Abstract
A surface acoustic wave (SAW) sensor with a microfluidic channel was studied to detect C-reactive protein (CRP). A piezoelectric lithium niobate substrate was used to examine the frequency response of the microfluidic SAW sensor. The amplitude (insertion loss) changes in the microfluidic SAW sensor were measured from the interaction of CRP/anti-CRP owing to mass variation. The fabricated microfluidic SAW sensor exhibited a detection limit of 4 ng/mL CRP concentration. A wide CRP concentration range (10 ng/mL to 0.1 mg/mL) can be detected by this sensor, which is higher than the existing CRP detection methods. A good linear relationship between the amplitude peak shift and CRP concentrations from 10 ng/mL to 0.1 mg/mL was obtained. The amplitude peak shifts in the sensor can be useful for estimating CRP concentration. This can be used as a biosensor to diagnose the risk of cardiovascular disease.
1. Introduction
In clinical applications, protein detection plays an important role in the diagnosis of diseases. Therefore, the effective detection of proteins is of utmost importance. For example, C-reactive proteins (CRP) could act as biomarkers of inflammation; the concentration of CRP increases a hundred times when chronic inflammation or cancers occur in the body. In normal human beings, the concentration of CRP is less than 1 μg/mL [1]. CRP detection is also useful for screening for cardiovascular diseases, hypertension, and diabetes, among others [2]. There are three categories for the evaluation of cardiovascular risk: concentrations of CRP lower than 1 μg/mL indicate low risk, 1–3 μg/mL indicates moderate risk, and 3–10 μg/mL indicates high risk [3]. Therefore, patients’ susceptibility to cardiovascular diseases can be identified at an early stage, which can then be controlled through appropriate lifestyle interventions and medicines. Patients with a higher CRP value indicate a higher adverse event risk. The concentration of CRP could be as high as 100 μg/mL or even higher when patients have bacterial infections, cancers, or other autoimmune diseases. Salvo et al. reported that CRP concentration is less than 8 µg/mL for normal clinical presentation of wounds [4]. Patients with and without wound complications have average CRP concentrations of ~35 µg/mL and ~9 µg/mL, respectively. When the CRP concentration is higher than 15 µg/mL, it is assumed to be a result of wound inflammation [5]. The patient’s wound in the proliferative and inflammatory phases had an average CRP concentration of 90 µg/mL, which then dropped to 5.2 µg/mL in the maturation phase [6]. Patients with trauma-related chronic wounds had an average CRP concentration of ~66 µg/mL [7]. CRP concentrations can reach 141 µg/mL when a patient’s wound is infected after surgically managed fractures [8]. Diabetic patients with foot ulcers have an infection threshold of a CRP concentration of 17 µg/mL [9]. Whether or not a diabetic patient with foot osteomyelitis has an additional amputation, the average CRP concentration is 95.6 µg/mL [10]. The patient can be healing and in remission when the CRP concentration is less than 40 µg/mL. Based on the above, a sensor with a sufficiently wide range of detection for CRP concentration is needed. However, very few sensors have a range sufficient to cover the clinically relevant range [11,12].
Recently, several techniques for measuring CRP concentrations have been introduced. Fluorescent immunosorbent assays have been used to detect CRP [13,14]. However, while these methods reported sufficient sensitivity, they were time consuming, expensive, and involved complicated detection procedures. To overcome these limitations, various biosensors based on electrochemical and optical methods [12,15,16,17,18,19,20,21,22] for CRP detection analysis have been reported. However, there is a need to develop a simple and highly efficient CRP detection technique for routine clinical testing. Currently, some applications of microelectromechanical system (MEMS) technology, such as biomedicine, analytical chemistry, and molecular biology, have attracted significant interest [23]. Microfluidics-based immunoassays offer significant advantages, including low-volume fluidic consumption, fast analysis, better process control, low fabrication costs, and a safe platform for chemicals [24]. Microfluidics has emerged from MEMS technology. It can control the flow of tiny amounts of liquids in a microfluidic system [25]. The reaction time and detection limit of these microfluidic biosensors can be minimized through innovative design.
Surface acoustic wave (SAW)-based microfluidics technology has been under exploration for the past decade. Studies have presented approaches for integrating SAW devices with microfluidics [26]. This integrated SAW biosensing has opened the path to achieving fast response times and low sample consumption. SAW-enabled microfluidic devices have been demonstrated to have many advantages, such as having simple fabrication and being compact and inexpensive devices [27]. These advantages allow SAW microfluidics to play an important role in various fields, including biology, chemistry, engineering, and medicine. SAWs are electroacoustic transducers that are easy to design and operate [28]. SAW sensors have established a powerful platform for use in chemical sensing or biosensing applications. In [29], an integrated microfluidic system was used to detect CRP. The detection process was performed automatically on this system. The total reaction time was less than 25 min, and the detection limit was reported at 12.5 ng/mL. In [30], a microfluidic system was reported to have rapid automatic detection of CRP. The method also automatically performs the whole detection process for CRP measurement. This developed microfluidic system can automatically complete the detection process within 30 min, and its detection limit is reported to be at 12.5 ng/mL. In [31], a lab-on-a-chip system was demonstrated to integrate a microfluidic chip and a guided-mode resonance (GMR) biosensor for CRP detection in whole blood samples. The developed GMR sensor has a 3.2 ng/mL limit of detection for CRP.
