radiotherapy for head and neck cancer: The role of dosimetric and clinical factors. Radiother. Oncol. 2012, 105, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Wopken, K.; Bijl, H.P.; van der Schaaf, A.; van der Laan, H.P.; Chouvalova, O.; Steenbakkers, R.J.H.M.; Doornaert, P.; Slotman, B.J.; Oosting, S.F.; Christianen, M.E.M.C.; et al. Development of a multivariable normal tissue complication probability (NTCP) model for tube feeding dependence after curative radiotherapy/chemo-radiotherapy in head and neck cancer. Radiother. Oncol. 2014, 113, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Dehing-Oberije, C.; De Ruysscher, D.; van der Weide, H.; Hochstenbag, M.; Bootsma, G.; Geraedts, W.; Pitz, C.; Simons, J.; Teule, J.; Rahmy, A.; et al. Tumor volume combined with number of positive lymph node stations is a more important prognostic factor than TNM stage for survival of non-small-cell lung cancer patients treated with (chemo)radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 1039–1044. [Google Scholar] [CrossRef]
- Wallington, M.; Saxon, E.B.; Bomb, M.; Smittenaar, R.; Wickenden, M.; McPhail, S.; Rashbass, J.; Chao, D.; Dewar, J.; Talbot, D.; et al. 30-day mortality after systemic anticancer treatment for breast and lung cancer in England: A population-based, observational study. Lancet Oncol. 2016, 17, 1203–1216. [Google Scholar] [CrossRef]
- Dehing-Oberije, C.; De Ruysscher, D.; van Baardwijk, A.; Yu, S.; Rao, B.; Lambin, P. The importance of patient characteristics for the prediction of radiation-induced lung toxicity. Radiother. Oncol. 2009, 91, 421–426. [Google Scholar] [CrossRef]
- Oberije, C.; Liao, Z.; De Ruysscher, D.; Tucker, S.; Lambin, P. Development and External Validation of a Model for Prediction of Radiation-Induced Dyspnea: An Approach combining Clinical Data with Information from Literature. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, S528. [Google Scholar] [CrossRef]
- Dehing-Oberije, C.; De Ruysscher, D.; Petit, S.; Van Meerbeeck, J.; Vandecasteele, K.; De Neve, W.; Dingemans, A.M.C.; El Naqa, I.; Deasy, J.; Bradley, J.; et al. Development, external validation and clinical usefulness of a practical prediction model for radiation-induced dysphagia in lung cancer patients. Radiother. Oncol. 2010, 97, 455–461. [Google Scholar] [CrossRef]
- Nalbantov, G.; Kietselaer, B.; Vandecasteele, K.; Oberije, C.; Berbee, M.; Troost, E.; Dingemans, A.-E.; van Baardwijk, A.; Smits, K.; Dekker, A.; et al. Cardiac comorbidity is an independent risk factor for radiation-induced lung toxicity in lung cancer patients. Radiother. Oncol. 2013, 109, 100–106. [Google Scholar] [CrossRef]
- Walsh, S.; van der Putten, W. A TCP model for external beam treatment of intermediate-risk prostate cancer. Med. Phys. 2013, 40, 031709. [Google Scholar] [CrossRef]
- Walsh, S.; Roelofs, E.; Kuess, P.; Lambin, P.; Jones, B.; Georg, D.; Verhaegen, F. A validated tumor control probability model based on a meta-analysis of low, intermediate, and high-risk prostate cancer patients treated by photon, proton, or carbon-ion radiotherapy. Med. Phys. 2016, 43, 734–747, Erratum in Med. Phys. 2016, 43, 5263. [Google Scholar] [CrossRef]
- Creutzberg, C.L.; van Stiphout, R.G.P.M.; Nout, R.A.; Lutgens, L.C.H.; Jürgenliemk-Schulz, I.M.; Jobsen, J.J.; Smit, V.T.H.B.; Lambin, P. Nomograms for Prediction of Outcome With or Without Adjuvant Radiation Therapy for Patients With Endometrial Cancer: A Pooled Analysis of PORTEC-1 and PORTEC-2 Trials. Int. J. Radiat. Oncol. Biol. Phys. 2015, 91, 530–539. [Google Scholar] [CrossRef]
- van Stiphout, R.G.P.M.; Lammering, G.; Buijsen, J.; Janssen, M.H.M.; Gambacorta, M.A.; Slagmolen, P.; Lambrecht, M.; Rubello, D.; Gava, M.; Giordano, A.; et al. Development and external validation of a predictive model for pathological complete response of rectal cancer patients including sequential PET-CT imaging. Radiother. Oncol. 2011, 98, 126–133. [Google Scholar] [CrossRef]
- Halilaj, I.; Chatterjee, A.; van Wijk, Y.; Wu, G.; van Eeckhout, B.; Oberije, C.; Lambin, P. Covid19Risk.ai: An open source repository and online calculator of prediction models for early diagnosis and prognosis of Covid-19. BioMed 2021, 1, 41–49. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).