Local Flexibility of a New Single-Ring Chaperonin Encoded by Bacteriophage AR9 Bacillus subtilis
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Purification
2.2. Cryo-EM Data Acquisition
2.3. Image Processing
2.4. Molecular Modeling
2.5. Small-Angle X-ray Scattering
3. Results
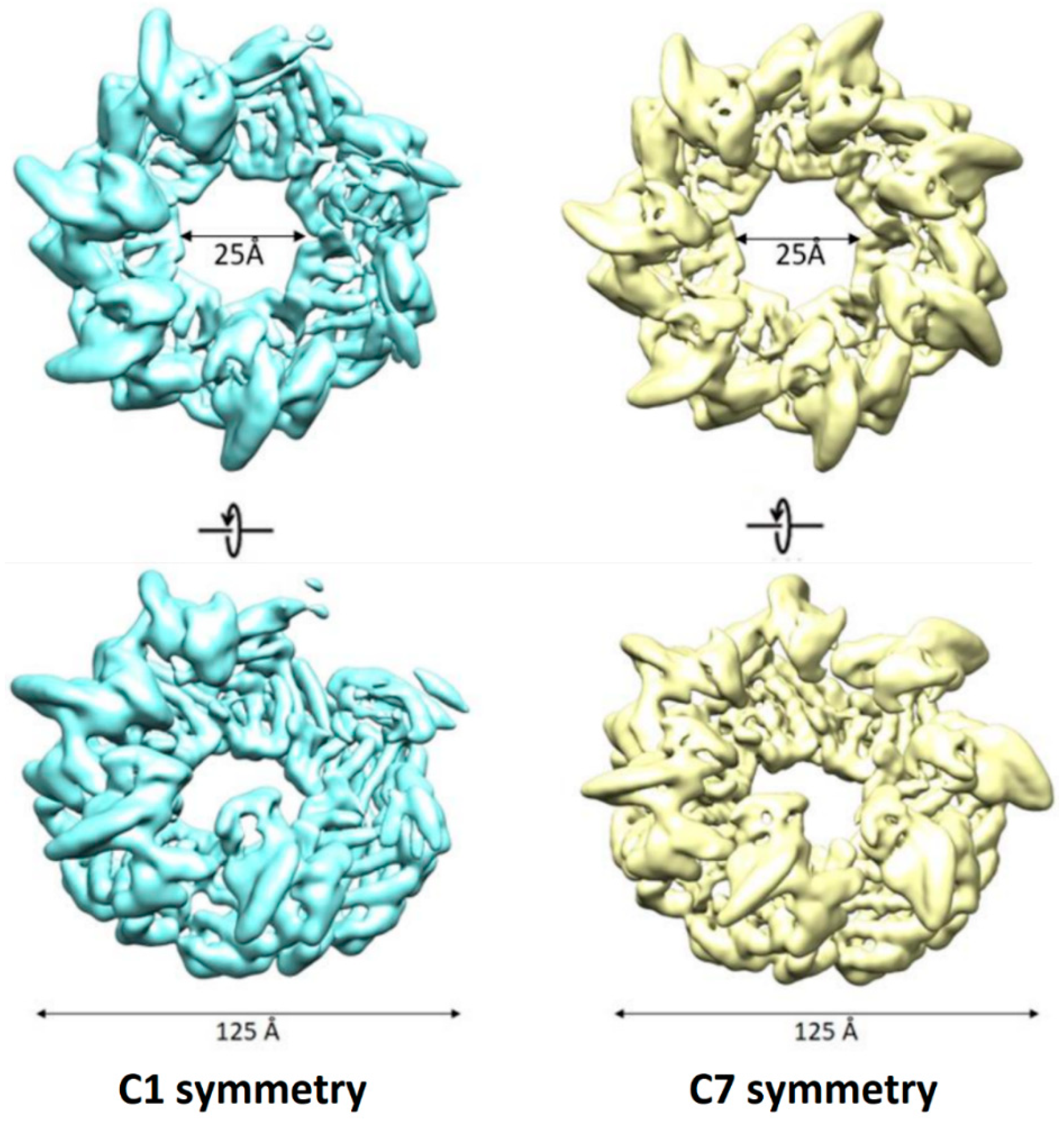
3.1. Apo-Form of AR9 Chaperonin Has a Flexible 3D Structure
3.2. Full-Atom Model of the Apo-Form of the AR9 Chaperonin Demonstrates More Stability than That of the OBP Chaperonin
3.3. Conformational Variability among the Subunits
3.4. Apo-AR9 Cryo-EM Structure Agrees with Solution Scattering Data
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vilasi, S.; Bulone, D.; Bavisotto, C.C.; Campanella, C.; Gammazza, A.M.; San Biagio, P.L.; Cappello, F.; de Macario, E.C.; Macario, A.J.L. Chaperonin of Group I: Oligomeric Spectrum and Biochemical and Biological Implications. Front. Mol. Biosci. 2018, 4, 99. [Google Scholar] [CrossRef]
- Horwich, A.L.; Fenton, W.A. Chaperonin-Assisted Protein Folding: A Chronologue. Q. Rev. Biophys. 2020, 53, e4. [Google Scholar] [CrossRef]
- Lopez, T.; Dalton, K.; Frydman, J. The Mechanism and Function of Group II Chaperonins. J. Mol. Biol. 2015, 427, 2919–2930. [Google Scholar] [CrossRef] [PubMed]
- Hertveldt, K.; Lavigne, R.; Pleteneva, E.; Sernova, N.; Kurochkina, L.; Korchevskii, R.; Robben, J.; Mesyanzhinov, V.; Krylov, V.N.; Volckaert, G. Genome Comparison of Pseudomonas Aeruginosa Large Phages. J. Mol. Biol. 2005, 354, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Kiljunen, S.; Hakala, K.; Pinta, E.; Huttunen, S.; Pluta, P.; Gador, A.; Lönnberg, H.; Skurnik, M. Yersiniophage ΦR1-37 Is a Tailed Bacteriophage Having a 270 Kb DNA Genome with Thymidine Replaced by Deoxyuridine. Microbiology 2005, 151, 4093–4102. [Google Scholar] [CrossRef]
- Cornelissen, A.; Hardies, S.C.; Shaburova, O.V.; Krylov, V.N.; Mattheus, W.; Kropinski, A.M.; Lavigne, R. Complete Genome Sequence of the Giant Virus OBP and Comparative Genome Analysis of the Diverse KZ-Related Phages. J. Virol. 2012, 86, 1844–1852. [Google Scholar] [CrossRef]
- Jang, H.B.; Fagutao, F.F.; Nho, S.W.; Park, S.B.; Cha, I.S.; Yu, J.E.; Lee, J.S.; Im, S.P.; Aoki, T.; Jung, T.S. Phylogenomic Network and Comparative Genomics Reveal a Diverged Member of the ΦKZ-Related Group, Marine Vibrio Phage ΦJM-2012. J. Virol. 2013, 87, 12866–12878. [Google Scholar] [CrossRef] [PubMed]
- Lavysh, D.; Sokolova, M.; Minakhin, L.; Yakunina, M.; Artamonova, T.; Kozyavkin, S.; Makarova, K.S.; Koonin, E.V.; Severinov, K. The Genome of AR9, a Giant Transducing Bacillus Phage Encoding Two Multisubunit RNA Polymerases. Virology 2016, 495, 185–196. [Google Scholar] [CrossRef]