In this paper, we report on a new microfluidic system that integrates with a SAW sensor for the detection of CRP. We used a shear horizontal-SAW (SH-SAW) substrate, which is an appropriate substrate material for successful device operation in liquids [32]. SAW biosensors coated with 2-methacryloyloxyethyl phosphorylcholine polymers were reported to have been used for the detection of CRP [33]. We will fabricate a microfluidic channel and integrate it with the SAW sensor on a lithium niobate (LiNbO3) substrate for detecting CRP.
2. Materials and Methods
2.1. Design and Fabrication
The SAW sensor is composed of an input interdigital transducer (IDT), a surface sensing area, and an output IDT on a piezoelectric substrate. The surface sensing area is located between the input and output IDTs. The input IDT converts electrical signals to surface acoustic waves. The surface acoustic waves propagate through a surface sensing area to an output IDT, which would be very sensitive to variations in the surface sensing area. The output IDT converts surface acoustic waves back to electrical signals. The output response greatly correlates to the corresponding variations on the surface sensing area. The detailed design of an IDT was described in our previous work [34]. A single, 500 µm thick, single-side polished, 64-degree Y-cut LiNbO3 substrate was used in this work. We designed a microfluidic channel to install in a SAW sensor, ensuring that measurements were more convenient and accurate. Polydimethylsiloxane (PDMS) was used as a structural material for the microfluidic module and was molded with an 8 mm thick polymethyl methacrylate (PMMA) piece. For manufacturing PDMS test pieces, a PDMS elastomer kit (Sylgard 184) was purchased from Dow Corning, and the base part A and the curing agent part B were mixed in a 10:1 weight ratio at room temperature. Then the mixed, uncured PDMS was degassed in a vacuum oven at low pressure by using a roughing pump for 45 min. After degassing, the PDMS mixture was poured into the PMMA molds, and the filled molds were cured using a high-temperature vacuum oven at a temperature of 125 °C. After the samples were all cured, they were cooled at room temperature for two hours prior to mold removal. As shown in Figure 1a, the microfluidic channel has a length of 4 mm and a width of 0.5 mm, while the liquid area in contact with the SAW sensing area is an elliptical area of 6 and 4 mm. In addition, the thickness of the channel at the bottom is 1 mm, and the distance from the surface is also 1 mm. Figure 1b,c shows a 3D model view of the microfluidic channel and the fabricated microfluidic module, respectively. We fabricated two microfluidic channels in one module. We could cut it in half to obtain one microfluidic channel for use as a conduction channel and for a surface sensing area reaction. The sensing area had a capacity of ~50 μL, and other liquid spaces totaled ~50 μL. The water pipes were connected at both ends of the channel. One end was connected with a thin syringe (total capacity: 1 mL) as the input end of the reaction volume, and the other end was fixed to the beaker. The beaker was used to collect the waste liquid when the solution was discharged after the reaction was completed.
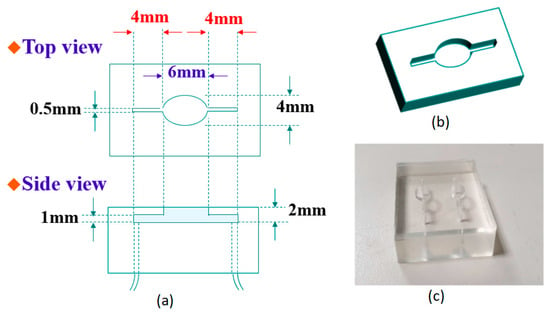
Figure 1.
(a) Detailed structure diagram of the microfluidic channel; (b) 3D model view of the microfluidic channel; (c) two fabricated microfluidic channels at one time.
2.2. Method of Immobilization
For the immobilization of the Au surface and the CRP/anti-CRP interaction process, the steps are described below:
The Au surface was first cleaned by UV/ozone and then treated with ethanol. Finally, the Au surface was dried with nitrogen gas.
- (a)
- For a self-assembled monolayer (SAM) formation on Au surfaces, 11-mercaptoundencanoic acid (11-MUA) in absolute ethanol (4 mM) was injected into the gold sensing area and incubated for 24 h [35]. After SAM formation, careful and complete rinsing of ethanol is necessary to get rid of the multilayer 11-MUA [36], and the residual ethanol is washed out by deionized water and dried by nitrogen (N2) gas. SAMs on the Au surface with 11-MUA provide the opportunity to modify the acid end group through other functionalized groups [37].
- (b)
- This reactive SAM layer was activated in situ by immobilizing the N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) reagents (volume ratio: 1:1 of 100 mM NHS and 400 mM EDC) for 20 min. The NHS-EDC activation approach is the most commonly used amine reactive cross-linking reagent because the main product of NHS-ester is the dominant intermediate product for immobilizing biomolecules containing free primary amino groups (e.g., anti-CRP in this study) via amide linkage [38,39]. After the activation process, the gold surface was washed with a sodium acetate buffer (10 mM, pH 5.0).
- (c)
- Immobilization of anti-CRP was performed in a sodium acetate buffer (10 mM, pH 5.0) for 24 h.
- (d)
- Blocking was done by incubating in ethanolamine-HCl (pH 8.5) solutions for 10 min, followed by washing and incubating in a phosphate-buffered saline (PBS) buffer.
- (e)
- The prepared CRP with the PBS buffer solution was injected over the sensing area and was allowed to react with anti-CRP to evaluate the interaction between them.
2.3. Method of Measurement
Figure 2 demonstrates the measurement system for the frequency response of the SAW sensors with the microfluidic channel. In order to clear the demonstration, a zoom-in on the SAW sensor and microfluidic module is presented on the right-hand side. After the gold film in the SAW sensor was connected to the anti-CRP molecule, the sensor was installed in the microfluidic system for the CRP/anti-CRP reaction. The maximum driving flow rate of the peristaltic pump used this time was 36.9 μL/min, and the experiment was set at 30 μL/min. Each experiment used a peristaltic pump to inject the CRP solution (the solvent was PBS) into the reaction chamber. At this time, the solution was in contact with the sensing area (gold film part) at the bottom of the reaction chamber. The contact probability of CRP molecules in the solution and anti-CRP depends on the diffusion rate of the CRP molecules in the solvent (PBS), and the control of the flow rate can keep the concentration gradient of the reaction area constant, which is very helpful for the quantitative analysis of biochemical experiments. In terms of experimental parameters, for each CRP solution, the injection time was set for 10 min and performed in a static flow (30 μL/min) to ensure that all the volumes of the CRP solution were the same. After the reaction, the surface of the gold film was washed twice with large amounts of PBS buffer to remove the CRP protein from the reaction, and then the specificity of CRP/anti-CRP was analyzed. Signal changes are caused by an increase in the number of bonds. In order to suppress noise generation during measurement, a PBS buffer was injected into the cavity. After the measurement was completed, the next stage of the experiment was carried out. The frequency response of the microfluidic SAW sensor was measured using a vector network analyzer. The frequency response of the microfluidic SAW sensor correlates to the corresponding mass and mechanical variations on the sensing surface.
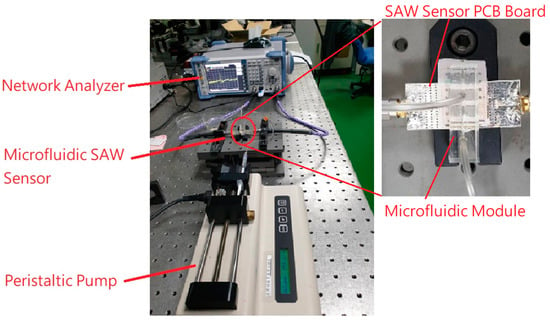
Figure 2.
The measurement system of the microfluidic SAW sensor with a zoom-in on the SAW sensor and microfluidic module.
3. Results and Discussion
Because SAW transducers convert electrical signals into acoustic signals, they propagate on the surface and interact with other structures (for example, gratings, filling layers, etc.) and reflect back. The interaction between the reflected wave and the incident sound wave produces a resonance peak. Therefore, whether the conduction effect is good or bad is closely related to the material of the element and the direction of the lattice arrangement. During the test, we sequentially injected air and the PBS buffer into the microchannels to detect the signal characteristics of the SAW sensing area under different environmental conditions. Figure 3 shows the resonant behavior of the SAW sensor with air and PBS injected into the microfluidic SAW device. The figure clearly shows the characteristic resonance frequency of SAW. The blank signal of each condition was measured five times, and the average value and standard deviation of the peak position were obtained, as listed in Table 1. The measurement results show that the average value of the resonance peak under the air condition (standard deviation in parentheses) is about −58.02 dB (0.0643 dB), and the PBS buffer is −50.76 dB (0.0228 dB). This confirms that the component can operate normally in a liquid environment. In addition, no contribution to the peak shift for the microfluidic module was observed when it was assembled into SAW sensors.