- Georgopoulos, C.P.; Hendrix, R.W.; Casjens, S.R.; Kaiser, A.D. Host Participation in Bacteriophage Lambda Head Assembly. J. Mol. Biol. 1973, 76, 45–60. [Google Scholar] [CrossRef]
- van der Vies, S.M.; Gatenby, A.A.; Georgopoulos, C. Bacteriophage T4 Encodes a Co-Chaperonin That Can Substitute for Escherichia Coli GroES in Protein Folding. Nature 1994, 368, 654–656. [Google Scholar] [CrossRef]
- Ang, D.; Richardson, A.; Mayer, M.P.; Keppel, F.; Krisch, H.; Georgopoulos, C. Pseudo-T-Even Bacteriophage RB49 Encodes CocO, a Cochaperonin for GroEL, Which Can Substitute for Escherichia Coli’s GroES and Bacteriophage T4′s Gp31. J. Biol. Chem. 2001, 276, 8720–8726. [Google Scholar] [CrossRef] [PubMed]
- Kurochkina, L.P.; Semenyuk, P.I.; Orlov, V.N.; Robben, J.; Sykilinda, N.N.; Mesyanzhinov, V.V. Expression and Functional Characterization of the First Bacteriophage-Encoded Chaperonin. J. Virol. 2012, 86, 10103–10111. [Google Scholar] [CrossRef] [PubMed]
- Semenyuk, P.I.; Orlov, V.N.; Sokolova, O.S.; Kurochkina, L.P. New GroEL-like Chaperonin of Bacteriophage OBP P. Fluorescens Suppresses Thermal Protein Aggregation in an ATP-Dependent Manner. Biochem. J. 2016, 473, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Bartolucci, C.; Lamba, D.; Grazulis, S.; Manakova, E.; Heumann, H. Crystal Structure of Wild-Type Chaperonin GroEL. J. Mol. Biol. 2005, 354, 940–951. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stanishneva-Konovalova, T.B.; Semenyuk, P.I.; Kurochkina, L.P.; Pichkur, E.B.; Vasilyev, A.L.; Kovalchuk, M.V.; Kirpichnikov, M.P.; Sokolova, O.S. Cryo-EM Reveals an Asymmetry in a Novel Single-Ring Viral Chaperonin. J. Struct. Biol. 2020, 209, 107439. [Google Scholar] [CrossRef]
- Bracher, A.; Paul, S.S.; Wang, H.; Wischnewski, N.; Hartl, F.U.; Hayer-Hartl, M. Structure and Conformational Cycle of a Bacteriophage-Encoded Chaperonin. PLoS ONE 2020, 15, e0230090. [Google Scholar] [CrossRef]
- Semenyuk, P.I.; Moiseenko, A.V.; Sokolova, O.S.; Muronetz, V.I.; Kurochkina, L.P. Structural and Functional Diversity of Novel and Known Bacteriophage-Encoded Chaperonins. Int. J. Biol. Macromol. 2020, 157, 544–552. [Google Scholar] [CrossRef]
- Zivanov, J.; Nakane, T.; Forsberg, B.B.O.; Kimanius, D.; Hagen, W.J.J.H.; Lindahl, E.; Scheres, S.H.W.H. New Tools for Automated High-Resolution Cryo-EM Structure Determination in RELION-3. elife 2018, 7, e42166. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic Correction of Beam-Induced Motion for Improved Cryo-Electron Microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Grant, T.; Grigorieff, N. Measuring the Optimal Exposure for Single Particle Cryo-EM Using a 2.6 Å Reconstruction of Rotavirus VP6. elife 2015, 4, e06980. [Google Scholar] [CrossRef]