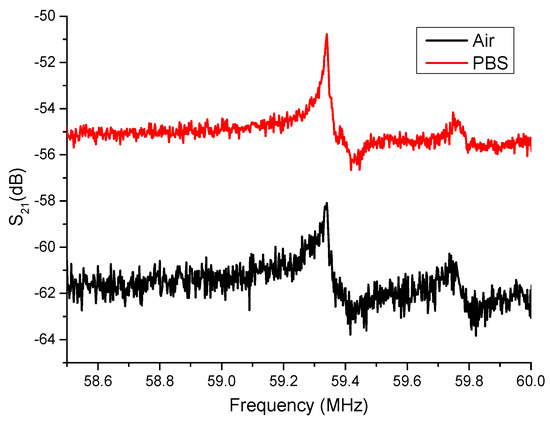
Figure 3.
The resonant behavior of the SAW sensor with air and PBS injected into the microfluidic SAW device.

Table 1.
The averaged resonance peak and their standard deviation of the microfluidic SAW sensor with air and PBS injection.
In the reaction solution, we configured CRP solutions with six concentrations: 10 ng/mL, 100 ng/mL, 1 µg/mL, 10 µg/mL, 100 µg/mL, and 1 mg/mL. The detected experiment is ordered from a low concentration to a high concentration. Figure 4 shows the amplitude response of the microfluidic SAW device with six concentrations: 10 ng/mL, 100 ng/mL, 1 µg/mL, 10 µg/mL, 100 µg/mL, and 1 mg/mL. Interestingly, the resonance frequency was not changed, and only the amplitude variation was observed. In the SAW sensor, the mass-loading effect generally changed the amplitude and the resonance frequency. The change in insertion loss and the lack of change in the resonance frequency for different CRP concentrations are unclear now. In Wang’s research [40], SH-SAW measurements with different weight concentrations of glycerol from 0% to 95% led to maximum frequency shifts of ~430 Hz, and their corresponding differences in refractive index and dielectric constant were around 0.133 and 35.7, respectively [41]. Compared to our study on CRP/anti-CRP, both physical properties are much less than the glycerol. The induced frequency shift will also be smaller than 430 Hz. Therefore, one possible reason is that the frequency shift is too small to be detectable in our measurement (<2 kHz). The figure clearly shows that, no matter what the CRP concentration is, it will not cause the frequency shift of the resonance peak. Generally, the mass-loading effect in a SAW filter changes the amplitude and the resonance frequency. Therefore, the reason why there is no frequency shift is still not clear. The evaluation of the repeatability of this integrated system is also essential to check the reliability. This was investigated by repeatedly injecting each concentration for measurement five times and was cleaned by PBS every time. The repeated experiment value and their peak value (insertion loss) varied only by 0.01% (standard deviation), thus confirming that the measurements have high repeatability.
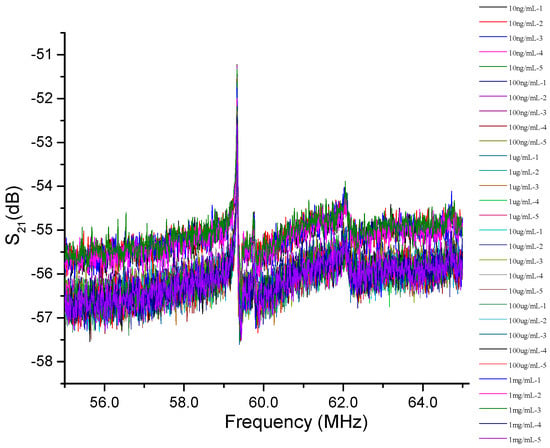
Figure 4.
The amplitude response of the microfluidic SAW sensor with six concentrations of 10 ng/mL, 100 ng/mL, 1 µg/mL, 10 µg/mL, 100 µg/mL, and 1 mg/mL.
Figure 5 shows the peak shift of the amplitude intensity measured five times at the same concentration for six different CRP concentrations. We can observe that regardless of the low concentration (10 ng/mL) or the high concentration (1 mg/mL), the reaction causes a decrease in the intensity peak. The higher the CRP concentration, the larger the peak shift in the amplitude intensity. A possible reason is that the repeated detection experiments can superimpose the coupling reaction of the CRP/anti-CRP on the surface of the gold film. This induces large mass variations and results in a continuous decrease in peak intensity. The higher CRP concentration causes a large mass variation. Therefore, it has a large amplitude intensity attenuation. Because various concentrations were tested five times in succession, five intensity peaks were obtained for each concentration. The peak shift in each concentration represents the quantitative situation of the CRP/anti-CRP coupling concentration. Therefore, we can estimate the CRP concentration from the peak shift. Figure 6 plots the relationship between amplitude attenuation (peak shift) and CRP concentrations, which clearly shows a good linear correlation between the peak shift and CRP concentrations from 10 ng/mL to 0.1 mg/mL, where the dark point is the mean value of the peak shift, and the error bar is the standard deviation for each CRP concentration. The peak shift at the CRP concentration of 1 mg/mL is larger than the other concentrations of CRP. Therefore, it was ruled out to obtain a good linear fitting. Detailed measurement data of this experiment are listed in Table 2 with measured CRP concentrations using the microfluidic system.
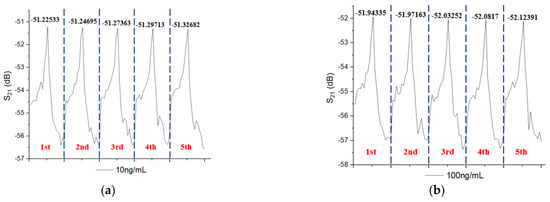
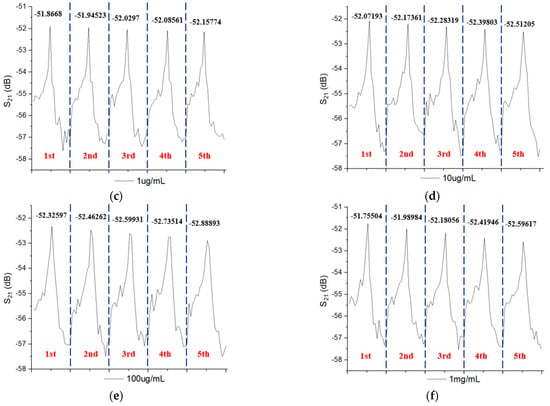
Figure 5.
The peak shift in amplitude intensity measured five times at the same concentration for six different CRP concentrations of (a) 10 ng/mL (b) 100 ng/mL, (c) 1 µg/mL, (d) 10 µg/mL, (e) 100 µg/mL, and (f) 1 mg/mL.
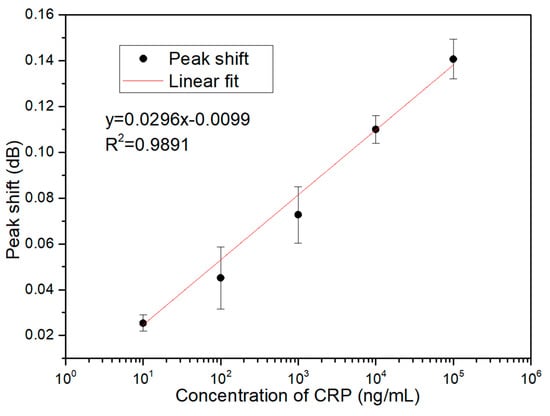
Figure 6.
The correlation of peak shift and CRP concentrations in the proposed microfluidic SAW sensor.

Table 2.
The peak shift and their standard deviation of the microfluidic SAW sensor for detecting different CRP concentrations.
Generally, the limit of detection (LOD) indicates the lowest amount that can be distinguished from the signal value of the analytical instrument in the sample, but it may not be able to quantify the correct value of the target object [42,43]. LOD is an important parameter of validation. Several approaches, such as the visual evaluation method, the signal-to-noise ratio method, and the calibration curve method, have been used to estimate the LOD. The visual evaluation method determines the LOD by analyzing samples with known analyte concentrations and determining the lowest concentration of analytes that can be quantified with acceptable accuracy and precision. This method is less frequently used due to the lack of a more rigorous verification method. The LOD may differ due to different cognitive detection limits. The signal-to-noise ratio method uses three times the signal-to-noise ratio to determine its LOD. This method can only be used in analysis methods that have noise background lines and peak height measurements. The calibration curve method is based on the standard deviation of the response and the slope of the calibration curve. The LOD was calculated as three times the standard deviation of the response divided by the slope of the calibration curve. This method is a widely accepted method owing to it being much easier and less time consuming in the calculation. It is most suitable when the analysis method does not involve background noise. In this study, the calibration curve method was used to estimate the LOD. For a linear calibration, the response y is assumed to be linearly related to the standard concentration x for a limited range of concentration. This can be expressed as y = a + bx. This model was used to calculate the LOD. The LOD is expressed as LOD = 3(σ/S), where σ is the standard deviation of the response, and S is the slope of the calibration curve. The standard deviation of the response was estimated by the standard deviation of either y-residuals or y-intercepts of the calibration lines, as listed in Table 3. The LOD values of the microfluidic SAW sensor calculated from the standard deviation of y-residuals and y-intercepts are 3.76 and 4.01 ng/mL, respectively.