- Zhang, K. Gctf: Real-Time CTF Determination and Correction. J. Struct. Biol. 2016, 193, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Merino, F.; Stabrin, M.; Moriya, T.; Antoni, C.; Apelbaum, A.; Hagel, P.; Sitsel, O.; Raisch, T.; Prumbaum, D.; et al. SPHIRE-CrYOLO Is a Fast and Accurate Fully Automated Particle Picker for Cryo-EM. Commun. Biol. 2019, 2, 218. [Google Scholar] [CrossRef] [PubMed]
- Croll, T.I. ISOLDE: A Physically Realistic Environment for Model Building into Low-Resolution Electron-Density Maps. Acta Crystallogr. D Struct. Biol. 2018, 74, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile Sample Environments and Automation for Biological Solution X-Ray Scattering Experiments at the P12 Beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef]
- Round, A.; Felisaz, F.; Fodinger, L.; Gobbo, A.; Huet, J.; Villard, C.; Blanchet, C.E.; Pernot, P.; McSweeney, S.; Roessle, M.; et al. BioSAXS Sample Changer: A Robotic Sample Changer for Rapid and Reliable High-Throughput X-Ray Solution Scattering Experiments. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 67–75. [Google Scholar] [CrossRef]
- Hajizadeh, N.R.; Franke, D.; Svergun, D.I. Integrated Beamline Control and Data Acquisition for Small-Angle X-Ray Scattering at the P12 BioSAXS Beamline at PETRAIII Storage Ring DESY. J. Synchrotron Radiat. 2018, 25, 906–914. [Google Scholar] [CrossRef]
- Franke, D.; Kikhney, A.G.; Svergun, D.I. Automated Acquisition and Analysis of Small Angle X-Ray Scattering Data. Nucl. Instrum. Methods Phys. Res. A 2012, 689, 52–59. [Google Scholar] [CrossRef]
- Blanchet, C.E.; Hermes, C.; Svergun, D.I.; Fiedler, S. A Small and Robust Active Beamstop for Scattering Experiments on High-Brilliance Undulator Beamlines. J. Synchrotron Radiat. 2015, 22, 461–464. [Google Scholar] [CrossRef]
- Svergun, D.I. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Svergun, D.I. Restoring Low Resolution Structure of Biological Macromolecules from Solution Scattering Using Simulated Annealing. Biophys. J. 1999, 76, 2879–2886. [Google Scholar] [CrossRef]