Table 3.
The standard deviation and LOD calculated from the y-residuals and y-intercepts of the calibration lines.
Although the variations for 10 ng/mL are close to the variations for 100 ng/mL, the average value for these concentrations is apparently different. For measuring CRP concentrations, one can measure them many times to get the average value. CRP concentrations can be estimated according to linearly fitting curves. Generally, CRP concentration in healthy humans is less than 1 µg/mL, but it can increase hundreds of times due to infections and other diseases. The clinically relevant range for CRP concentrations is at 1–200 µg/mL [4]. The proposed microfluidic SAW sensor, with the capability of a wide detection range of CRP concentrations, can cover this clinical range. Further, as listed in Table 3, the calculated LOD value of the microfluidic SAW sensor is about 4 ng/mL. This low LOD value of the microfluidic SAW sensor is high enough for clinical diagnosis applications.
4. Conclusions
SAW sensors with microfluidic channels were investigated for CRP detection. We observed that the shift in amplitude peak increases with increasing CRP concentration due to the mass-loading effect. The peak shift exhibited a good linear relation to CRP concentrations, from 10 ng/mL to 0.1 mg/mL. The wide detection range of CRP concentrations demonstrates the potential for use in clinical diagnosis. In addition, the detectable CRP concentration of 4 ng/mL in the microfluidic SAW sensor is sufficiently high for clinical diagnosis applications.
Author Contributions
Conceptualization, C.-L.T. and M.-J.J.; methodology, Y.-C.L., C.-L.T. and M.-J.J.; validation, Y.-C.L., M.S., C.-W.C. and Y.-H.L.; investigation, Y.-C.L., M.S., C.-W.C. and Y.-H.L.; writing—original draft preparation, M.S.; writing—review and editing, M.-J.J.; supervision, L.-B.C., S.-F.H. and C.-S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Chang Gung University and Chang Gung Memorial Hospital, Grant Numbers BMRPA52, CMRPD2G0291, and CMRPD2G0292, and Grant Number MOST 109-2221-E-182-003-MY2 from the Ministry of Science and Technology, Taiwan, ROC.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pepys, M.B.; Hirschfield, G.M. C-reactive protein: A critical update. J. Clin. Investig. 2003, 111, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Myers, G.L.; Rifai, N.; Tracy, R.P.; Roberts, W.L.; Alexander, R.W.; Biasucci, L.M.; Kimberly, M.M. CDC/AHA workshop on markers of inflammation and cardiovascular disease: Application to clinical and public health practice: Report from the laboratory science discussion group. Circulation 2004, 110, e545–e549. [Google Scholar] [CrossRef]
- Yeh, T.H.; Willerson, J.T. Coming of age of C-reactive protein using inflammation markers in cardiology. Circulation 2003, 107, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Salvo, P.; Dini, V.; Kirchhain, A.; Janowska, A.; Oranges, T.; Chiricozzi, A.; Lomonaco, T.; Di Francesco, F.; Romanelli, M. Sensors and biosensors for c-reactive protein, temperature and ph, and their applications for monitoring wound healing: A review. Sensors 2017, 17, 2952. [Google Scholar] [CrossRef]
- Legendre, C.; Debure, C.; Meaume, S.; Lok, C.; Golmard, J.L.; Senet, P. Impact of protein deficiency on venous ulcer healing. J. Vasc. Surg. 2008, 48, 688–693. [Google Scholar] [CrossRef]
- Cerveró-Ferragut, S.; López-Riquelme, N.; Martín-Tomás, E.; Massa-Domínguez, B.; Pomares-Vicente, J.; Soler-Pérez, M.; Sánchez-Hernández, J.F. Quantitative analysis of blood cells and inflammatory factors in wounds. J. Wound Care 2017, 26, 121–125. [Google Scholar] [CrossRef]
- Liu, T.; Yang, F.; Li, Z.; Yi, C.; Bai, X. A Prospective Pilot Study to EvaluateWound Outcomes and Levels of Serum C-reactive Protein and Interleukin-6 in the Wound Fluid of Patients with Trauma-related Chronic Wounds. Ostomy Wound Manag. 2014, 60, 30–37. [Google Scholar]
- Chapman, G.; Holton, J.; Chapman, A. A threshold for concern? C-reactive protein levels following operatively managed neck of femur fractures can detect infectious complications with a simple formula. Clin. Biochem. 2016, 49, 219–224. [Google Scholar] [CrossRef]
- Jeandrot, A.; Richard, J.L.; Combescure, C.; Jourdan, N.; Finge, S.; Rodier, M.; Corbeau, P.; Sotto, A.; Lavigne, J.P. Serum procalcitonin and C-reactive protein concentrations to distinguish mildly infected from non-infected diabetic foot ulcers: A pilot study. Diabetologia 2008, 51, 347–352. [Google Scholar] [CrossRef]
- Van Asten, S.A.; Jupiter, D.C.; Mithani, M.; La Fontaine, J.; Davis, K.E.; Lavery, L.A. Erythrocyte sedimentation rate and C-reactive protein to monitor treatment outcomes in diabetic foot osteomyelitis. Int. Wound J. 2017, 14, 142–148. [Google Scholar] [CrossRef]
- Magliulo, M.; De Tullio, D.; Vikholm-Lundin, I.; Albers, W.M.; Munter, T.; Manoli, K.; Palazzo, G.; Torsi, L. Label-free C-reactive protein electronic detection with an electrolyte-gated organic field-effect transistor-based immunosensor. Anal. Bioanal. Chem. 2016, 408, 3943–3952. [Google Scholar] [CrossRef]
- Yen, Y.-K.; Lai, Y.-C.; Hong, W.-T.; Pheanpanitporn, Y.; Chen, C.-S.; Huang, L.-S. Electrical detection of c-reactive protein using a single freestanding, thermally controlled piezoresistive microcantilever for highly reproducible and accurate measurements. Sensors 2013, 13, 9653–9668. [Google Scholar] [CrossRef]
- Yang, S.F.; Gao, B.Z.; Tsai, H.Y.; Fuh, C.B. Detection of c-reactive protein based on a magnetic immunoassay by using functional magnetic and fluorescent nanoparticles in microplates. Analyst 2014, 139, 5576–5581. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, B.; Chen, M.; Jiang, T.; Zhou, D.; Huang, J.; Fu, W. Sensitive and rapid quantification of C-reactive protein using quantum dot-labeled microplate immunoassay. J. Trans. Med. 2012, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Mitsakakis, K.; Gizeli, E. Detection of multiple cardiac markers with an integrated acoustic platform for cardiovascular risk assessment. Anal. Chim. Acta 2011, 699, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bing, X.; Wang, G. Label free c-reactive protein detection based on an electrochemical sensor for clinical application. Int. J. Electrochem. Sci. 2017, 12, 6304–6314. [Google Scholar] [CrossRef]
- Thangamuthu, M.; Santschi, C.; J.F. Martin, O. Label-free electrochemical immunoassay for c-reactive protein. Biosensors 2018, 8, 34. [Google Scholar] [CrossRef]
- Vance, S.A.; Sandros, M.G. Zeptomole detection of c-reactive protein in serum by a nanoparticle amplified surface plasmon resonance imaging aptasensor. Sci. Rep. 2014, 4, 5129. [Google Scholar] [CrossRef]
- Vashist, S.K.; Schneider, E.; Luong, J.H. Surface plasmon resonance-based immunoassay for human c-reactive protein. Analyst 2015, 140, 4445–4452. [Google Scholar] [CrossRef]
- Wang, W.; Mai, Z.; Chen, Y.; Wang, J.; Li, L.; Su, Q.; Li, X.; Hong, X. A labelfree fiber optic spr biosensor for specific detection of c-reactive protein. Sci. Rep. 2017, 7, 16904. [Google Scholar]
- Kim, N.; Kim, D.-K.; Cho, Y.-J. Development of indirect-competitive quartz crystal microbalance immunosensor for c-reactive protein. Sens. Actuat. B Chem. 2009, 143, 143,444–448. [Google Scholar] [CrossRef]
- Gao, K.; Cui, S.; Liu, S. Development of an electrochemical quartz crystal microbalance-based immunosensor for c-reactive protein determination. Int. J. Electrochem. Sci. 2018, 13, 812–821. [Google Scholar] [CrossRef]
- Reyes, D.R.; Iossifidis, D.; Auroux, P.A.; Manz, A. Micro total analysis systems. 1. Introduction, theory, and technology. Anal. Chem. 2002, 74, 2623–2636. [Google Scholar] [CrossRef]
- Luo, J.K.; Fu, Y.Q.; Milne, W.I. Acoustic Wave Based Microfluidics and Lab-on-a-Chip. In Modeling and Measurement Methods for Acoustic Waves and for Acoustic Microdevices; Beghi, M.G., Ed.; IntechOpen: London, UK, 2013; Available online: https://www.intechopen.com/books/modeling-and-measurement-methods-for-acoustic-waves-and-for-acoustic-microdevices/acoustic-wave-based-microfluidics-and-lab-on-a-chip (accessed on 28 August 2013). [CrossRef]