- Svergun, D.; Barberato, C.; Koch, M.H.J. CRYSOL—A Program to Evaluate X-Ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Tria, G.; Mertens, H.D.T.; Kachala, M.; Svergun, D.I. Advanced Ensemble Modelling of Flexible Macromolecules Using X-Ray Solution Scattering. IUCrJ 2015, 2, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Yang, D.; LaRonde-LeBlanc, N.; Lorimer, G.H. Crystal Structure of a GroEL-ADP Complex in the Relaxed Allosteric State at 2.7 A Resolution. Proc. Natl. Acad. Sci. USA 2013, 110, E2958–E2966. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.T.; Konarev, P.V.; Svergun, D.I. New Developments in the ATSAS Program Package for Small-Angle Scattering Data Analysis. J. Appl. Crystallogr. 2012, 45, 342–350. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Kurochkina, L.P.; Semenyuk, P.I.; Sokolova, O.S. Structural and Functional Features of Viral Chaperonins. Biochemistry 2022, 87, 1–9. [Google Scholar] [CrossRef]
- Sun, Z.; Scott, D.J.; Lund, P.A. Isolation and Characterisation of Mutants of GroEL That Are Fully Functional as Single Rings. J. Mol. Biol. 2003, 332, 715–728. [Google Scholar] [CrossRef]
- Illingworth, M.; Ramsey, A.; Zheng, Z.; Chen, L. Stimulating the Substrate Folding Activity of a Single Ring GroEL Variant by Modulating the Cochaperonin GroES. J. Biol. Chem. 2011, 286, 30401–30408. [Google Scholar] [CrossRef]
- Illingworth, M.; Salisbury, J.; Li, W.; Lin, D.; Chen, L. Effective ATPase Activity and Moderate Chaperonin–Cochaperonin Interaction Are Important for the Functional Single-Ring Chaperonin System. Biochem. Biophys. Res. Commun. 2015, 466, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.L.; Cowan, N.J. A Single Ring Is Sufficient for Productive Chaperonin-Mediated Folding In Vivo. Mol. Cell 1998, 2, 93–99. [Google Scholar] [CrossRef]
- Yan, X.; Shi, Q.; Bracher, A.; Miličić, G.; Singh, A.K.; Hartl, F.U.; Hayer-Hartl, M. GroEL Ring Separation and Exchange in the Chaperonin Reaction. Cell 2018, 172, 605–617.e11. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Llorente, Y.; Jebara, F.; Patra, M.; Malik, R.; Nisemblat, S.; Chomsky-Hecht, O.; Parnas, A.; Azem, A.; Hirsch, J.A.; Ubarretxena-Belandia, I. Structural Basis for Active Single and Double Ring Complexes in Human Mitochondrial Hsp60-Hsp10 Chaperonin. Nat. Commun. 2020, 11, 1916. [Google Scholar] [CrossRef] [PubMed]
- Leisi, E.V.; Barinova, K.V.; Kudryavtseva, S.S.; Moiseenko, A.V.; Muronetz, V.I.; Kurochkina, L.P. Effect of Bacteriophage-Encoded Chaperonins on Amyloid Transformation of α-Synuclein. Biochem. Biophys. Res. Commun. 2022, 622, 136–142. [Google Scholar] [CrossRef]
- Danziger, O.; Rivenzon-Segal, D.; Wolf, S.G.; Horovitz, A. Conversion of the Allosteric Transition of GroEL from Concerted to Sequential by the Single Mutation Asp-155→Ala. Proc. Natl. Acad. Sci. USA 2003, 100, 13797–13802. [Google Scholar] [CrossRef]
- Meyer, A.S.; Gillespie, J.R.; Walther, D.; Millet, I.S.; Doniach, S.; Frydman, J. Closing the Folding Chamber of the Eukaryotic Chaperonin Requires the Transition State of ATP Hydrolysis. Cell 2003, 113, 369–381. [Google Scholar] [CrossRef]
- Kanzaki, T.; Ushioku, S.; Nakagawa, A.; Oka, T.; Takahashi, K.; Nakamura, T.; Kuwajima, K.; Yamagishi, A.; Yohda, M. Adaptation of a Hyperthermophilic Group II Chaperonin to Relatively Moderate Temperatures. Protein Eng. Des. Sel. 2010, 23, 393–402. [Google Scholar] [CrossRef]
- Ishida, R.; Okamoto, T.; Motojima, F.; Kubota, H.; Takahashi, H.; Tanabe, M.; Oka, T.; Kitamura, A.; Kinjo, M.; Yoshida, M.; et al. Physicochemical Properties of the Mammalian Molecular Chaperone HSP60. Int. J. Mol. Sci. 2018, 19, 489. [Google Scholar] [CrossRef]
- Spinello, A.; Ortore, M.G.; Spinozzi, F.; Ricci, C.; Barone, G.; Gammazza, A.M.; Piccionello, A.P. Quaternary Structures of GroEL and Naïve-Hsp60 Chaperonins in Solution: A Combined SAXS-MD Study. RSC Adv. 2015, 5, 49871–49879. [Google Scholar] [CrossRef]
- Kachala, M.; Valentini, E.; Svergun, D.I. Application of SAXS for the Structural Characterization of IDPs. Adv. Exp. Med. Biol. 2015, 870, 261–289. [Google Scholar] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokolova, O.S.; Pichkur, E.B.; Maslova, E.S.; Kurochkina, L.P.; Semenyuk, P.I.; Konarev, P.V.; Samygina, V.R.; Stanishneva-Konovalova, T.B. Local Flexibility of a New Single-Ring Chaperonin Encoded by Bacteriophage AR9 Bacillus subtilis. Biomedicines 2022, 10, 2347. https://doi.org/10.3390/biomedicines10102347
Sokolova OS, Pichkur EB, Maslova ES, Kurochkina LP, Semenyuk PI, Konarev PV, Samygina VR, Stanishneva-Konovalova TB. Local Flexibility of a New Single-Ring Chaperonin Encoded by Bacteriophage AR9 Bacillus subtilis. Biomedicines. 2022; 10(10):2347. https://doi.org/10.3390/biomedicines10102347
Chicago/Turabian StyleSokolova, Olga S., Evgeny B. Pichkur, Ekaterina S. Maslova, Lidia P. Kurochkina, Pavel I. Semenyuk, Petr V. Konarev, Valeriya R. Samygina, and Tatiana B. Stanishneva-Konovalova. 2022. "Local Flexibility of a New Single-Ring Chaperonin Encoded by Bacteriophage AR9 Bacillus subtilis" Biomedicines 10, no. 10: 2347. https://doi.org/10.3390/biomedicines10102347
APA StyleSokolova, O. S., Pichkur, E. B., Maslova, E. S., Kurochkina, L. P., Semenyuk, P. I., Konarev, P. V., Samygina, V. R., & Stanishneva-Konovalova, T. B. (2022). Local Flexibility of a New Single-Ring Chaperonin Encoded by Bacteriophage AR9 Bacillus subtilis. Biomedicines, 10(10), 2347. https://doi.org/10.3390/biomedicines10102347