- Laser, D.J.; Santiago, J.G. A review of micropumps. J. Micromech. Microengi. 2004, 14, R35. [Google Scholar] [CrossRef]
- Länge, K.; Blaess, G.; Voigt, A.; Götzen, R.; Rapp, M. Integration of a surface acoustic wave biosensor in a microfluidic polymer chip. Biosens. Bioelectron. 2006, 22, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Li, P.; Lin, S.C.S.; Stratton, Z.S.; Nama, N.; Guo, F.; Slotcavage, D.; Mao, X.; Shi, J.; Costanzo, F.; et al. Surface acoustic wave microfluidics. Lab Chip 2013, 13, 3626–3649. [Google Scholar] [CrossRef]
- Go, D.B.; Atashbar, M.Z.; Ramshani, Z.; Chang, H.C. Surface acoustic wave devices for chemical sensing and microfluidics: A review and perspective. Anal. Methods 2017, 9, 4112–4134. [Google Scholar] [CrossRef]
- Yang, Y.N.; Lin, H.I.; Wang, J.H.; Shiesh, S.C.; Lee, G.B. An integrated microfluidic system for C-reactive protein measurement. Biosens. Bioelectron. 2009, 24, 3091–3096. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.B.; Chen, Y.H.; Lin, H.I.; Shiesh, S.C.; Lee, G.B. An integrated microfluidic system for fast, automatic detection of C-reactive protein. Sens. Actuat. B Chem. 2011, 157, 710–721. [Google Scholar] [CrossRef]
- Tsai, M.Z.; Hsiung, C.T.; Chen, Y.; Huang, C.S.; Hsu, H.Y.; Hsieh, P.Y. Real-time CRP detection from whole blood using micropost-embedded microfluidic chip incorporated with label-free biosensor. Analyst 2018, 143, 503–510. [Google Scholar] [CrossRef]
- Länge, K.; Rapp, B.E.; Rapp, M. Surface acoustic wave biosensors: A review. Anal. Bioanal. Chem. 2008, 391, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Pomowski, A.; Baricham, C.; Rapp, B.E.; Matern, A.; Länge, K. Acoustic biosensors coated with phosphorylcholine groups for label-free detection of human C-reactive protein in serum. IEEE Sens. J. 2015, 15, 4388–4392. [Google Scholar] [CrossRef]
- Jeng, M.-J.; Sharma, M.; Li, Y.-C.; Lu, Y.-C.; Yu, C.-Y.; Tsai, C.-L.; Huang, S.-F.; Chang, L.-B.; Lai, C.-S. Surface Acoustic Wave Sensor for C-Reactive Protein Detection. Sensors 2020, 20, 6640. [Google Scholar]
- Stettner, J.; Frank, P.; Griesser, T.; Trimmel, G.; Schennach, R.; Gilli, E.; Winkler, A. A Study on the Formation and Thermal Stability of 11-MUA SAMs on Au(111)/Mica and on Polycrystalline Gold Foils. Langmuir 2009, 25, 1427–1433. [Google Scholar] [CrossRef]
- Daniel, K.; Gregor, W.; Piotr, C.; Andreas, T.; Christof, W. A comprehensive study of self-assembled monolayers of anthracenethiol on gold: Solvent effects, structure, and stability. J. Am. Chem. Soc. 2006, 128, 1723–1732. [Google Scholar]
- Frey, B.L.; Corn, R.M. Covalent Attachment and Derivatization of Poly(L-lysine) Monolayers on Gold Surfaces As Characterized by Polarization-Modulation FT-IR Spectroscopy. Anal. Chem. 1996, 68, 3187–3193. [Google Scholar] [CrossRef]
- Wang, C.; Yan, Q.; Liu, H.-B.; Zhou, X.H.; Xiao, S.J. Different EDC/NHS Activation Mechanisms between PAA and PMAA Brushes and the Following Amidation Reactions. Langmuir 2011, 27, 12058–12068. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zheng, H.N.; Jiang, C.; Lia, K.; Xiao, S.J. EDC/NHS activation mechanism of polymethacrylic acid: Anhydride versus NHS-ester. RSC Adv. 2015, 5, 69939–69947. [Google Scholar] [CrossRef]
- Wang, T. Optimization and Characterization of Integrated Microfluidic Surface Acoustic Wave Sensors and Transducers. 2016 Graduate Theses and Dissertations. Available online: http://scholarcommons.usf.edu/etd/6153 (accessed on 10 November 2020).
- Physical Properties of Glycerine and its Solutions; Glycerine Producers’ Association: New York, NY, USA, 1963; Available online: https://www.aciscience.org/docs/Physical_properties_of_glycerine_and_its_solutions.pdf (accessed on 10 November 2020).
- Sengul, U. Comparing determination methods of detection and quantification limits for aflatoxin analysis in hazelnut. J. Food Drug Anal. 2016, 24, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Gupta, V.B. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2011, 2, 21–25. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).